Abstract
Motivation: Metabolomics is a rapidly evolving field that holds promise to provide insights into genotype–phenotype relationships in cancers, diabetes and other complex diseases. One of the major informatics challenges is providing tools that link metabolite data with other types of high-throughput molecular data (e.g. transcriptomics, proteomics), and incorporate prior knowledge of pathways and molecular interactions.
Results: We describe a new, substantially redesigned version of our tool Metscape that allows users to enter experimental data for metabolites, genes and pathways and display them in the context of relevant metabolic networks. Metscape 2 uses an internal relational database that integrates data from KEGG and EHMN databases. The new version of the tool allows users to identify enriched pathways from expression profiling data, build and analyze the networks of genes and metabolites, and visualize changes in the gene/metabolite data. We demonstrate the applications of Metscape to annotate molecular pathways for human and mouse metabolites implicated in the pathogenesis of sepsis-induced acute lung injury, for the analysis of gene expression and metabolite data from pancreatic ductal adenocarcinoma, and for identification of the candidate metabolites involved in cancer and inflammation.
Availability: Metscape is part of the National Institutes of Health-supported National Center for Integrative Biomedical Informatics (NCIBI) suite of tools, freely available at http://metscape.ncibi.org. It can be downloaded from http://cytoscape.org or installed via Cytoscape plugin manager.
Contact: metscape-help@umich.edu; akarnovs@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Large-scale omics studies have been successful at revealing differences in gene expression, protein and metabolite abundance and post-translational protein modifications, thus providing different level views of the molecular processes that lead to disease phenotypes. Molecular pathway databases and metabolic maps that contain computationally predicted and literature-derived information provide a path to connecting these views together. While many tools have been developed for the analysis of gene expression and proteomics data, to date there are few that allow the user to analyze metabolomics data and to link them with other omics data. High-quality genome scale metabolic reconstructions represent a critical component in developing such a tool. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was one of the first database to provide information about biological pathways in conjunction with gene data from a range of different organisms (Kanehisa, 2006). Over 50 organism-specific metabolic reconstructions were published, providing excellent sources of information about metabolic pathways, genes encoding metabolic enzymes, reactions catalyzed by them and the compounds that participate in these reactions (Duarte et al., 2007; Hao et al., 2010; Ma et al., 2007; Romero et al., 2005; Sigurdsson et al., 2010).
In addition to the information about the components of metabolic pathways, databases like KEGG, BioCyc and SMPD provide data visualization in the form of individual pathway charts or the overall view of metabolic pathways (Frolkis et al., 2010; Kanehisa, 2006; Romero et al., 2005). The pathway charts are actively used by many researchers to help interpret their data, formulate new hypotheses and present the results. However, as the data input volume and complexity grow, such manual analysis is becoming increasingly difficult. Efforts have been made to visualize the experimental data over the static pathways charts (Garcia-Alcalde et al., 2011; Paley and Karp, 2006) or make these charts interactive (Junker et al., 2006; Klukas and Schreiber, 2010). One recently developed tool, Paintomics, provides the ability to load gene expression and metabolite measurements and visualize them over KEGG pathway maps (Garcia-Alcalde et al., 2011). A more interactive tool, Vanted, has been developed for the exploration of experimental metabolomics data in the context of metabolic pathways (Junker et al., 2006; Klukas and Schreiber, 2010). Although developed for plants, it can be used for any data: users can load KEGG maps or build their own pathways. The recently published metabolomics pathways analysis tool MetPA also allows visualization of experimental data in the context of metabolic pathways (Xia and Wishart, 2010) and uses several statistical methods to perform pathway enrichment analysis conceptually similar to gene set enrichment analysis methods (Dennis et al., 2003; Draghici et al., 2007; Subramanian et al., 2005).
Pathways analysis and visualization has become an integral part of biological interpretation of omics experiments. However, one of the inherent limitations of pathway-based visualization is that both genes and metabolites can be part of multiple pathways. In order to understand the overall effect of an altered gene or metabolite, the user must go through multiple pathways and understand the connections among them. An alternative is building a network with genes/metabolites as nodes, where each node is unique and nodes from multiple pathways can be linked together. Such networks provide an easy way to connect multiple pathways and build gene/compound centric maps enabling quick data exploration and logical well-informed hypothesis generation.
We present here a new, substantially updated version of our previously developed tool Metscape (Gao et al., 2010) that provides functionality for creating pathway and network level views and analyzing several types of experimental data. Metscape is a plugin for Cytoscape (Shannon et al., 2003), a widely used open-source network analysis and visualization tool. It allows users to upload lists of metabolites and genes with experimental measurements; identify related genes, metabolites, reactions, enzymes and pathways; build and analyze the networks of genes and metabolites; and visualize the changes in experimental data over time or experimental conditions. The tool also allows users to identify and visualize enriched pathways from expression profiling data.
