Abstract
Motivation: Flux balance analysis (FBA) is a well-known technique for genome-scale modeling of metabolic flux. Typically, an FBA formulation requires the accurate specification of four sets: biochemical reactions, biomass metabolites, nutrients and secreted metabolites. The development of FBA models can be time consuming and tedious because of the difficulty in assembling completely accurate descriptions of these sets, and in identifying errors in the composition of these sets. For example, the presence of a single non-producible metabolite in the biomass will make the entire model infeasible. Other difficulties in FBA modeling are that model distributions, and predicted fluxes, can be cryptic and difficult to understand.
Results: We present a multiple gap-filling method to accelerate the development of FBA models using a new tool, called MetaFlux, based on mixed integer linear programming (MILP). The method suggests corrections to the sets of reactions, biomass metabolites, nutrients and secretions. The method generates FBA models directly from Pathway/Genome Databases. Thus, FBA models developed in this framework are easily queried and visualized using the Pathway Tools software. Predicted fluxes are more easily comprehended by visualizing them on diagrams of individual metabolic pathways or of metabolic maps. MetaFlux can also remove redundant high-flux loops, solve FBA models once they are generated and model the effects of gene knockouts. MetaFlux has been validated through construction of FBA models for Escherichia coli and Homo sapiens.
Availability: Pathway Tools with MetaFlux is freely available to academic users, and for a fee to commercial users. Download from: biocyc.org/download.shtml.
Contact: mario.latendresse@sri.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Flux balance analysis (FBA) is a methodology (Orth et al., 2010; Thiele and Palsson, 2010) for constructing genome-scale, steady-state models of metabolic networks. It has a variety of applications from evaluation of potential growth media for an organism to prediction of phenotypes of knockout mutants. However, current FBA technology has a number of limitations. The development of FBA models is extremely time consuming, requiring 12–24 months (Thiele and Palsson, 2010). Once developed, models are typically communicated using cryptic data files that make the models difficult for other parties to comprehend and evaluate. The fluxes calculated by a model are also difficult to comprehend—interpreting the long list of fluxes produced from an FBA computation to understand the flux levels of reactions within the network can be very time consuming.
We present a new software technology for generating FBA models that accelerates model development and produces models that are easier to inspect and comprehend. Our approach, based on a new tool called MetaFlux, couples FBA with pathway databases (DBs) via the Pathway Tools (Karp et al., 2010) software environment, of which MetaFlux is a part.
The first phase of model development is to infer a metabolic reaction list from an annotated genome sequence. Although some groups appear to still perform this process in a manual fashion (Thiele and Palsson, 2010), Pathway Tools has performed this process automatically for many years (Dale et al., 2010; Paley and Karp, 2002). It maps the enzyme names, EC numbers and Gene Ontology terms found in an annotated genome to reactions in the MetaCyc DB (Caspi et al., 2010). The resulting reaction list (and predicted metabolic pathways and pathway hole fillers) are stored in the form of a Pathway/Genome Database (PGDB).
MetaFlux provides a novel completion method for accelerating the second phase of model development, in which the reaction list, plus associated nutrient, secretion,and biomass metabolite sets, are converted to a functional FBA model. By completion we mean the software suggests components (e.g. reactions and nutrients) to add to a model to render the model feasible. A model is feasible if the linear optimizer used to solve the system of equations of which an FBA model is comprised, can find a non-zero solution to those equations. Intuitively, for an FBA model to be feasible, it means that the metabolic network can produce all compounds in the biomass equation from the nutrients. The completion method reduces the time-consuming work of meticulously refining the network of reactions, the set of biomass metabolites and the selection of appropriate metabolites as nutrients and secretions (e.g. byproducts, toxins and signaling molecules), which are needed to produce a feasible FBA model.
Genome-scale metabolic network models typically contain hundreds of reactions, and are typically missing reactions in their early formulations, since most genome-scale networks are derived from genome annotations that are themselves incomplete. Similarly, the initially formulated set of nutrient and secreted compounds may be incomplete. Any of the preceding omissions can result in an infeasible FBA model. The MetaFlux gap-filler suggests changes to the reaction network [an approach pioneered by (Kumar et al., 2007)], and to the nutrients and secretions, that will complete the model to make it feasible.
In addition, the initially formulated biomass metabolite set could contain metabolites that cannot be produced even after the reaction network, nutrients and secretions have been extended by the gap-filler. Rather than simply report that the model is infeasible, the MetaFlux gap-filler identifies the maximal subset of biomass metabolites that can be produced, thus focusing the user's model refinement work on the unproducible metabolites.
One way to use MetaFlux to correct an infeasible model is as follows. In practice, a user can start with a very simple model (a fixed-part) that is trivially feasible (e.g. no biomass is produced) and use MetaFlux to complete it to produce the maximal subset of biomass metabolites coming from the infeasible model by adding a minimum number of additional nutrients, secretions and reactions from try-sets. Metaflux is often used in that manner by starting from a fixed-part that is feasible and adding necessary and useful components from user-specified try-sets to obtain a feasible model with a maximal set of biomass metabolites. This approach is productive as a feasible model is always obtained, whereas starting with a fixed-part that is infeasible might not create a feasible model.
A related problem is that the user might want to assess, if additional biomass metabolites are considered for addition to the model, what is the maximal set of those additional metabolites that can be added while maintaining a feasible model. For example, in Lee et al. (2009), the authors did a laborious search for the set of metabolites that could be added to their biomass reaction. Answering the preceding question using other FBA software requires an exponential number of trials if all subsets are tried, whereas MetaFlux can answer this question in one trial.
Furthermore, our approach facilitates the comprehension of FBA models, because the PGDB containing the FBA model can be published on the Web (e.g. see BioCyc.org) where the user can explore the FBA model using a wide range of query and visualization tools (such as to visualize metabolites, reactions, pathways and their connections to the genome). Comprehension of predicted metabolic fluxes can be enhanced by painting those fluxes onto a metabolic network diagram and onto pathway diagrams. Comparison of FBA models is facilitated by the use of controlled vocabularies for metabolites, reactions and pathways across multiple pathway DBs (and the associated FBA models). In addition, Pathway Tools contains model validation tools including a reaction-balanced checker and a tool for identifying dead-end metabolites (Karp et al., 2010).
