Abstract
As microbial ecologists take advantage of high-throughput analytical techniques to describe microbial communities across ever-increasing numbers of samples, the need for new analysis tools that reveal the intrinsic spatial patterns and structures of these populations is crucial. Here we present SitePainter, an interactive graphical tool that allows investigators to create or upload pictures of their study site, load diversity analyses data and display both diversity and taxonomy results in a spatial context. Features of SitePainter include: visualizing α -diversity, using taxonomic summaries; visualizing β -diversity, using results from multidimensional scaling methods; and animating relationships among microbial taxa or pathways overtime. SitePainter thus increases the visual power and ability to explore spatially explicit studies.
Availability: https://sourceforge.net/projects/sitepainter
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: antoniog@colorado.edu, Rob.Knight@colorado.edu
1 INTRODUCTION
As sequencing capacity increases, microbial community analysis is undergoing a revolution in throughput. Where it was once a monumental task just to analyze microbial communities in a handful of samples, we can now process thousands of samples in a single sequencing run (Caporaso et al., 2011). Accordingly, instead of describing those communities in a few samples collected under a few specific conditions, we can now perform comprehensive spatial sampling, exploring interactions among members of the microbial communities and revealing large-scale spatial dynamics (Brodie et al., 2007; Caporaso et al., 2011; DeLong 2009; Gilbert et al., 2010; Gonzalez et al., 2011).
However, the power of these analyses is often limited by our ability to visualize the data. When analyzing biogeographical data, researchers would like to know where specific microbes live, how abundant they are and whether there are any notable patterns relating microbial taxa to each other or to environmental conditions. Traditional ordination methods for visualizing large numbers of microbial communities such as Principal Coordinates Analysis (PCoA) (Gower and Legendre, 1986) are extremely useful for reducing the dimensionality of vast multivariate datasets, but the patterns are often unclear, especially when the results do not map easily onto the sampling structure. For example, it may not be clear that all the points that cluster together at one end of an axis are on the surface of a complex shape without closely examining the sample identifiers (Fig. 1C1).
Fig. 1.
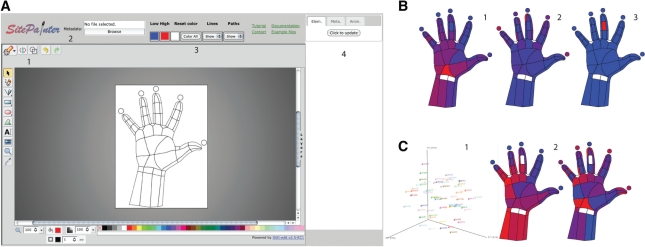
SitePainter illustration of microbial community patterns in a human hand. (A) SitePainter user interface: (1) file image manipulation, (2) metadata loading and processing menu, (3) coloring scheme and (4) interactive menu. (B) The α -diversity analysis showing relative abundance of bacterial taxa on the hand: low values in blue an high values in red (1) Gammaproteobacteria, (2) Bacilli and (3) Actinomycetales. (C) The β -diversity analysis showing overall similarities and differences among samples: (1) 3D PCoA axis where each point represents a sample and each sample is colored independently and (2) first two axes of the PCoA analysis, where similar colors represent samples similar to each other along a given axis in the abstract ordination space, with low values in blue and high values in red.
To address this key barrier to microbial community research, we developed SitePainter that provides compelling visualizations that allow the researcher to better explore spatial patterns in microbial communities. SitePainter allows the user to draw or import representative images for biogeographical sites [in the example, we show a human hand (Fig. 1B–C), although any picture from micro- to landscape-scale is possible] and regions within that site (e.g. a finger, palm, finger tips, etc.). Once the biogeographical image is drawn, the user can import tab-delimited text files, such as those produced by Quantitative Insights Into Microbial Community (QIIME) (Caporaso et al., 2010) to map onto their image. For example, it is often useful to plot the α -diversity (the number of species present in each sample), taxonomy summaries (the abundance of particular kinds of species in the sample) and/or β -diversity (the overall similarities and differences among samples).