We demonstrate the utility of Metscape 2 with three examples. The first example involves using Metscape to analyze the 1H-NMR unbiased metabolomics data from patients with sepsis-induced acute lung injury (ALI) and from a mouse model of ALI. The second describes the analysis of metabolite and gene expression data from human pancreatic adenocarcinoma. In the third example, we show how Metscape can be used to guide the search for metabolites with potential involvement in cancer and inflammation.
2 METHODS
The architecture of Metscape 2 has been completely redesigned compared with Metscape 1 (Gao et al., 2010) and a number of new features have been added. Metscape 2 is based on standard three-tier architecture described in detail in Supplementary Material.
The Metscape plugin supports two types of queries. The user can: (i) supply a list of genes and/or compounds; or (ii) select one of the canonical metabolic pathways. The query is handed off to the Metabws service that returns the list of gene–enzyme–reaction–compound relations, which can then be viewed as: (i) a compound–reaction–enzyme–gene (CREG) network, (ii) a compound–reaction (CR) network, (iii) a compound–gene (CG) network or (iv) a compound (C) network.
Metscape 2 uses human metabolic networks. Moreover, it automatically performs mouse/rat to human homology mapping. This is done by the homolog mapping service that maps rat and mouse genes to their corresponding human homologs. All other services use human data.
In summary, Metscape 2 enables the use of both gene expression data and metabolite data as experimental data sources, provides access to the gene set enrichment tool LRpath and has the ability to load user supplied concepts. Animation of time series compound data is supported with an updated user interface.
3 RESULTS
First, we describe the features of Metscape 2. Next we demonstrate several potential workflows with three applications. We show that Metscape was useful in visualizing the data, linking them to prior knowledge of metabolic pathways and helpful for generating new hypotheses, thereby contributing to the overall understanding of the underlying biological processes.
3.1 Metscape 2 user interface features and workflows
3.1.1 User interface features
Metscape is a tool for interactive exploration and visualization of experimental metabolomics and gene expression data in the context of human metabolic networks. In addition to the new plugin architecture, the Metscape 2 interface was significantly changed compared with Metscape 1 (Gao et al., 2010) and many new features were added. The most prominent new feature is the ability to enter gene expression data and examine them in the context of metabolic networks. Two most common types of outputs from microarray or RNA-Seq experiments are (i) lists of genes that are differentially expressed under certain experimental conditions and (ii) lists of pathways, Gene Ontology terms and other concepts that are significantly enriched with genes from an experimental dataset. Metscape 2 allows users to enter both types of data.
In addition to the C and CR network graphs represented in Metscape 1 (named according to the types of nodes shown in a network), we added two types of network graphs—CREG and CG. In CREG graphs, metabolites (or compounds), reactions, enzymes and genes are represented as nodes and the relationships among them are represented as edges; the CG graphs have two types of nodes—compounds and genes, and the edges represent both reactions and enzymes (Supplementary Fig. S1).
The main objective of visualizing experimental data in biological networks is to provide the context that enables data interpretation and leads to generation of new hypotheses. This can be facilitated by providing annotations and links to various data sources. Metscape has several ways to display additional information for the nodes and edges in a given network. First, additional information can be displayed in the data panel at the bottom of the screen by selecting appropriate node and edge attributes, e.g. compound name, gene description, reaction equation (see Supplementary Table S1 for the complete list of attributes). Second, by double clicking on any node or edge, users can display additional information in the Results panel on the right side of the screen (Supplementary Fig. S3). The results panel contains information about compounds, reactions, enzymes, genes, links to external databases such as PubChem and KEGG and to the literature via the Metab2Mesh tool (http:/metab2mesh.ncibi.org, National Center for Integrative Biomedical Informatics). We illustrate the use of many of these features in the three example workflows below.
3.1.2 Metscape 2 workflows
A Metscape session can be started by loading a list of compounds with or without experimental measurements. Users can upload a file, directly enter KEGG compound IDs or copy and paste a list of IDs from a clipboard. Once the compounds have been entered, Metscape will attempt to map them to internal IDs. If any of the input data were not mapped, their IDs will be reported in the Missing Data window (Supplementary Fig. S2c). At the next step, the user can select the network type and either choose to build the network from input data, or select a pathway from the dropdown list, and then proceed to build the network. The resulting network graph will include the query compounds plus any compounds, reactions, enzymes and genes (depending on the network type selected) that participate in the same reactions as the query compounds. If a pathway was selected, the network graph for that pathway will be displayed with experimental data visualized for the relevant nodes. If a C network graph is selected, the edges will be drawn between ‘seed’ compounds and their neighboring compounds. In addition to standard Cytoscape operations, Metscape offers several extra features, including building subnetworks, pathways filtering, expanding a currently displayed network and displaying additional information for a set of selected nodes, as described above.
Alternatively, the user can start with a list of differentially expressed genes, a list of enriched concepts or both. The detailed description of the input file formats is available from the Metscape web page and Metscape user manual (http://metscape.ncibi.org/metscape2/help.html). The concept file can be generated using any previously described gene set enrichment analysis program such as GSEA (Subramanian et al., 2005) or LRpath (Sartor et al., 2009) from gene expression data. The user has an option to do gene set enrichment testing from within Metscape. If this option is selected, a directional test against KEGG pathways with default parameters will be performed. LRpath uses logistical regression to identify the gene sets that are enriched with the query genes (http://lrpath.ncibi.org).