2 SYSTEMS AND METHODS
This article focuses on the generation of FBA models via a method we call Multiple gap-filling. Gap-filling (Kumar et al., 2007; Orth and Palsson, 2010; Reed et al., 2006) is the process of completing the reaction network of an organism, by adding reactions from a reference DB, to produce a set of biomass metabolites. In general, such a completion might be infeasible (meaning the linear optimizer still finds no solution even after gap-filling) since some biomass metabolites might not be producible even by adding all reactions from the reference DB. When the completion is infeasible, it is necessary to modify the biomass reaction to remove some of its metabolites and retry the gap-filling process. Doing so manually is very time consuming as it is potentially necessary to try all subsets of the biomass metabolites. Similarly for nutrients and secretions, it is sometimes necessary to add nutrients and secretions to obtain a feasible FBA model, but manual exploration of all combinations will be tedious.
The objective function of an FBA model is not always the maximization of the biomass. For example, ATP production is sometimes the objective function to maximize. Nevertheless, we consider that all objective functions can be expressed by a set of metabolites. Such a set of metabolites can be represented in MetaFlux as the ‘biomass’.
Multiple gap-filling is an extension of gap-filling where an FBA model is generated by simultaneously computing minimal completions of the reactions in the reaction network, the metabolites for the biomass reaction, the nutrients and the secretions.
Feasibility of an FBA model is the most fundamental aspect to maintain. Without a feasible FBA model, no fluxes can be obtained. Therefore, our approach is to start with the smallest feasible model and complete it as much as possible by maintaining feasibility. Indeed, starting with an infeasible model is problematic since it is not guaranteed that a feasible model will be obtained even via multiple gap-filling. By starting with a feasible model, it is possible to guarantee that a feasible completed model will be obtained. For example, an FBA model that is not required to produce any metabolite in the biomass is trivially feasible. Starting with such a model, it might be possible to complete it, and still maintain feasibility, by adding metabolites to the biomass. This might require adding reactions to the reaction network and/or adding metabolites as nutrients or secretions. This process is essentially the approach of multiple gap-filling.
In general, our approach uses try-sets and fixed-sets. The fixed-sets are the elements of the model of which we are the most confident: the current set of reactions in the reaction network of the organism, the most likely set of metabolites in the biomass reaction, and the likely sets of metabolites for nutrients and secretions. These sets form the initial FBA model. As mentioned in the previous paragraph, the set of metabolites for the biomass might be empty. It is even recommended to begin with an empty biomass fixed-set to ensure feasibility of the initial model.
Similarly, there are four try-sets corresponding to the fixed-sets. These try-sets are supplied by the user as sources from which the software can complete the FBA model. The try-set for reactions, simply called the try-reactions set, is a reference DB of reactions. In the experimental results provided in this article, we used MetaCyc (Caspi et al., 2010; Karp and Caspi, 2011) as a reference DB for reactions. MetaCyc version 15.0 contains 9200 metabolic reactions. Try-sets are also provided for biomass components, nutrients and secretions. In all, four fixed-sets and four try-sets are provided by the user.
In summary, the approach is to start with properly chosen try-sets, that can, without major impediments, contain extraneous elements, and iteratively move the appropriate elements from the try-sets to the fixed-sets as suggested by MetaFlux. The fixed-sets form the final FBA models.
3 ALGORITHM
We present the mathematical formulation to complete an FBA model, formulated as a mixed integer linear program given the try-sets and fixed-sets. We assume that all reactions have been processed such that all generic reactions have been transformed into one or several instantiated reactions (Section 4.1). All unbalanced reactions are also removed as discussed in Section 4.1.
The mixed integer linear programming (MILP) formulation has a fixed-part and a try-part. The fixed-part consists of four fixed-sets: the reactions R, the nutrient metabolites N, the secretion metabolites S and the biomass metabolites B. Similarly, in general, four corresponding try-sets are given: the try-reactions Rt, the try-nutrient metabolites Nt, the try-secretion metabolites St and the try-biomass metabolites Bt.
The sets R and Rt contain only unidirectional reactions. That is, if a reversible reaction is present in the model, two reactions, of opposite direction, are used to represent it. Therefore, in a solution, all reaction fluxes are zero or positive. Notice that the set Rt contains not only the try-reactions from a reference DB, but also, if requested by the user, all reversed forms of irreversible reactions from the reference DB and/or the PGDB of the organism. This approach supports exploration of reversed reactions from the PGDB and from the reference DB.
A binary variable is associated with each element of Rt, Nt, St and Bt. We denote by V the set of binary variables. Each binary variable controls the presence or absence of the corresponding reaction or metabolite in the model. In the following, we formulate the constraints based on these binary variables to control the completion of a model, that is, to formulate a MILP that performs multiple gap-filling.
Since we are using MILP, all constraints must be linear. That is, all constraints are of the form ∑cixi = bi where the ci and bi are constants and xi are variables. For each reaction r (rt for try-reaction), a continuous variable fr (frt for try-reaction rt) is introduced to represent its flux. Each reaction is unidirectional, so that the value of the flux is zero or positive. An frt variable is conditionally bounded by its binary variable srt. That is, assuming that all flux reactions are bounded above by the constant b, then the following constraint is added to the MILP formulation for each try-reaction rt:
This constraints the flux of the try-reaction to be zero, if the binary variable srt is zero, essentially not adding try-reaction rt to the model; otherwise, when srt is one, the flux of the try-reaction rt can be any positive value bounded by b. Furthermore, since the objective function (see below) has the term wrtsrt where the weight wrt is non-zero and negative, srt will be set to one only if rt is non-zero since when rt is zero the objective function could be made trivially higher by setting srt to zero. So, the variable frt, representing the flux of try-reaction rt, is non-zero if and only if srt is one.
The formulation for a try-nutrient, a try-secretion or a try-biomass metabolite is similar to a try-reaction. In fact, a nutrient n is essentially an exchange reaction from nothing to n; likewise for a secretion s, it is a reaction from s to nothing. Therefore, try-nutrients, try-secretions and try-biomass metabolites can be encoded in the MILP formulation as special try-reactions.