2 SITEPAINTER
SitePainter is a software package with an interactive user interface, implemented with HTML and Javascript, which accepts Scalable Vector Graphics (SVG) images and annotates them with per sample data defined by the user. As an example of usage, we show the biogeography of the human hand and assign bacterial information based on QIIME-formatted files. The main work areas of SitePainter are: (i) file image manipulation that allows the user to load and save SVG images; (ii) the metadata loading and processing menu, where the user can select the metadata (tab-delimited format) to be displayed in combination with the images; (iii) the coloring scheme, where the user can select how to represent low and high values, reset the image color or show/hide the display of lines and absent paths, to show more compelling images; and (iv) the interactive menu that allows the user to work with previous selections to find patterns that may not have been apparent when using other visualization tools and techniques (Fig. 1A).
To illustrate the effectiveness of SitePainter, we show the results of a study in which different regions from a subject's hand were sampled for microbes. The subject's hand was outlined, and each region divided into smaller areas that were then sampled using cotton swabs dipped in saline solution. Using SitePainter, we were able to rapidly deduce which bacteria were in different regions of the hand, along with a graphical representation of microbial abundances by processing the sequence data with QIIME (Caporaso et al., 2010) and mapping the abundance of each taxon onto the hand image (Fig. 1B). Using this display, we can immediately see that Gammaproteobacteria is present in the palm, Bacilli are more visible toward the fingertips and Actinomycetales is present in only one area of the hand. This feature thus allows the user to easily visualize the distribution and abundance of microbes across sites.
Going beyond the abundances of individual microbial taxa, SitePainter also allows the user to view similarities and differences at the whole-community level by loading the PCoA axis of the microbial data to reveal patterns that are not obvious in the PCoA plot but are immediately obvious when displayed in the context of the site itself (Fig. 1C: compare left panel to the right two panels. In this case, we can see a clear gradient from the thumb and index fingers to the left-bottom corner of the hand; additionally we can see that the distal phalanges are similar, maybe due to constant contact).
In addition to analyses of microbial taxa, as shown here, SitePainter can also be used to interpret the abundance of genes or pathways across sites for metabolic studies. Several further examples can be found in the Human Microbiome Project web site: http://hmpdacc.org/sp/. These examples analyze the entire Human Microbiome Project (Peterson et al., 2009; Turnbaugh et al., 2007) dataset at several taxonomic levels, and at several different levels of functional classification for shotgun metagenomic reads. We expect that this will be a useful community resource for those trying to interpret the complexities of the human microbiome.
3 CONCLUSIONS
SitePainter provides a user-friendly tool that allows rapid, clear visualization of microbial data on arbitrary user-supplied images. Additionally, it contains several user interface features, such as rapid switching between taxa/coordinates and animations of e.g., diversity overtime. These features facilitate fast exploratory analyses of spatial data. We believe that SitePainter will have a large impact on the field, especially in the case of diseases with complex spatial structure such as psoriasis and ulcerative colitis, and also in large-scale environmental sampling projects such as the Earth Microbiome Project (Gilbert et al., 2010).
ACKNOWLEDGEMENTS
We thank Dirk Gevers, Curtis Huttenhower, Owen White, Lita Proctor and Jesse Zaneveld for feedback and assistance with the HMP implementation of SitePainter, and Dan Sisco, Clotilde Teiling and Chinnappa Kodira at 454 Life Sciences and personnel at the company for carrying out the sequencing associated with this project.
Funding: National Institutes of Health (Grants 5-R01-HL0090480 & 3-R01-HG004872); the Crohns and Colitis Foundation of America (Grant WU-09-72/2905035N); The Bill & Melinda Gates Foundation (Grant WU-09-187/51678); and the Howard Hughes Medical Institute.
Conflict of Interest: none declared.
REFERENCES
- Brodie E.L., et al. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl Acad. Sci. USA. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E.F. The microbial ocean from genomes to biomes. Nature. 2009;459:200–206. doi: 10.1038/nature08059. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., et al. The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic’ study of seasonal and diel temporal variation. PLoS One. 2010;5:e15545. doi: 10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J.A., et al. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand. Genomic Sci. 2010;3:243–248. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., et al. Our microbial selves: what ecology can teach us. EMBO Rep. 2011;12:775–784. doi: 10.1038/embor.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower J.C., Legendre P. Metric and Euclidean properties of dissimilarity coefficients. J. Classification. 1986;3:5–48. [Google Scholar]
- Peterson J., et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]