The third possibility is to load gene and compound data in parallel. Once the data are loaded, the user can choose one of the four network types and proceed to build a network. Networks in Metscape are built according to the following general rules. If only genes are used as input, then all the enzymes, reactions and compounds that match those genes are used to build the network. If a concept file is provided, genes from that file will be used as input. If a concept file is not provided, all genes from gene file are used as input. In this case, it is advisable to load a smaller set of genes (e.g. the most significant differentially expressed genes). If both genes and compounds are used as input, then only CREG couplings that match both the input lists are used.
Table 1 compares features of Metscape 2 with four other programs for the analysis and visualization of experimental data in the context of metabolic networks: Vanted (Junker et al., 2006; Klukas and Schreiber, 2010), MetPA (Xia and Wishart, 2010), Omics viewer (Paley and Karp, 2006) and Paintomics (Garcia-Alcalde et al., 2011). Like other programs, Metscape uses node size, color and border to visualize trends in the experimental data. However, unlike any other program, Metscape provides an easy way to connect the experimental genes/compounds into a single network. We illustrate the Metscape features and workflows in the following sections.
Table 1.
Comparison of Metscape to selected metabolic pathway analysis software
| Feature | Metscape | Vanted | MetPA | Omics viewer | Paintomics |
|---|---|---|---|---|---|
| Connected to a pathway database? | Yes | No | Yes | Yes | Yes |
| Can import experimental metabolomics data? | Yes | Yes | Yes | Yes | Yes |
| Can import experimental gene expression data? | Yes | Yes | No | Yes | Yes |
| Interactive? | Yes | Yes | Yes | No | No |
| Single pathway view? | Yes | Yes | Yes | Yes | Yes |
| Multiple pathways view? | Yes | No | No | Yes | No |
| Network view? | Yes | No | No | No | No |
| Search by ID/name | Yes | N/A | Yes | Yes | Yes |
| Nodes redundant? | No | Yes | Yes | Yes | Yes |
| Supports building custom pathways? | No | Yes | No | No | No |
| Access to network analysis and statistical analysis tools? | Yesa | Yes | Yes | No | No |
aVia other Cytoscape plugins.
3.2 Metscape analysis of metabolomics data from sepsis-induced ALI
In a recently published 1H-NMR metabolomics study of sepsis-induced ALI (Stringer et al., 2011), 40 plasma metabolites were identified in lipophilic and hydrophilic fractions of each sample. From these, 28 compounds were mapped to KEGG IDs. Quantitative analysis revealed the difference in levels of a number of metabolites including total glutathione, adenosine, phosphatidylserine and sphingomyelin. These changes reflect complex pathology and provide evidence for the involvement of such processes as oxidant stress (glutathione), energy balance (adenosine), apoptosis (phosphatidylserine) and endothelial barrier function (sphingomyelin) in sepsis-induced ALI.
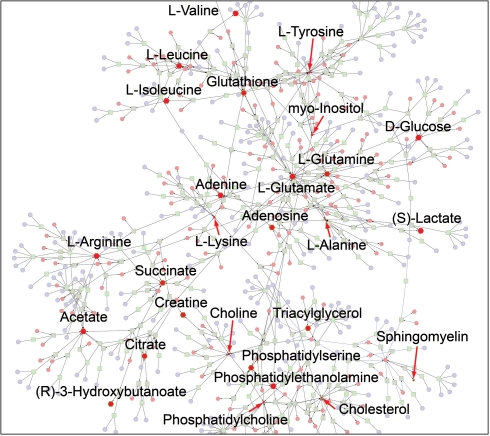
Further analysis showed that eight additional compounds changed significantly [P<0.02, false discovery rate (FDR) <5%] in ALI, compared with healthy subjects. The file containing the list of KEGG IDs, fold change and P-values adjusted for multiple comparisons was loaded into Metscape (Supplementary Table S2). As noted above, Metscape 2 supports four types of network graphs. We first created a CREG graph to obtain an overview of all components of the sepsis-induced ALI network. The resulting network consisted of two components: (i) a small sphingomyelin subnetwork and (ii) a large subnetwork that contained the rest of the experimental compounds. We were able to connect the two components with a single expand operation by adding the nodes related to the compound N-acylsphingosine (C00195) (Fig. 1).
Fig. 1.
A fully connected network of ALI metabolites detected in the sepsis-induced ALI experiment. Metabolites with experimental data are shown in red. The size of the nodes represents the direction of the change. Red arrows point to downregulated compounds.
This type of graph also provided a complete list of metabolic pathways (Supplementary Table S3) and genes related to experimental metabolites. To examine the data further, we built a compound network. It consisted of two major subnetworks combining all statistically significant differentiating compounds. The first subnetwork included adenosine, phosphatidylserine, sphingomyelin, triacylglycerol and cholesterol; the second contained citrate, glutamine, alanine, creatine, succinate, 3-hydroxybutanoate and glutathione. The shortest path between five compounds in the first subnetwork included the reactions from three pathways such as glycerophospholipid metabolism, phosphatidylinositol phosphate metabolism and purine metabolism.