Notice that the try-biomass metabolites are independent of each other; that is, each try-biomass metabolite has its own binary variable. On the other hand, in a typical FBA formulation, the biomass metabolites are represented as one biomass reaction, not one reaction per biomass metabolite. Therefore, to have a non-zero flux for the biomass, each metabolite of the biomass reaction must be produced by the model. This constraint alone is very strong and can likely make an FBA model infeasible. In contrast, the MILP formulation avoids such a strong constraint.
The general objective of the desired FBA model is specified by the user. This objective can take many different forms: (i) generate as many biomass metabolites as possible suggesting to add a minimum number of reactions or (ii) generate a maximum number of secreted metabolites by suggesting to add a minimum number of reactions, and more. These objectives can be specified by providing numerical coefficients, called weights and denoted wi, for each type of try-reaction, try-secretion, try-nutrient and try-biomass. Therefore, the MILP objective function to maximize is
| (1) |
As mentioned, the value of the weight coefficient wi depends on the object controlled by binary variable si. To be more precise, the weight depends on the type of component (e.g. nutrient) associated with the binary variable si: this weight could be for a nutrient, secretion or biomass metabolite; or one of the weights for a reaction (e.g. taxonomic range of a reaction from the reference DB). We use different weight parameters for the reactions from the reference DB (MetaCyc) depending on the following conditions:
the reaction is in the taxonomic range of the PGDB;
the reaction is outside the taxonomic range of the PGDB;
the reaction's taxonomic range is unknown ;
the reverse reaction is used; and
the reaction is spontaneous.
A reversed reaction from the PGDB has also its own weight. Notice that the weights of reactions do not take into account known metabolic pathways, although this approach could be explored in future work. The selection of appropriate weights is further discussed in Section 3.3. This objective function has an additional term, not shown here, to avoid high fluxes in loops as described in Section 3.2. The next subsection describes another term that could be added to the objective function.
3.1 Biomass and the objective function
A variation of the objective function (1) is the following:
| (2) |
The term wBfB was added to Equation (1). The variable fB represents the flux of the biomass reaction composed of the fixed biomass metabolites and wB a user-given weight (typically a positive integer). Notice that the fluxes of the try-biomass metabolites do not contribute to fB.
One possible scenario in the use of such an objective function would be to answer questions like: which reactions could be added to increase the flux of the biomass reaction? Or could some nutrients be used to increase the flux of the biomass reaction?
Such questions can be answered by selecting appropriate weights for the reactions to add versus the weight wB. For example, selecting wB=2 (that is, a gain of 2 units per one unit of flux) and a weight of −20 (that is, a cost of 20 units) for any reaction to add would allow adding one reaction for each increase of 10 units in the fB flux. The term wB fB can easily be deactivated by selecting the weight wB to be zero. Notice that the term fB is typically the objective function of an FBA formulation that has no try-sets. That is, fB is the typical objective of an FBA formulation to maximize.
3.2 Loops and unbounded fluxes
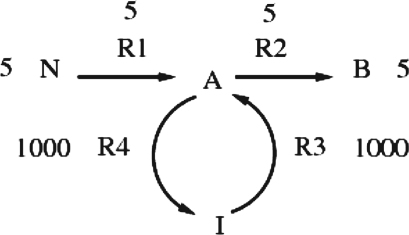
It can often be observed that the formulation of Section 3 produces a solution with high fluxes assigned to many reactions. These fluxes are close to their upper bound and many do not contribute to the biomass (example: Fig. 1). Some reactions might contribute some fluxes to the biomass while others might simply be set to a zero flux and no change to the biomass would occur. These high fluxes are often caused by cycles in the network, and are called loops by some authors (Pinchuk et al., 2010; Price et al., 2006; Smallbone and Simeonidis, 2009).
Fig. 1.
Hypothetical reactions with high fluxes but contributing zero flux to the biomass. This is a very simple reaction network where each reaction has only one reactant and one product. Metabolite N is the nutrient and metabolite B is the sole metabolite in the biomass. Metabolite I is some other intermediate metabolite. Reactions R1 and R2 contribute to the biomass with a flux of 5. Reactions R3 and R4 have a very high flux of 1000 but they do not contribute to the biomass, although they do produce metabolite A.
Loop fluxes are a significant impediment when analyzing FBA results, because they mislead the user into thinking that these reactions are major contributors to biomass production. For example, in our Escherichia coli FBA model we often observed tens of loop reactions. Therefore, it is preferable to limit the fluxes of these reactions to biologically reasonable values.
The MILP formulation of Section 3 does not provide a solution with accurate absolute fluxes, but rather it provides accurate relative fluxes. By relative fluxes, we mean that the correct ratios of the biomass metabolite fluxes are found, but not necessarily the correct absolute fluxes. Therefore, it is not necessary to apply a technique that would control the unbounded high fluxes and at the same time find the exact absolute fluxes. The following term is added to the objective function 1 to remove loops, with the biological justification that an organism does not need to produce more metabolites, and/or in greater quantities, if these metabolites are not used to either produce more metabolites, and/or in greater quantities, in the biomass or as secretion.
| (3) |
That is, the sum of the reaction fluxes (including any reactions added to the model) is added to the objective function scaled with a negative factor. This term minimizes the fluxes while still maximizing the general objective. These reactions do not include the virtual reactions representing the biomass metabolites, nutrients or secretions. Since the objective function is maximized and the overall term is negative, the fluxes will be minimized. The factor ch is typically a small positive value <1. It reduces the importance of the term ∑rifri compared with the other terms of the objection function so that adding a reaction to increase other gains is not detrimental.
In summary, the general objective function is obtained by adding terms 2 and 3 giving
| (4) |
where wB is a weight (integer), fB is the flux (real value) of the biomass reaction, si is a boolean variable, V is the set of all boolean variables, wi is a weight (integer), ri is any reaction, ch is a small non-negative constant (typically <1) and fri is the flux (real value) of reaction ri.
3.3 Selecting values for the weights
The selection of the appropriate weight values in the MILP formulation of Section 3 is key in solving specific goals. We present some common goals and discuss weight selection for them. In general, we use the term weight to mean either a cost (a negative weight) or a gain (a positive weight). All weights are integers.
