Abstract
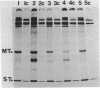
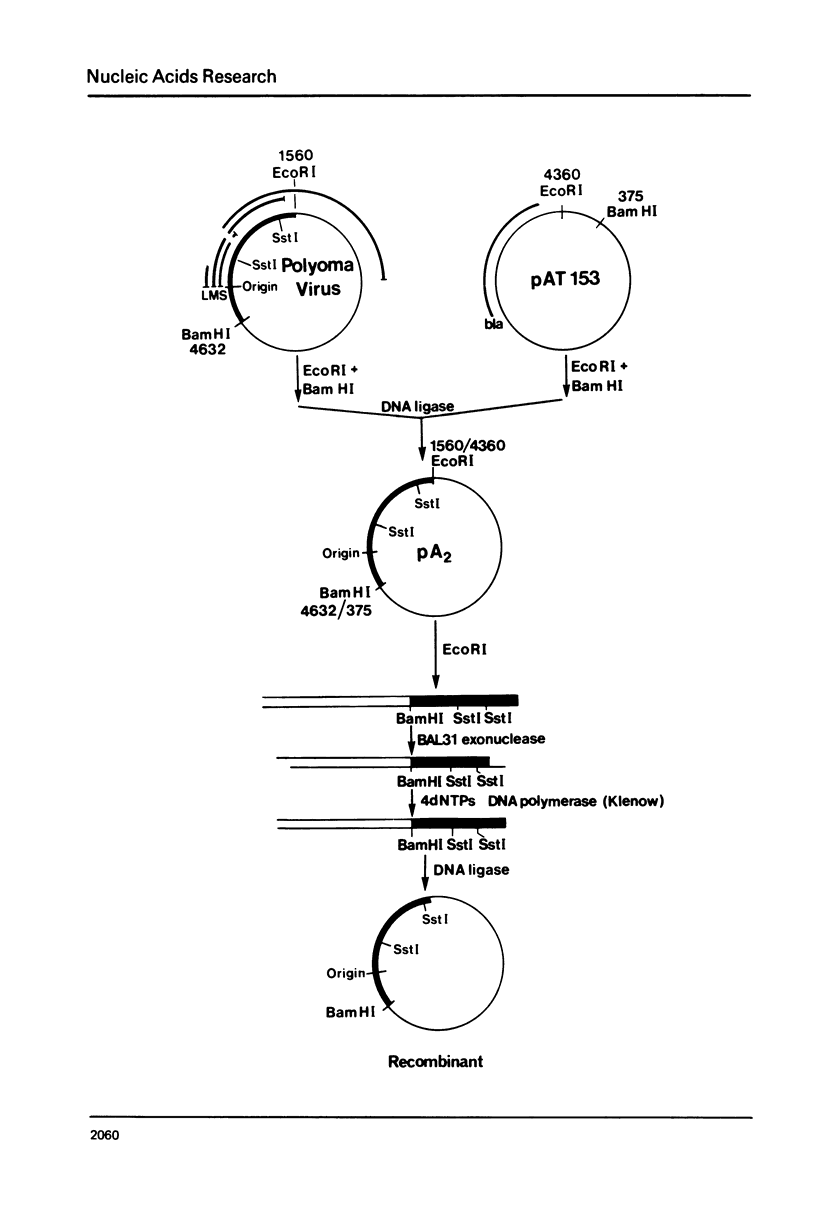
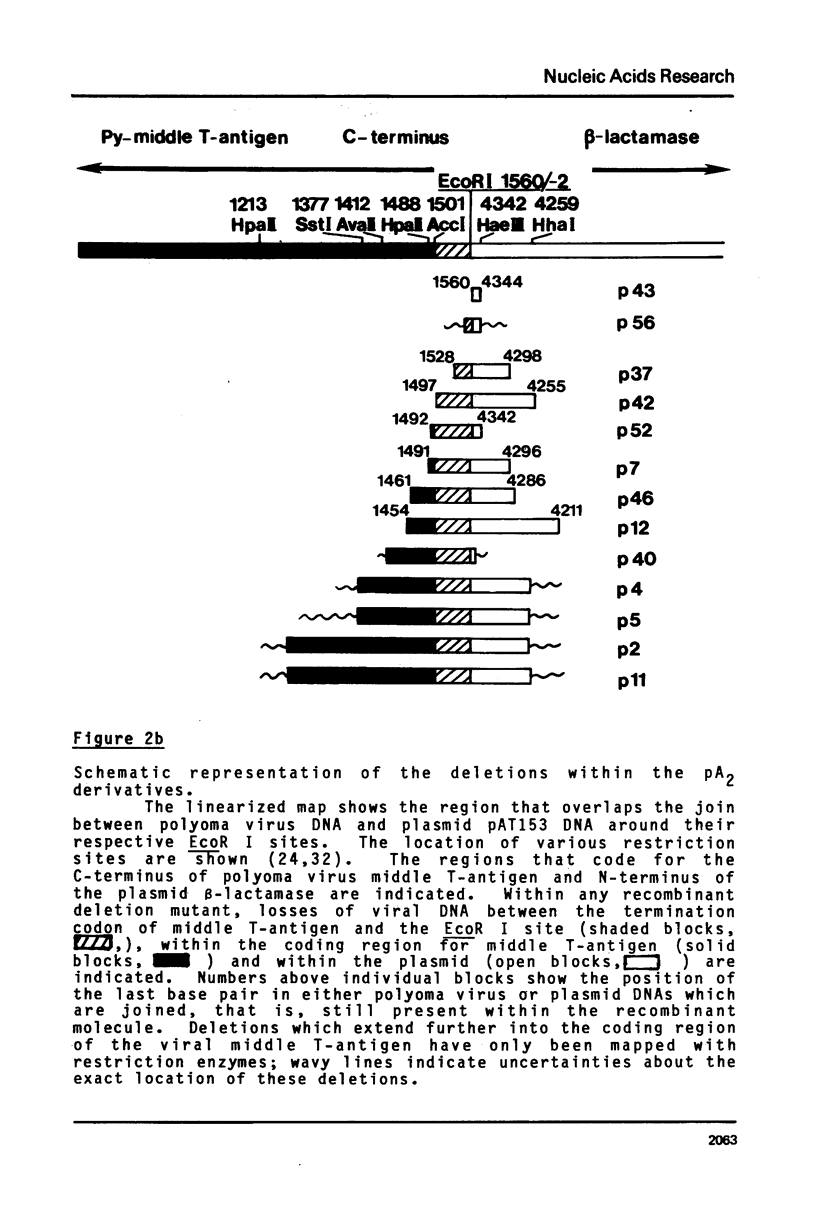
Deletions of polyoma virus DNA around the region that codes for the C-terminus of the viral middle T-antigen were created using a transforming fragment (BamH I/EcoR I) of viral DNA cloned in the plasmid vector pAT153. These species were recloned and assayed for their ability to transform Rat-1 cells in culture. Our results showed that whereas the DNA sequence between the presumed translational termination codon for the viral middle T-antigen and the single viral EcoR I site could be removed with no apparent effect on transformation, the removal of the termination codon itself or any amino acid coding sequences of this protein caused a drastic decrease in the transforming ability of the DNA. Transfection of Rat-1 cells with plasmids that contained viral DNA with deletions which corresponded to the last fourteen or more amino acids of the middle T-antigen never gave rise to cellular transformation.