The first subnetwork exemplifies the derangement of lipid metabolism that occurs in critically ill patients and often results in hypocholesterolemia which in turn may be associated with illness severity and a poor prognosis (Chiarla et al., 2004; Dunham and Chirichella, 2011). The changes in metabolites associated with the glycerophospholipid and phosphatidylinositol phosphate metabolism (e.g. sphingomyelin, phosphatidylserine) and modest increase in total glycerolphospholipids in ALI patients compared with healthy controls (1.1 versus 0.88, P=0.135) are likely related to severe inflammation accompanied by cellular injury and apoptosis characteristic of sepsis-induced ALI (Kagan et al., 2004; Tyurina et al., 2010). These inflammatory processes also involve oxidant stress, which contributes to the loss of antioxidant homeostasis. This is evidenced by the metabolites of the second subnetwork that is associated with glutathione (a potent antioxidant) and glutamine (an abundant amino acid and precursor of glutathione). In acute oxidant stress, reduced glutathione (GSH) is rapidly converted to its oxidized form (GSSG). Since 1H-NMR cannot differentiate GSSG and GSH, our finding of increased glutathione is most likely due to GSSG, which increases in sepsis and oxidant stress (Andresen et al., 2008; Biswas and Rahman, 2009). The biological consequence of increased GSSG includes activation of cellular apoptotic pathways and subsequent cell death.
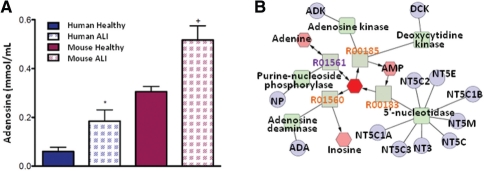
We further demonstrate the utility of Metscape for integrating metabolomics results from human and model organisms. As mentioned above, Metscape can map mouse and rat data to human metabolic networks though homolog mapping. The experimental model of IL-1-β + TNF-α -induced lung injury was previously employed to assay NMR-detectable metabolites in lung tissue (Serkova et al., 2008). Principal component analysis (PCA) showed a derangement in energy homeostasis. Subsequent analysis of these data using quantitative metabolomics revealed several metabolites that were different between control and IL-1-β + TNF-α-treated mice. One of these is adenosine, which was increased in human ALI plasma samples (Fig. 2A). Adenosine is a nucleotide composed of adenine linked to ribose and is a major molecular; component of ADP, AMP and ATP, as well as nucleic acids. Its elevation in sepsis-induced ALI is most likely due to cellular stress-induced release of ATP that is rapidly metabolized by ectonucleotidases (Eckle et al., 2007).
Fig. 2.
Adenosine is elevated in both human ALI and a mouse model of ALI. (A) Human data are plasma from healthy controls (n = 6) and sepsis-induced ALI (n = 13), P = 0.02; mouse data are lung tissue obtained from untreated controls (n = 4) and 6 h after IL-1β + TNF-α-induced lung injury (n = 4), P = 0.01. Data are mean+SEM. (B) CREG network for adenosine (shown as red hexagon).
Initially, increased extracellular adenosine has a protective role in the lungs because it improves barrier function, enhances alveolar fluid clearance and reduces inflammation (Kreindler and Shapiro, 2007; Lucas et al., 2009; Matthay, 2002). Conversely, adenosine also orchestrates signaling that leads to deterioration of the lungs. This includes the promotion of angiogenesis, enhanced production of matrix proteins and propagation of inflammation (Zhou et al., 2009). Collectively, these remodeling processes could lead to long-term lung dysfunction and disease such as pulmonary fibrosis.
Two key enzymes that regulate adenosine production and metabolism are adenosine deaminase (EC.3.5.4.4) and ecto-5′-nucleotidase (EC3.1.3.5). Adenosine deaminase converts adenosine to inosine (Fig. 2B). Levels of both enzymes are altered in patients with lung inflammation. The expression of ADA1, ADA2 and CD73, the genes that encode these two enzymes, is also altered in patients with chronic lung disease. Building Metscape metabolic network for the mouse data produced a list of mouse genes encoding metabolic enzymes related to adenosine and other metabolites of interest that can be investigated in further experiments (Supplementary Table S4).
3.3 Metscape analysis of the metabolomics and gene expression data from human pancreatic ductal adenocarcinoma
Pancreatic ductal adenocarcinoma (PDAC) is one of the deadliest cancers, primarily due to poor treatment response, which can mostly be attributed to tumor heterogeneity. A number of expression profiling studies of whole tissue and microdissected PDAC performed over the last decade have contributed to our understanding of the molecular nature of this disease. Several recent large-scale studies of micro dissected and whole tissue samples helped to delineate the differences in therapeutic response and clinical outcome of different PDA subtypes (Badea et al., 2008; Collisson et al., 2011). Badea et al. profiled paired tumor and normal pancreatic tissue samples from 36 PDAC patients using Affymetrix U133 plus 2.0 whole-genome microarray and identified 239 genes that were upregulated in tumor samples (P<10−14 and fold change >2). Further analysis of these genes showed the enrichment in TGF-β target genes and genes involved in epithelial–mesenchymal transition.