Favoring biomass metabolites:
a popular goal is to produce as many biomass metabolites as possible from the given try-sets. For such a goal, the gain on each biomass metabolite should be set to a value larger than the cost of adding several reactions, nutrients and secretions. For example, if the cost of adding any reaction is set to 10 (that is, a negative weight of −10), and we accept adding as many as 20 reactions to produce one metabolite in the biomass, the gain on each metabolite of the biomass should be set to at least 200 (weight 200).
In the extreme case, we might be interested in making sure that any number of reactions could be added to produce any metabolite. In that case, the gain (positive weight) for adding one biomass metabolite should be greater to the sum of the costs of adding all possible reactions from the reference DB.
Favoring secreted metabolites:
there are many possible solutions to an FBA problem, i.e. alternative sets of reactions with non-zero fluxes that produce the biomass metabolites. But the reactions that secrete a metabolite could be considered more biologically correct than those that do not secrete any metabolite for the same biomass produced. In such a case, the reactions producing these secretions should be favored in the model.
The general MILP formulation 3 allows any values for the weights, in particular for the secretions: we could favor secretions by selecting a strictly positive value for their weight, say 5. This positive weight would have the effect of favoring reactions that produce the secreted metabolites, not only the added reactions from the reference PGDB, but also reactions from the organism. Indeed, any reaction set to a non-zero flux that could produce an excess of the secreted metabolite would increase the objective function.
4 IMPLEMENTATION
MetaFlux is tightly integrated with Pathway Tools (Karp et al., 2010). A user interface controls execution of both the gap-filler and the FBA model solver. MetaFlux automatically generates an MILP or linear program (for the gap-filler or FBA model solver, respectively) from a PGDB, as a file in .lp format. MetaFlux invokes the SCIP solver on that file, retrieves the solution and produces a report file that lists all suggestions of the gap-filler, and the predicted reaction fluxes.
The user can request that predicted fluxes be painted on the Cellular Overview (Latendresse and Karp, 2011), which is an organism-specific metabolic map diagram generated by Pathway Tools from an PGDB. The range of flux values is mapped to a color scale, and each reaction that carries a flux is assigned an appropriate color value from that scale. Animation can be used for the visual comparison of different flux states, e.g. of fluxes resulting from growth under different nutrients. Fluxes can also be displayed on individual pathway diagrams.
Since the PGDB is the FBA model, FBA models developed with MetaFlux are readily inspectible using the wide range of query and visualization tools in Pathway Tools (Karp et al., 2010), which can run as both a desktop application and as a web server. Users can easily look up individual reactions or metabolites; metabolite pages list all reactions and pathways in which the metabolite participates, and provide the chemical structure and alternative names for the metabolite. Pathway pages depict the organization of reactions into metabolic pathways. The E.coli and human FBA models described in the next sections are accessible via the EcoCyc.org and HumanCyc.org websites, and through the Supplementary Material.
Another advantage of constructing FBA models within the Pathway Tools environment is that such models are based on controlled vocabularies of metabolites and reactions. Most metabolite and reaction objects within each PGDB were computationally derived from the MetaCyc DB by the PathoLogic program. That derivation process assigns the same DB object identifiers to a given metabolite and reaction in every PGDB, thus facilitating comparison of PGDBs and FBA models from multiple organisms using the large number of Pathway Tools comparison operations.
Pathway Tools includes a variety of editing tools for interactively updating a PGDB including a reaction editor, a chemical structure editor and a pathway editor (Karp et al., 2010). In addition, Pathway Tools can export a PGDB to SBML format.
4.1 Preprocessing of reactions
Instantiation of generic reactions:
many enzymatic reactions are written in the biomedical literature as ‘generic reactions’ whose metabolites include one or more compound classes (e.g. ‘a carbohydrate’). Pathway Tools can faithfully represent such generic reactions and the corresponding compound classes and instances, and generic reactions are used extensively in MetaCyc and other PGDBs because of the brevity with which they represent a family of reactions.
However, generic reactions introduce difficulties in network computations such as FBA. Consider the following hypothetical generic reaction, where X is a metabolite class containing the instances x1, x2 and x3, and Y is a class containing the instances y1 and y2.
R1:X+H2O=Y
Imagine that the only valid instantiation of R1 is
R2:x1+H2O=y1
Imagine further that the PGDB contained this additional reaction:
R3:a+b=x1
Software that naively traversed the metabolic network by searching for literal matches between the reactants and products of reactions would not detect that the same metabolite can be both a product of R3 and a reactant of R1. Therefore, we developed a preprocessing stage that generates instantiated forms of generic reactions, in which appropriate compound instances have been substituted for compound classes. Successfully instantiated reactions are included in the generated FBA model that is sent to the solver.
The instantiation code enumerates all possible ways of combining the instances of X with those of Y, and generates corresponding reaction instances by substituting each class with one of its instances. If, for a given instance in X, there is exactly one instance in Y that leads to a mass balanced reaction, then an instantiated reaction structure is created for this combination of instances. This instantiated reaction is added to the FBA model (but is not permanently stored in the PGDB as a new frame). If more than one instance in Ys leads to a mass balanced equation for a given instance in Xs, then the situation is considered ambiguous, and no instantiated reaction is generated.
Note that for instantiations to succeed, it is key that the appropriate instance metabolites exist in the PGDB, and that they have been correctly classified under the compound classes used in the generic reactions. We are curating our compound hierarchy on an ongoing basis, to improve the success rate of this procedure.
Polymerization pathways are handled separately. These are often involved in fatty acid metabolism, where a series of elongation steps are chained together. For a limited set of polymerization pathways, Pathway Tools generates instantiated reactions, with as many as eight monomer units added. The chemical formula of a monomer unit is automatically inferred from the one reaction in the polymer pathway that contains the polymerization step.
To include other cross-linked and complex compounds like glycans, we recommend defining representative compounds that stand for common chemical fragments within the lipopolysaccharide network, and to formulate reactions and pathways based on those representatives.
Unbalanced reactions:
unbalanced reactions should not be used in an FBA model. An unbalanced reaction could create an infeasible model or a model that generates incorrect fluxes, because they do not follow the law of conservation of mass. Therefore, during the preprocessing phase, unbalanced reactions are removed from the fixed-reactions set and try-reactions set.