Full text
PDF







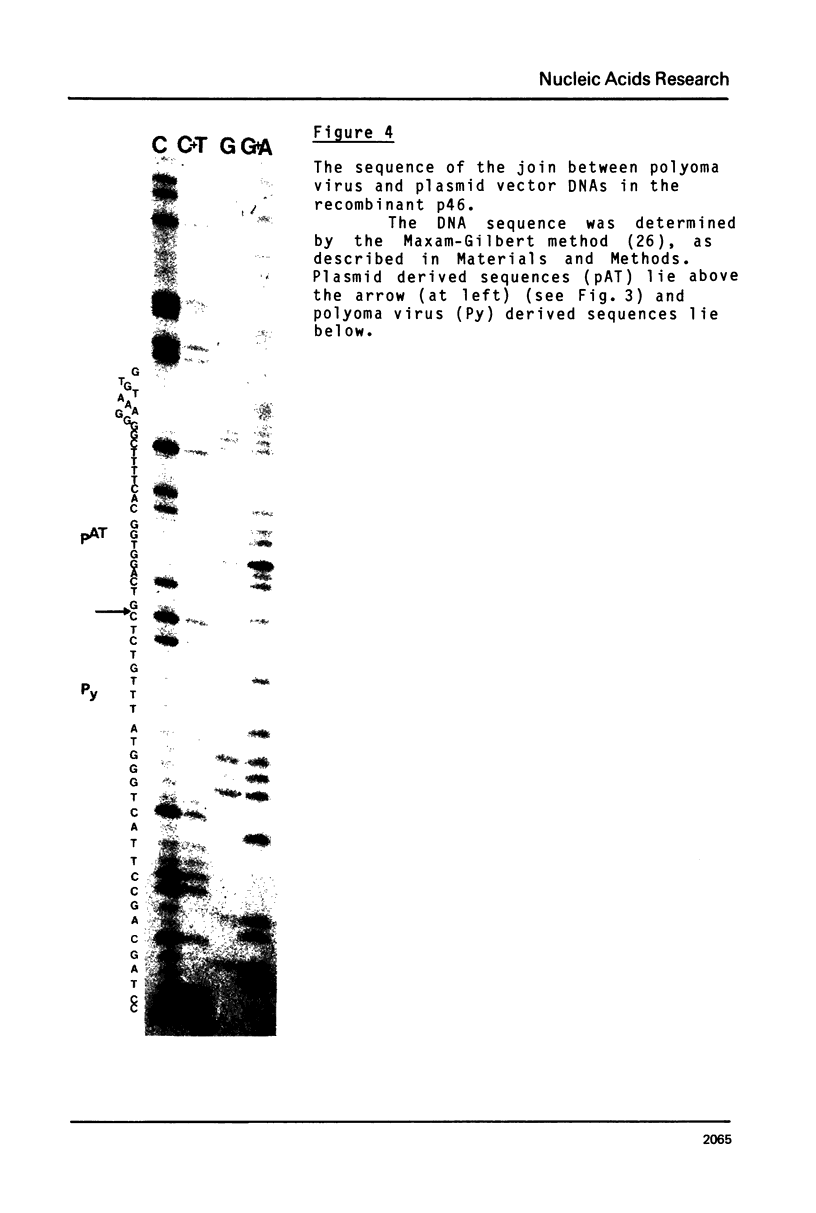
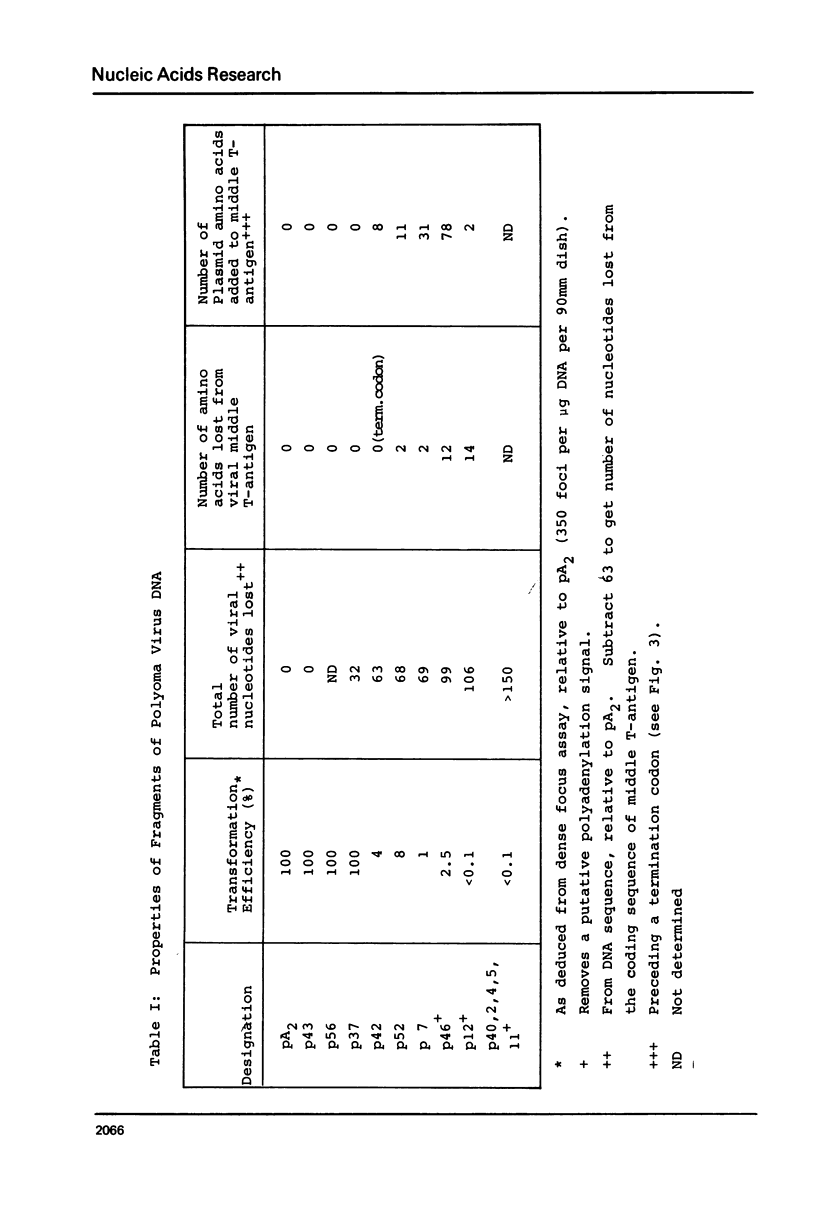







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Bastin M., Bourgaux-Ramoisy D., Bourgaux P. Biological properties of polyoma DNA fragments cloned in plasmid pBR322. J Gen Virol. 1980 Sep;50(1):179–184. doi: 10.1099/0022-1317-50-1-179. [DOI] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen B. Virus-specific early RNA in 3T6 cells infected by a tsA mutant of polyoma virus. Virology. 1978 Mar;85(1):222–230. doi: 10.1016/0042-6822(78)90426-9. [DOI] [PubMed] [Google Scholar]
- Della Valle G., Fenton R. G., Basilico C. Polyoma large T antigen regulates the integration of viral DNA sequences into the genome of transformed cells. Cell. 1981 Feb;23(2):347–355. doi: 10.1016/0092-8674(81)90130-6. [DOI] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke B., Eckhart W. Polyoma gene function required for viral DNA synthesis. Virology. 1973 Sep;55(1):127–135. doi: 10.1016/s0042-6822(73)81014-1. [DOI] [PubMed] [Google Scholar]
- Freeman A. E., Gilden R. V., Vernon M. L., Wolford R. G., Hugunin P. E., Huebner R. J. 5-Bromo-2'-deoxyuridine potentiation of transformation of rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2415–2419. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Ostrander D. A., Hodnett J. L., Legerski R. J., Robberson D. L. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975 Sep;2(9):1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Ito Y., Novak U., Spurr N., Dilworth S., Smolar N., Pollack R., Smith K., Rifkin D. B. Early mutants of polyoma virus (dl8 and dl23) with altered transformation properties: is polyoma virus middle T antigen a transforming gene product? Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):271–283. doi: 10.1101/sqb.1980.044.01.031. [DOI] [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel M. A., Simmons D. T., Hourihan S. L., Rowe W. P., Martin M. A. Interrupting the early region of polyoma virus DNA enhances tumorigenicity. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3713–3716. doi: 10.1073/pnas.76.8.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N. Polyoma virus T antigens expressed in transformed cells: significance of middle T antigen in transformation. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):149–157. doi: 10.1101/sqb.1980.044.01.017. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
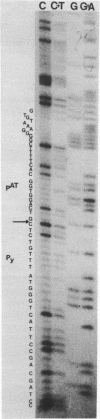
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. L., Chowdhury K., Martin M. A., Israel M. A. Polyoma large tumor antigen is not required for tumorigenesis mediated by viral DNA. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1336–1340. doi: 10.1073/pnas.77.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Dilworth S. M., Griffin B. E. Coding capacity of a 35 percent fragment of the polyoma virus genome is sufficient to initiate and maintain cellular transformation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3278–3282. doi: 10.1073/pnas.77.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K., Welch W. J., Sefton B. M., Esch F. S., Ling N. C. Vesicular stomatitis virus glycoprotein is anchored in the viral membrane by a hydrophobic domain near the COOH terminus. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3884–3888. doi: 10.1073/pnas.77.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Griffin B. E. Polyoma virus. The early region and its T-antigens. Nucleic Acids Res. 1979 Oct 25;7(4):839–857. doi: 10.1093/nar/7.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]