Expression profiling studies contributed significantly to the understanding of underlying molecular mechanisms of pancreatic cancer and resulted in improved classification of the tumor subtypes. However, the lack of early diagnostic markers still remains a problem. Proteomics and metabolomics have the potential to provide additional biological insight for solving this problem. Lucal et al. found elevated levels of purine nucleoside phosphorylase and its metabolites (both products and substrates) in tumor tissue, pancreatic juice and blood of tumor patients (Lucas et al., 2009). There are several metabolomics studies in human and animal models of PDAC (Bathe et al., 2011; Fang et al., 2007; Ouyang et al., 2011; Urayama et al., 2010). The LC–MS study by Urayama et al. identified a number of potential differentiating metabolites including several amino acids (N-methylalanine, lysine, glutamine, phenylalanine), arachidonic acid, several lipids [lysoPC (18:2), PC (34:2), PE (26:0)] and bile acids (tauroursodeoxycholic acid, taurocholic acid, deoxycholylglycine and cholylglycine).
In these published studies, no attempts were made to link any of the metabolomics findings with the results of expression profiling studies of PDAC. Metscape represents the next generation of programs that enable integration and comparisons of different types of experimental data. Here we demonstrate the utility of Metscape for bringing together gene expression and metabolite data and deriving a better understanding of the underlying metabolic processes associated with pancreatic cancer.
Gene expression data from 36 paired tumor and normal pancreatic tissue samples obtained by Badea et al. (2008) were downloaded from the GEO database (GSE15471). Previously published analysis of these data involved identification of differentially expressed genes with rather stringent cutoff (P<9×10−12) and enrichment analysis with L2L, a tool that compares the user submitted list to the predefined gene sets (e.g. gene lists from other experiments, GO categories, etc.) (Newman and Weiner, 2005).
Initially, we submitted this dataset to LRpath, which is a logistic regression-based method and does not require a significance cutoff. LRpath analysis against GO and KEGG databases revealed 274 enriched gene sets with FDR <0.01 (Supplementary Table S5). Among the most significant upregulated concepts were GO categories related to RNA and DNA metabolism, RNA splicing, cell cycle, cell adhesion, regulation of apoptosis, NF-kappaB cascade protein catabolism and immune response and 11 KEGG pathways, including ECM-receptor interaction, focal adhesion, small cell lung cancer, proteasome, cell cycle and B cell receptor signaling. TGF-β signaling identified in the original publication was confirmed as one of the upregulated pathway (P=0.01). The downregulated GO categories included sensory perception of various stimuli, G-protein signaling, cyclic nucleotide mediated signaling, calcium, potassium and sodium ion transport, and fatty acid oxidation. The most significant downregulated pathways were neuroactive ligand–receptor interaction, taste transduction, calcium signaling pathway, glycine, serine and threonine metabolism, nitrogen metabolism, glyoxylate and dicarboxylate metabolism.
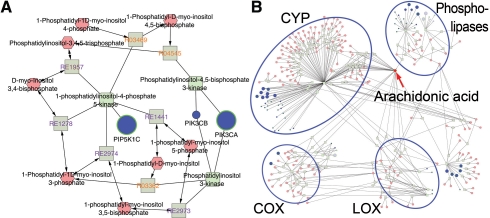
To examine the data in metabolic context, the list of Entrez gene IDs, P-values adjusted for FDR and log fold change values, the list of metabolites with fold change and P-values identified by Urayama et al. (2010) and the LRpath results against the KEGG database were loaded into Metscape and the networks were created. We first built a CREG network, which resulted in one large subnetwork with 5429 nodes and a number of smaller subnetworks. Metscape provides a quick and convenient way to explore large networks and focus on different data subsets of interest. This can be achieved by selecting one or more pathways or concepts from the pathway or concept filter tabs at the bottom of the screen. The main difference between the two filters is that the concept filter is populated with concepts from an input concept file and pathway filter is populated with metabolic pathways from the Metscape database. Figure 3A shows the subnetwork for focal adhesion, one of the top enriched KEGG pathways. It contains 15 genes, 9 of which have q<0.01, and 16 compounds. Most genes in this subnetwork encode kinases and phosphatases that have been implicated in various cancers, e.g. met proto-oncogene (hepatocyte growth factor receptor), platelet-derived growth factor receptor beta (PDGFRB), protein phosphatase 1 (PPP1CA), phosphoinositide-3-kinase (PIK3CA and PIK3CB) and phosphatidylinositol-4-phosphate 5-kinase (PIP5K1C). The latter enzyme, PIP5K1C, phosphorylates phosphatidylinositol 4-phosphate and converts it to phosphatidylinositol 4,5-bisphosphate. A recent study in Drosophila showed that an increased level of phosphatidylinositol 4-phosphate resulted in increased