Since a single unbalanced reaction could generate an incorrect FBA model, the process of balance-checking reactions is stringent: if it cannot be determined with certainty that a reaction is balanced, that reaction is not included in the model. There are reactions for which the preprocessing could not determine if they were really unbalanced (e.g. if chemical structures are missing for their substrates), but they are still not included in the model.
The log file contains a list of all reactions that were not included in the model due to their unbalanced state. The balance state of a reaction can be verified within the reaction editor.
4.2 Gene and reaction knockouts
MetaFlux can solve FBA models in the context of single and multiple knockouts of genes and reactions. The user specifies a set of n genes and/or k reactions and the number of g>0 genes and r>0 reactions that are simultaneously ‘removed’ from the FBA model. Each g–r combination of the subsets of genes and reactions constitute a reduced FBA model for one knockout experiment. That is, the total number of knockout experiments is 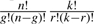 . Removing a gene implies that zero, one or several reactions might become inactive in the model. There might be no reaction or several reactions becoming inactive since knocking out one gene takes into account protein complexes as well as isozymes. MetaFlux solves the corresponding reduced FBA model typically showing growth or no-growth of the organism, although partial growth is also possible.
. Removing a gene implies that zero, one or several reactions might become inactive in the model. There might be no reaction or several reactions becoming inactive since knocking out one gene takes into account protein complexes as well as isozymes. MetaFlux solves the corresponding reduced FBA model typically showing growth or no-growth of the organism, although partial growth is also possible.
Genome-wide sets of single- and multiple-knockouts for genes and reactions can be succinctly specified using keywords. For example, all metabolic genes, that is, all genes whose product catalyzes at least one metabolic reaction, can be specified with one keyword.
MetaFlux outputs the results of all knockout experiments into a single solution file listing the genes that were knocked out, the reactions made inactive and the flux of the biomass reaction (i.e. the objective function). The user can also request that a file be produced for each knockout experiment that lists the fluxes of all the reactions in the corresponding reduced FBA model.
5 DISCUSSION
5.1 Application to Homo Sapiens
We have applied our FBA modeling tool to the HumanCyc PGDB. HumanCyc was created in 2003 (Romero et al., 2004) from the complete Human proteome extracted from GenBank. After a hiatus of several years, curation of HumanCyc resumed in 2009, and the accuracy of the HumanCyc metabolic network has benefited from this modeling exercise. The manual aspects of this model-building project involved ~4 weeks of full-time work. The resulting FBA model is available as HumanCyc version 15.5, and in the Supplementary Materials.
In addition to MetaFlux, we used the dead-end metabolite tool, available in Pathway Tools, to identify compounds that are only reactants, or only products, of HumanCyc reactions. Such compounds are necessarily excluded from FBA models due to balance constraints. Therefore, any compounds that are truly only reactants in a metabolic network must be obligate nutrients (such as essential amino acids or vitamins); we added some input dead-ends to the fixed set of input nutrients as needed to generate all biomass metabolites with no added reaction. Similarly, dead-end metabolites that are only products in HumanCyc reactions cannot be further metabolized by the metabolic network and must be added to the secretion set to balance the FBA model. This was done for two dead-end metabolites in the HumanCyc model (see File 1 in Supplementary Material).
The gap-filling analysis of the HumanCyc metabolic network proposed insertion of a number of reactions (which in some cases constituted entire pathways) from the MetaCyc DB for inclusion in HumanCyc to enable production of biomass metabolites. We researched these reactions in the experimental literature and found that many of the proposed reactions had been observed experimentally, which we added to HumanCyc.
Examples of added reactions are EC 5.4.2.7, which is mediated by a phosphodeoxyribomutase encoded by the PGM2 gene. We added a metabolic pathway for choline degradation that includes reactions catalyzed by a choline dehydrogenase encoded by CHDH (EC 1.1.99.1) and a betaine aldehyde dehydrogenase encoded by ALDH7A1 (EC 1.2.1.8). We also inserted reactions previously absent from HumanCyc that are known to occur experimentally but with no known enzymes. These include EC 2.4.2.23 catalyzed by an as yet unidentified deoxyuridine phosphorylase; an entire pathway for palmitoleic acid biosynthesis, including EC 1.4.99.–, catalyzed by a palmitoyl desaturase; and EC 3.1.2.14 catalyzed by a putative palmitoleayl thioesterase.
We also modified reaction directions as suggested by the gap-filler and supported by literature research. These include the addition of the reverse reaction direction for existing reactions: EC 2.4.2.1, a purine nucleoside phosphorylase encoded by the gene PNP, which acts upon multiple substrates; EC 1.17.4.1, an ADP reductase encoded by gene RRM; EC 1.5.1.3, a dihydrofolate reductase (DHFR); and EC 5.3.1.1, a triosephosphate isomerase (TPI1).
The transformation of HumanCyc into a working FBA model was an iterative process that involved >30 computational experiments. Those experiments identified biomass components that could not be produced because the required inputs were absent, such as pantothenic acid, Vitamin B5 required for enzymatic co-factors and missing essential nutrients such as choline. Other problems encountered involved errors in human curation of the network model, such as enzymes curated as existing in the wrong compartment (example: HMGCoA Reductase in peroxisome but not cytosol).
An interesting result occurred when MetaFlux made multiple attempts to add or reverse reactions to produce dimethylglycine formerly called vitamin B15 or B16 (Graber et al., 1981). This compound is not considered a classical vitamin, because no deleterious effects have been found when absent from the diet, but is used as a nutritional supplement to enhance athletic performance (Gray and Titlow, 1982). It is unclear where dimethylglycine is produced in human metabolism other than degradation of choline which is an essential nutrient (Haubrich and Gerber, 1981; Skiba et al., 1982). Dimethylglycine is in food and is consumed as a nutrient (Huang et al., 2008), and since the modeling program requires more of it to balance the metabolic network, this may be the case for the biological system as well.