Hedgehog signaling (Yavari et al., 2010). Phosphatidylinositols (PI) are lipid constituents of the plasma and organelle membranes. Different phosphorylated versions of PI have been shown to regulate cytoskeletal organization, signal transduction, and membrane and protein trafficking (Skwarek and Boulianne, 2009). This example demonstrates that Metscape provides a quick way to identify the compounds that are associated with the metabolic genes of interest. Currently, there are no experimental metabolomics data for any of the PIs in PDAC samples; these compounds could be targeted for follow-up experiments. Metscape also allowed us to examine available experimental metabolomics data coupled with relevant expression profiling data for PDAC. Figure 3B shows the network for arachidonic acid, one of the compounds that were increased in PDAC plasma samples. Arachidonic acid is a component of phospholipids. The products of its metabolism, eicosanoids, have been implicated in various diseases including cancers (Panigrahy et al., 2010). Three families of enzymes that control the three main branches of arachidonic acid metabolism belong to cyclooxygenase (COX), lipoxygenase (LOX) and cytochrome P450 (CYP) families. Two members of the COX family, PTGS1 and PTGS2, were upregulated in PDAC samples (q=0.026 and 0.005, respectively), whereas three of four genes encoding LOX enzymes were downregulated (q<0.002). Of the 28 genes encoding various CYP enzymes, the 14 most significant genes (q<0.05) were found to be downregulated; 8 of these were part of the enriched concept ‘Metabolism of xenobiotics by cytochrome’. Interestingly, most of the genes encoding phospholipases responsible for releasing arachidonic acid from phospholipids were also downregulated and eight of those were also part of the glycine, serine and threonine metabolism concept. In summary, this analysis suggests that the increased level of arachidonic acid is not likely to be the result of increased release from phospholipids. Since genes encoding CYP and LOX enzymes are downregulated, the decreased activity of these two branches of arachidonic acid metabolism could be responsible for the accumulation of this metabolite.
Fig. 3.
Metscape analysis of PDAC gene expression and metabolite data. (A) Focal adhesion was identified from gene expression data by LRpath analysis as the most significant concept. Parts of the subnetwork containing phosphoinositide-3-kinase (PIK3CA and PIK3CB) and phosphatidylinositol-4-phosphate 5-kinase (PIP5K1C) are shown. Gene node border color is green if the gene was significant (q<0.05). (B) Arachidonic acid network. COX, cyclooxygenases; LOX, lipoxygenases; CYP, cytochrome P450 monooxygenases.
3.4 Building and exploring the network of inflammation-related metabolites potentially involved in cancers
One of the documented risk factors for developing cancer is chronic inflammation. For example, it has been shown that there is a link between pancreatic adenocarcinoma and chronic pancreatitis (Dinarello, 2006). Chronic inflammatory disease has the potential to evolve toward neoplasia. Miron et al. found increased levels of TNF-α and IL-6 in serum of patients with chronic pancreatitis and pancreatic adenocarcinoma compared with healthy controls (Miron et al., 2010). Their results suggested a pathogenic role for chronic inflammation in pancreatic carcinogenesis. If small molecule metabolic markers common to cancers and inflammation are identified, they could potentially be used as early markers of disease, helping to understand the underlying molecular pathways and lead to new therapies.
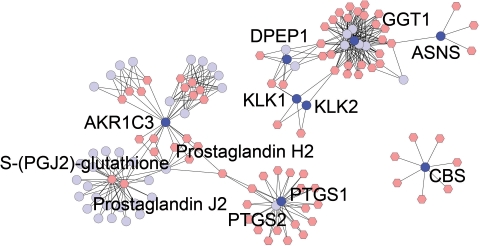
We used Metscape in combination with the previously developed CentiScape plugin for Cytoscape (Scardoni et al., 2009) to reconstruct and annotate the metabolic networks, identify related nodes and pathways and guide our search for metabolites with potential involvement in inflammation and cancers. We started by searching the literature and publicly available HMDB database (http://hmdb.ca) to generate a list of 83 metabolites involved in inflammation (Supplementary Table S6). This list was used to query Metscape and to build a CREG network which resulted in a large, but not fully connected, network. The smaller isolated network components were expanded using the Metscape expand function and a fully connected network graph with 499 nodes and 1400 non-redundant binary interactions was created. Supplementary Table S7 provides a complete list of pathways for this network. Notably, these pathways are involved in generating lipid-derived pro-inflammatory mediators, mainly related to arachidonic acid metabolism, and in generating intermediates leading to proinflammatory oxidative stress. Metscape provides two ways to access the pathway information. In addition to the pathway filter tab described above, pathways are displayed in the data panel as attributes of reaction nodes (or reaction edges in C network graphs).
We then used CentiScape to categorize nodes of this network according to their individual topological relevance. Specifically, we calculated the centrality indexes (Centroid, Betweenness and Eccentricity) for all nodes in order to be able to better focus the analysis on the most important nodes. The genes with the Centroid and Betweenness centrality scores that exceeded the network average were extracted and annotated using literature-derived information and the subset of genes involved both in inflammation and in cancer development. This subset of genes was then used to further query Metscape and reconstruct the corresponding metabolic network (Fig. 4 and Supplementary Tables S8 and S9). We hypothesize that the resulting network contains metabolites and genes that are potentially relevant both in cancer development as well as in inflammation generation. This example demonstrates that Metscape functionality can easily be enhanced and expanded by using it in combination with other Cytoscape plugins.
Fig. 4.
The network of the candidate metabolites and genes involved in both inflammation and cancers. Only few ‘hub’ genes and metabolites are labeled.
4 DISCUSSION
Visualization of genes, enzymes, metabolites, pathways and their relationships proved is an important step in biological interpretation of high-throughput omics experiments and understanding molecular mechanisms of diseases. The ability to link different types of biological data and examine them within the same framework further enhances this understanding. While there is an abundance of tools for visualizing molecular pathways, few currently offer the level of interactivity desired by many researchers. We have presented a freely available software application, Metscape 2, that provides the framework for integrative analysis of metabolomics and gene expression data and facilitates interactive data exploration. It maps user-submitted data to gene/metabolite concepts stored in the internal database, retrieves the relationships for the mapped concepts and generates the network graphs, where experimental data are highlighted with the number of visual features, including node color, size and border. One of the important Metscape 2 features is the ability to generate network graphs using both user-defined sets of input nodes and the set of canonical metabolic pathways. This unique feature, together with the ability to link isolated network graphs by using the expand function, makes Metscape an important addition to the set of existing visualization tools. Several types of networks graphs available in Metscape provide different level views of the data. For example, the CREG networks provide the most accurate representation of the relationships between compounds, reactions, enzymes and genes, while CG networks enable a high-level overview of the data.
Another unique feature of Metscape 2 is the ability to perform gene set enrichment testing and visualize the enriched concepts in metabolic networks graphs. The number of reliable high-resolution gene expression profiling data sets far exceeds those available for metabolites. Therefore, computational algorithms that attempt to predict metabolite changes from gene expression or proteomics data are particularly important (Zelezniak et al., 2010). The Metscape framework can be easily expanded to accommodate such algorithms in the future. It also provides a straightforward way to incorporate other types of data such as flux through reactions.
We have demonstrated the utility of Metscape 2 for analyzing the unbiased metabolomics data from plasma samples of patients with sepsis-induced ALI and have shown that the networks created by Metscape were useful for inferring the relationships between metabolites, genes and pathways and helpful in generating new hypotheses that can be tested experimentally. We also showed that Metscape can be used to analyze gene expression data from PDAC samples and link them to plasma metabolites from PDAC patients. The biological interpretation of such datasets is complicated by the fact that gene expression data were obtained using tumor tissue while the metabolites were measured in plasma, which represents a physiological ‘average’ of the whole organism. Careful experimental validation will be required to test our computationally derived hypotheses. Finally, we demonstrated how Metscape functionality can be augmented by using one of the previously developed Cytoscape plugins, CentiScape.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the members of the National Center for Integrative Biomedical Informatics (NCIBI) for testing the software and providing valuable input on improving the user interface. We thank Paul Trombley for his help in preparing the figures.
Funding: National Institutes of Health (U54DA21519); Michigan Diabetes Research and Training Center Pilot and Feasibility Grant (to A.K.); Fondazione Cariveron (to C.L.); NIHP30ES017885 (to M.A.S. and G.S.O.); Michigan Nutrition Obesity Research Center (5P30DK089503) and 5R01DK079084 (to C.B. and B.A.).
Conflict of Interest: none declared.