From a simple nutrient set of nine essential amino acids, four vitamins, glucose, glutamine, pyruvate, phosphate and choline, the HumanCyc FBA model is able to produce the 11 non-essential amino acids, the ribo and deoxy-ribo nucleotides, four enzymatic cofactors, complex membrane lipids and steroid hormones. The resulting model produces 52 biomass metabolites (see Table 1 and File 1 in Supplementary Material). The fluxes can be visualized by clicking the following region of text to invoke BioCyc Omics Viewer using File 1 in Supplementary Material.
Table 1.
Reaction counts for each PGDB
| PGDB | Reactions in PGDB | Reactions in model | Reactions with flux |
|---|---|---|---|
| HumanCyc | 1721 | 2411 | 241 |
| EcoCyc | 1330 | 1888 | 370 |
| MetaCyc | 6750 | 13 920 | NA |
Column 2: number of metabolic reactions. Column 3: number of reactions in the FBA model, after instantiation of generic reactions and converting reversible reactions into two unidirectional reactions. Column 4: number of reactions that carried flux in a solution of the model. The MetaCyc statistics are relevant because they show the number of reactions considered by the gap-filler. The MetaCyc ‘Reactions in Model’ cell includes forward and backward directions of every MetaCyc reaction since the gap-filler considers reversed reactions.
5.2 Application to E.coli
EcoCyc (Keseler et al., 2011) is a highly curated PGDB of the well-studied Gram-negative bacterium E.coli K-12 MG1655. Although EcoCyc has a long curation history, this is our attempt to produce an FBA model from the EcoCyc PGDB.
We started with an initial set of biomass compounds containing only core metabolites needed for making proteins and nucleic acids, and a few cofactors like Coenzyme A, NAD+ and NADP+. All biomass compounds were placed into the try-set. Nutrients of a minimal medium with glucose as a carbon source were placed into the try-nutrient set. The ATP synthase reaction was explicitly added to the model, which otherwise did not contain transport reactions, although electron transfer reactions are included. We used the gap-filler to suggest addition of reactions from MetaCyc, and to suggest reversal of reactions, from both EcoCyc and MetaCyc, using default values for the weights. After initial fixes that allowed the core metabolites to be produced, we incrementally added more compounds to the biomass.
A common problem we encountered was related to reactions involving protein metabolites. If the chemically modified forms of a protein lack chemical structures in the PGDB, then the mass balance of its reactions cannot be determined, and such reactions were filtered from the model. To remedy these cases, adequate protein structures were devised and added, and the reactions were properly balanced. One example was the reaction ADPREDUCT-RXN, whose metabolites are reduced and oxidized thioredoxin; once adjustments were made, the biomass compound dATP was produced by the model. Another example was reaction 1.8.4.8-RXN, which also needed reduced and oxidized thioredoxin to produce sulfite, on the path ultimately to l-cysteine. This biomass compound was produced through gap-filler addition of reactions from MetaCyc that involved sulfite. Balancing this reaction eliminated the unnecessary reaction additions. A third example was reaction RXN0-882.
The gap-filler suggested adding >140 reactions to EcoCyc to produce the minor biomass constituent spermidine. It turned out that a waste product of spermidine synthesis in EcoCyc is S-methyl-5-thio-d-ribose, which is a metabolic dead-end because EcoCyc contained no transporter for this compound (it is known to be secreted from the cell). The existence of the dead-end inhibited utilization of the existing pathway in EcoCyc, thus the gap-filler saw the need to add a large number of alternative reactions. Adding S-methyl-5-thio-d-ribose to the secretions set resolved this problem. In future work, it would be useful to investigate whether automatically adding all dead-end metabolites to the try-secretions set would make it easier to find these cases.
Extending the biomass to include lipid products involved significant work. To demonstrate the principle, we focused on producing lipids containing fatty acids with 16 carbons. The main barrier was the proper curation of the generic reactions and the classifications of lipid instances, such that generic reactions were instantiated correctly. The Pathway Tools command ‘Show pathway's instantiated reactions’ is very useful for debugging generic reactions.
Once the gap-filler no longer added reactions to the model, we converted the model into a true FBA model, without any try-sets. To ensure that the TCA cycle carried flux under the aerobic growth condition we tested, we had to remove several reactions explicitly from the model, involving the glyoxalate bypass and citrate lyase. These reactions are normally disabled by cellular regulation. The resulting FBA model produces 58 biomass metabolites (see Table 1 and File 2 in Supplementary Material). The fluxes can be visualized by clicking the following region of text to invoke the BioCyc Omics Viewer using File 2 in Supplementary Material.
Many of the encountered problems in preparing an FBA model for EcoCyc took between half a day and a day to analyze and resolve. Overall, it took 4 weeks of work to construct the model, which is available as EcoCyc version 15.5, and in Supplementary Materials. For this PGDB, the reaction gap-filler did not help, as EcoCyc was already well curated in the sense of containing the required reactions, such that no truly missing reactions are needed to be imported from MetaCyc. Most fixes involved repairs to existing reactions. The gap-filler component that was very useful was determination of which biomass compounds could not be produced at a given point in time.
We validated the E.coli model, and MetaFlux in general, by using the gene knockout component of MetaFlux to simulate the effects of single-gene deletions in E.coli in glucose minimal medium. As our gold standard for E.coli knockout phenotypes we used the Supplementary Material from Feist et al. (2007), specifically, the 238 genes that were listed as essential under glucose minimal medium experimental conditions, and the remaining 1022 non-essential genes. This approach allows a direct comparison of the accuracy of the Feist et al. FBA model iAF1260 (accuracy = 92% for 1260 genes; accuracy = 90.6% for the same 873 genes assessed in our model below) with our FBA model on the same gold standard. Our model had an accuracy of 86.1% for the 873 genes that our model shared with the gold standard [151 true positives (TPs), 601 true negatives (TNs), 59 false positives (FPs), and 62 false negatives (FNs); accuracy is defined as (TP+TN)/(TP+TN+FP+FN)]. We find the accuracy of our model to be acceptable, particularly given that the model of Feist et al. has been under development for many years. Please see File 3 in Supplementary Material for a table of all the genes and their experimental and predicted essentialities.