REFERENCES
- Andresen M., et al. Lipoperoxidation and protein oxidative damage exhibit different kinetics during septic shock. Mediators Inflamm. 2008;2008:168652. doi: 10.1155/2008/168652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea L., et al. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- Bathe O.F., et al. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol. Biomarkers Prev. 2011;20:140–147. doi: 10.1158/1055-9965.EPI-10-0712. [DOI] [PubMed] [Google Scholar]
- Biswas S.K., Rahman I. Environmental toxicity, redox signaling and lung inflammation: the role of glutathione. Mol. Aspects Med. 2009;30:60–76. doi: 10.1016/j.mam.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarla C., et al. The relationship between plasma cholesterol, amino acids and acute phase proteins in sepsis. Amino Acids. 2004;27:97–100. doi: 10.1007/s00726-004-0064-x. [DOI] [PubMed] [Google Scholar]
- Collisson E.A., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G. Jr, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dinarello C.A. The paradox of pro-inflammatory cytokines in cancer. Cancer Metastasis Rev. 2006;25:307–313. doi: 10.1007/s10555-006-9000-8. [DOI] [PubMed] [Google Scholar]
- Draghici S., et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte N.C., et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham C.M., Chirichella T.J. Attenuated hypocholesterolemia following severe trauma signals risk for late ventilator-associated pneumonia, ventilator dependency, and death: a retrospective study of consecutive patients. Lipids Health Dis. 2011;10:42. doi: 10.1186/1476-511X-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle T., et al. Identification of ectonucleotidases CD39 and CD73 in innate protection during acute lung injury. J. Immunol. 2007;178:8127–8137. doi: 10.4049/jimmunol.178.12.8127. [DOI] [PubMed] [Google Scholar]
- Fang F., et al. Discrimination of metabolic profiles of pancreatic cancer from chronic pancreatitis by high-resolution magic angle spinning 1H nuclear magnetic resonance and principal components analysis. Cancer Sci. 2007;98:1678–1682. doi: 10.1111/j.1349-7006.2007.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolkis A., et al. SMPDB: the small molecule pathway database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., et al. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics. 2010;26:971–973. doi: 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alcalde F., et al. Paintomics: a web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27:137–139. doi: 10.1093/bioinformatics/btq594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao T., et al. Compartmentalization of the Edinburgh Human Metabolic Network. BMC Bioinformatics. 2010;11:393. doi: 10.1186/1471-2105-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker B.H., et al. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan V.E., et al. Oxidative lipidomics of apoptosis: redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free Radic. Biol. Med. 2004;37:1963–1985. doi: 10.1016/j.freeradbiomed.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klukas C., Schreiber F. Integration of -omics data and networks for biomedical research with VANTED. J. Integr. Bioinform. 2010;7:112. doi: 10.2390/biecoll-jib-2010-112. [DOI] [PubMed] [Google Scholar]
- Kreindler J.L., Shapiro S.D. Lung turns to AA (adenosine analogues) to dry out. Nat. Med. 2007;13:406–408. doi: 10.1038/nm0407-406. [DOI] [PubMed] [Google Scholar]
- Lucas R., et al. Regulators of endothelial and epithelial barrier integrity and function in acute lung injury. Biochem. Pharmacol. 2009;77:1763–1772. doi: 10.1016/j.bcp.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay M.A. Alveolar fluid clearance in patients with ARDS: does it make a difference? Chest. 2002;122:340S–343S. doi: 10.1378/chest.122.6_suppl.340s. [DOI] [PubMed] [Google Scholar]
- Miron N., et al. Proinflammatory cytokines: an insight into pancreatic oncogenesis. Roum. Arch. Microbiol. Immunol. 2010;69:183–189. [PubMed] [Google Scholar]
- Newman J.C., Weiner A.M. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 2005;6:R81. doi: 10.1186/gb-2005-6-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang D., et al. Metabolomic profiling of serum from human pancreatic cancer patients using (1)H NMR spectroscopy and principal component analysis. Appl. Biochem. Biotechnol. 2011;165:148–154. doi: 10.1007/s12010-011-9240-0. [DOI] [PubMed] [Google Scholar]
- Paley S.M., Karp P.D. The Pathway Tools cellular overview diagram and Omics Viewer. Nucleic Acids Res. 2006;34:3771–3778. doi: 10.1093/nar/gkl334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahy D., et al. Cytochrome P450-derived eicosanoids: the neglected pathway in cancer. Cancer Metastasis Rev. 2010;29:723–735. doi: 10.1007/s10555-010-9264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor M.A., et al. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardoni G., et al. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serkova N.J., et al. Utility of magnetic resonance imaging and nuclear magnetic resonance-based metabolomics for quantification of inflammatory lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;295:L152–L161. doi: 10.1152/ajplung.00515.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson M.I., et al. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst. Biol. 2010;4:140. doi: 10.1186/1752-0509-4-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skwarek L.C., Boulianne G.L. Great expectations for PIP: phosphoinositides as regulators of signaling during development and disease. Dev. Cell. 2009;16:12–20. doi: 10.1016/j.devcel.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Stringer K.A., et al. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma (1)H-nuclear magnetic resonance quantitative metabolomics and computational analysis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011;300:L4–L11. doi: 10.1152/ajplung.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina Y.Y., et al. Oxidative lipidomics of hyperoxic acute lung injury: mass spectrometric characterization of cardiolipin and phosphatidylserine peroxidation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2010;299:L73–L85. doi: 10.1152/ajplung.00035.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urayama S., et al. Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun. Mass. Spectrom. 2010;24:613–620. doi: 10.1002/rcm.4420. [DOI] [PubMed] [Google Scholar]
- Xia J., Wishart D.S. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- Yavari A., et al. Role of lipid metabolism in smoothened derepression in hedgehog signaling. Dev. Cell. 2010;19:54–65. doi: 10.1016/j.devcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelezniak A., et al. Metabolic network topology reveals transcriptional regulatory signatures of type 2 diabetes. PLoS Comput. Biol. 2010;6:e1000729. doi: 10.1371/journal.pcbi.1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., et al. Enhanced airway inflammation and remodeling in adenosine deaminase-deficient mice lacking the A2B adenosine receptor. J. Immunol. 2009;182:8037–8046. doi: 10.4049/jimmunol.0900515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.