5.3 Discussion summary
Overall, we found that development of FBA models using MetaFlux was considerably shorter than times traditionally cited for FBA model development: 4 weeks for EcoCyc and 4 weeks for HumanCyc. However, an exact comparison of development times is tricky. On one hand, the development of HumanCyc would have been shorter had we begun that project with the current version of MetaFlux, because some of its most valuable debugging and report tools were developed toward the end of the project as a result of our model-development experiences from HumanCyc. On the other, our FBA models have not undergone as much validation as some published models (Feist et al., 2007), and both EcoCyc and HumanCyc had undergone curation before we converted them to FBA models. The PathoLogic component of Pathway Tools can further shorten model development times relative to the procedure in Thiele and Palsson (2010) because it automatically infers the metabolic reactions of an organism from its annotated genome.
Although many reaction insertions and reversals proposed by the gap filler were correct and helpful in our development efforts, we note that a significant number (~50%) of the gap-filler-proposed reactions were considered to be unlikely by our curators, due to either lack of support in the experimental literature or reactions that were clearly outside their taxonomic range (e.g. reactions that occur only in plants).
Henry et al. (2010) recently published a method for high-throughput generation of FBA models that includes automated gap filling of reaction insertions and of reaction directions for tens of genomes. Given our experience that a significant fraction of reaction insertions suggested by the gap-filler are incorrect, we must question the trustworthiness of a purely automated approach. We acknowledge the possibility that differences in details of our algorithms or reference reaction DBs would yield different gap-filler accuracies. Future work could compare our approaches on a given genome.
An additional reason that we believe that some manual intervention will be required for some time for the development of high-quality FBA models is that the automated method used in Henry et al. (2010) to estimate the biomass composition of a given organism is approximate and should be supplemented by manual addition of biomass compounds identified through experimental work, which in turn will often lead to a need to manually supplement the reaction network of the organism if the needed reactions are not present in the reference reaction DB from which the gap-filler draws.
Solving, to optimality, gap-filler MILP problems using SCIP often took <10 min on a 3.0 GHz Intel/4 GB workstation, although a few runs required close to 1 h of processing. SCIP was chosen, among other non-commercial solvers, since it has good performance for solving MILP formulations. In particular, our experience shows that it is very often much faster, on at least the type of formulations used in MetaFlux, than the Gnu Linear Programming Kit (GLPK). SCIP is also free for academic users.
Limitations of our approach include the following. Our reaction gap-filler has limited flexibility regarding compartmentation; reactions can be gap-filled into the same compartment as the reaction occupied in MetaCyc, but cannot be shifted arbitrarily among compartments—this issue will be remedied in a subsequent release. Although use of the Cellular Overview to inspect fluxes predicted by MetaFlux was useful in our projects, shortcomings of the approach include that reactions suggested for addition by the gap-filler are not present in the Cellular Overview of a given PGDB (since those reactions are not yet part of the PGDB), making it difficult to understand the connections of such reactions to the metabolic network. And because one reaction can occur in multiple places in the Cellular Overview, it can be confusing to see an isolated reaction within a single pathway that carries flux (that reaction may carry flux within a different pathway). Another issue is that a given generic reaction in the Cellular Overview can be instantiated to multiple instance reactions, each of which can carry a different flux value, but the Cellular Overview does not contain separate lines for each instance reaction.
5.4 Related work
Other FBA software packages include the COBRA Toolbox (Becker et al., 2007; Bordbar et al., 2011), Acorn (Sroka et al., 2011), SimPheny (Mahadevan et al., 2006), SurreyFBA (Gevorgyan et al., 2010), FASIMU (Hoppe et al., 2011), BioMet Toolbox (Cvijovic et al., 2010), CycSim (Le Fèvre et al., 2009), WEBcoli (Jung et al., 2009) (only for E.coli) and Model SEED (Henry et al., 2010). The capabilities of these systems are as follows. Ability to model essential genes and reactions: COBRA, FASIMU, BioMet, SimPheny, Acorn, Model SEED, CycSim, MetaFlux. Flux variability analysis: COBRA, FASIMU, Acorn. Inference of reactome and metabolic pathways from genome: Model SEED, Pathway Tools. Gap-filling (Kumar et al., 2007): Model SEED, COBRA, MetaFlux. Multiple gap-filling: MetaFlux. Visualization of fluxes onto automatically generated layouts of full metabolic maps and individual pathways: FASIMU, SimPheny, CycSim, WEBcoli, Acorn, SurreyFBA, COBRA, MetaFlux/Pathway Tools.
GapFill (Kumar et al., 2007) was the first program for filling gaps in FBA models. Its gap-filling strategies include inserting new reactions (from to a reference DB such as MetaCyc), reversing the directionality of existing reactions, and adding transport reactions between compartments or the external space. GapFill does not gap-fill a biomass reaction as it assumes that the biomass reaction is fixed and forms one complete reaction. Therefore, if no subset of reactions can complete the network such that the biomass reaction has a non-zero flux, GapFill will not produce suggestions, whereas MetaFlux will identify the subset of biomass metabolites that can be produced. In addition, GapFill does not postulate changes to the nutrients or secreted compounds as MetaFlux does.
A unique aspect of Model SEED is its capability to infer the biomass composition of an organism. In contrast, MetaFlux enables an iterative generation process of FBA models integrated into a complete tool for navigating, querying and modifying PGDBs. Model SEED uses a gap-filling algorithm similar to that of GapFill.
High fluxes due to loops have been reported by many other researchers, and solutions to detect and remove them have been proposed (Price et al., 2006; Smallbone and Simeonidis, 2009). These approaches apply to solving an FBA model and not for solving MILP as done in this work. We have applied a simple technique of minimizing the reaction fluxes to remove these high fluxes.
The GrowMatch program reconciles an FBA model with experimental predictions (Kumar and Maranas, 2009). It uses gene knockout experimental data to correct an FBA model based on growth/no-growth mismatches between in silico prediction and in vivo experimental data. It does not generate FBA models per se, although it can be used to validate a model and help correct a biomass reaction.
5.5 Final conclusion
MetaFlux is a new tool that can increase the speed to construct FBA models for PGDBs. It is well integrated in Pathway Tools, which offers other tools to navigate, modify and analyze PGDBs. In particular, flux values can color metabolic maps available in Pathway Tools.
When compared with all other FBA software tools known to us, MetaFlux has a unique capability to simultaneously suggest modifications to the four essential sets describing an FBA model, namely, the set of reactions, the set of metabolites of the biomass reaction, the set of nutrients and the set of secretions.
We have shown the capabilities of MetaFlux on two DBs, EcoCyc and HumanCyc. This experience demonstrated the applicability of MetaFlux on two complex DBs and the rapidity it provided to construct FBA models.
ACKNOWLEDGEMENTS
We thank Ranjan Srivastava for extensive discussions and advice regarding FBA methodology.
Funding: Award numbers (GM080746, U24GM077678, and GM092729) from the National Institute of General Medical Sciences. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Conflict of Interest: The first, second and third co-authors receive royalties from commercial users of pathway tools.
Footnotes
†Present address: Department of Pharmacology and Clinical Neuroscience, Umeå University, Umeå, Sweden.
REFERENCES
- Becker S.A., et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat. Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- Bordbar A., et al. COBRA Toolbox 2.0. Protocol Exchange. 2011 doi:10.1038/protex.2011.234. [Google Scholar]
- Caspi R., et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvijovic M., et al. Biomet toolbox: genome-wide analysis of metabolism. Nucleic Acids Res. 2010;38(Suppl. 2):W144–W149. doi: 10.1093/nar/gkq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J.M., et al. Machine learning methods for metabolic pathway prediction. BMC Bioinformatics. 2010;11:15. doi: 10.1186/1471-2105-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A., et al. A genome-scale metabolic reconstruction for Escherichia coli K– 12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007;3:121–138. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevorgyan A., et al. SurreyFBA: a command line tool and graphics user interface for constraint based modelling of genome scale metabolic reaction networks. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq679. doi:10.1093/bioinformatics/btq679. [DOI] [PubMed] [Google Scholar]
- Graber C.D., et al. Immunomodulating properties of dimethylglycine in humans. J. Infect. Dis. 1981;143:101–105. doi: 10.1093/infdis/143.1.101. [DOI] [PubMed] [Google Scholar]
- Gray M.E., Titlow L.W. The effect of pangamic acid on maximal treadmill performance. Med. Sci. Sports Exerc. 1982;14:424–427. doi: 10.1249/00005768-198206000-00003. [DOI] [PubMed] [Google Scholar]
- Haubrich D.R., Gerber N.H. Choline dehydrogenase. assay, properties and inhibitors. Biochem. Pharmacol. 1981;30:2993–3000. doi: 10.1016/0006-2952(81)90265-3. [DOI] [PubMed] [Google Scholar]
- Henry C.S., et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- Hoppe A., et al. Fasimu: flexible software for flux-balance computation series in large metabolic networks. BMC Bioinformatics. 2011;12:28. doi: 10.1186/1471-2105-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., et al. Manipulation of sinapine, choline and betaine accumulation in arabidopsis seed: towards improving the nutritional value of the meal and enhancing the seedling performance under environmental stresses in oilseed crops. Plant Physiol. Biochem. 2008;46:647–654. doi: 10.1016/j.plaphy.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Jung T., et al. WEbcoli: an interactive and asynchronous web application for in silico design and analysis of genome-scale e.coli model. Bioinformatics. 2009;25:2850–2852. doi: 10.1093/bioinformatics/btp496. [DOI] [PubMed] [Google Scholar]
- Karp P., Caspi R. A survey of metabolic databases emphasizing the MetaCyc family. Arch. Toxicol. 2011;85:1015–1033. doi: 10.1007/s00204-011-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P., et al. Pathway Tools version 13.0: Integrated software for pathway/genome informatics and systems biology. Brief. Bioinformatics. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler I.M., et al. Ecocyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.S., Maranas C.D. GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput. Biol. 2009;5:e1000308. doi: 10.1371/journal.pcbi.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.S., et al. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latendresse M., Karp P.D. Web-based metabolic network visualization with a zooming user interface. BMC Bioinformatics. 2011 doi: 10.1186/1471-2105-12-176. doi: 10.1186/1471-2105-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fèvre F., et al. CycSim — an online tool for exploring and experimenting with genome-scale metabolic models. Bioinformatics. 2009;25:1987–1988. doi: 10.1093/bioinformatics/btp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-S., et al. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple staphylococcus aureus genomes identify novel antimicrobial drug targets. J. Bacteriol. 2009;191:4015–4024. doi: 10.1128/JB.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R., et al. Characterization of metabolism in the fe(iii)-reducing organism geobacter sulfurreducens by constraint-based modeling. Appl. Environ. Microbiol. 2006;72:1558–1568. doi: 10.1128/AEM.72.2.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Palsson B.O. Systematizing the generation of missing metabolic knowledge. Biotechnol. Bioeng. 2010;107:403–412. doi: 10.1002/bit.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., et al. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley S., Karp P. Evaluation of computational metabolic-pathway predictions for H. pylori. Bioinformatics. 2002;18:715–724. doi: 10.1093/bioinformatics/18.5.715. [DOI] [PubMed] [Google Scholar]
- Pinchuk G.E., et al. Constraint-based model of shewanella oneidensis mr-1 metabolism: a tool for data analysis and hypothesis generation. PLoS Comput. Biol. 2010;6:e1000822. doi: 10.1371/journal.pcbi.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N.D., et al. Candidate states of Helicobacter pylori's genome-scale metabolic network upon application of Loop Law thermodynamic constraints. Biophys. Theory Model. 2006;90:3919–3928. doi: 10.1529/biophysj.105.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.L., et al. Systems approach to refining genome annotation. Proc. Natl Acad. Sci. USA. 2006;103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2004;6:1–17. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiba W.E., et al. Human hepatic methionine biosynthesis. purification and characterization of betaine:homocysteine s-methyltransferase. J. Biol. Chem. 1982;257:14944–14948. [PubMed] [Google Scholar]
- Smallbone K., Simeonidis E. Flux balance analysis: a geometric perspective. J. Theor. Biol. 2009;258:311–315. doi: 10.1016/j.jtbi.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Sroka J., et al. Acorn: a grid computing system for constraint based modeling and visualization of the genome scale metabolic reaction networks via a web interface. BMC Bioinformatics. 2011;12:196. doi: 10.1186/1471-2105-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I., Palsson B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]