Abstract
Chlamydia trachomatis, a pathogen responsible for diseases of significant clinical and public health importance, remains poorly characterized because of its intractability to routine molecular genetic manipulation. We have developed a combinatorial approach to rapidly generate a comprehensive library of genetically defined mutants. Chemical mutagenesis, coupled with whole-genome sequencing (WGS) and a system for DNA exchange within infected cells, was used to generate Chlamydia mutants with distinct phenotypes, map the underlying genetic lesions, and generate isogenic strains. As a result, we identified mutants with altered glycogen metabolism, including an attenuated strain defective for type II secretion. The coupling of chemically induced gene variation and WGS to establish genotype–phenotype associations should be broadly applicable to the large list of medically and environmentally important microorganisms currently intractable to genetic analysis.
Keywords: genetics, genomics, lateral gene transfer, obligate pathogens
The obligate intracellular bacterium Chlamydia trachomatis accounts for >2.8 million genital tract infections per year in the United States, with associated clinical presentations such as pelvic inflammatory disease, ectopic pregnancies, and infertility (1), and an associated healthcare cost of >$2 billion annually (2). In addition, ocular Chlamydia infections are responsible for ∼1.3 million cases of blinding trachoma in sub-Saharan Africa, the Middle East, and Asia, where the disease is endemic (3).
Infection begins with the attachment of an elementary body (EB), the infectious form of the bacteria, to epithelial cells (4). Once intracellular, the EB transitions to the reticulate body (RB) form and replicates within a membrane-bound vacuole termed an “inclusion.” Later in infection, RBs redifferentiate into the EB form, which are then released into the extracellular space to initiate new rounds of infection (5). Chlamydia is not amenable to routine manipulation with standard molecular genetic tools (e.g., shuttle plasmids, transposons, transducing phages) that have been central to most studies in bacterial pathogenesis (6). Consequently, the extent to which individual Chlamydia genes contribute to the evasion of innate immunity, nutrient acquisition, developmental transitions, or other processes important for the pathogen's survival within a eukaryotic host is unclear.
Here, we describe a method in which chemical mutagenesis is coupled to genome sequencing and a system of DNA exchange among Chlamydia strains to identify mutations responsible for distinct phenotypes, including mutations in previously uncharacterized virulence factors.
Results
EMS Generates Chlamydia Mutants with Stable Plaque Morphotypes.
To establish an experimental genetic system for C. trachomatis, we first generated mutants by treating Chlamydia-infected Vero cells with the alkylating agent ethyl methyl sulfonate (EMS). We assessed the optimal levels of mutagenesis by determining the decrease in the number of infectious progeny and the frequency of rifampin-resistant (RifR) strains emerging as a result of point mutations in the β subunit of RNA polymerase (Fig. S1) (7). We then isolated clonal populations of mutants by infecting Vero cell monolayers at limiting dilutions, followed by a soft agar overlay, and incubating for 7–10 d until visible plaques formed. We observed a diversity of plaque morphologies among mutagenized bacteria, including clumped (Clmp), honey-comb shaped (Hcm), and large granular (Grn) plaques (Fig. S2A). A common plaque morphotype also included small plaques (Spq) (Fig. S2B). These phenotypes were generally stable because plaque-purified mutants amplified in cell culture retained their altered plaque morphologies upon reinfection of Vero cell monolayers.
Whole-Genome Sequencing Establishes a Correlation Between Mutations in the Glycogen-Branching Enzyme GlgB and a Granular Plaque Morphology.
We devised a strategy to perform a genetic analysis of mutants with diverse plaque morphotypes. In brief, we: (i) defined the spectrum of EMS-induced gene lesions by whole genome sequencing (WGS); (ii) identified mutated genes shared by phenotypically similar mutants; and (iii) performed coinfections between wild-type and mutant strains to generate recombinants wherein particular gene lesions could be linked to a phenotype (Fig. 1). To test the validity of this approach, we characterized four Grn mutants generated in a RifR background (Fig. 2A). By WGS, we determined that the number of lesions per genome ranged from 3 to 20 C/G to T/A transitions, as expected for EMS-induced mutations. Three of the four Grn mutants harbored nonsynonymous mutations in glgB, which encodes a glycogen-branching enzyme (Fig. 2B). Glycogen is a branched polymer of α1,4-linked glucose chains interconnected by α1,6 linkages (8). Chlamydia encodes multiple enzymes for glycogen metabolism, including polymer-branching and -debranching activities (9). The glgB mutation in Grn1 leads to an R305C amino acid change and maps to a region of GlgB conserved among prokaryotes and eukaryotes (Fig. 2C and Fig. S3). The mutation in Grn2 (P606L) is in a region of GlgB conserved among the Chlamydiales, whereas Grn3 had two mutations, E597K and D722N, in a less conserved region. Given the nature of the amino acid changes in Grn1 and Grn2 mutants, we postulated that these changes significantly alter the expression or the function of this enzyme. Consistent with this prediction, the Escherichia coli glgB gene bearing analogous mutations failed to complement a glgB deletion strain (Fig. S4), suggesting that unstable or loss-of-function mutations in GlgB are responsible for the formation of plaques with large granular deposits.
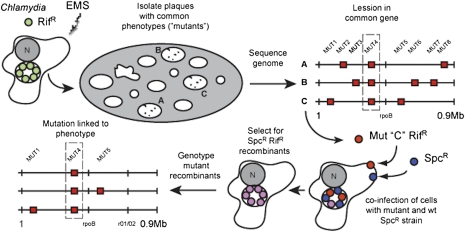
Fig. 1.
Strategy for forward genetic analysis and recombination-based mapping in Chlamydia. Rifampin resistant (RifR) C. trachomatis was mutagenized with EMS during its replicative stage and the resulting infectious progeny were on Vero cell monolayers until visible plaques formed. The genomes of mutants sharing a common plaque morphotype were sequenced to identify common genetic lesions. To establish linkage between these gene lesions and plaque morphology, recombinant RifR SpcR strains were selected in the presence of rifampin and spectinomycin after coinfection of HeLa cells with RifR mutants and wild-type SpcR strains (“crosses”). The segregation of individual mutations present in the parental mutant strain among the recombinant progeny displaying the desired morphotype was determined by targeted DNA sequencing.
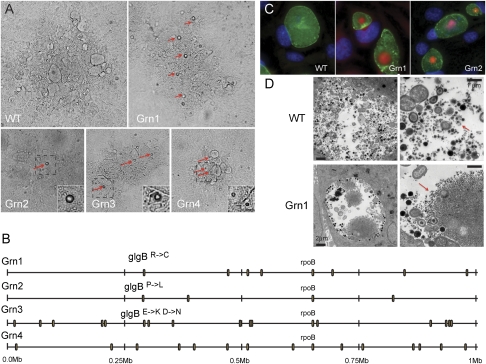
Fig. 2.
Identification of mutagen-induced gene lesions that lead to glycogen aggregation in Chlamydia. (A) Four examples of Chlamydia plaques displaying the accumulation of granular aggregates (Grn), a common morphotype found among plaques from EMS-derived Chlamydia mutants. Red arrows indicate light-refracting granules (see insets). (B) Whole-genome sequencing of four Grn mutants identified mutations in glgB, coding for a glycogen-branching enzyme, in three Grn mutants. These mutations lead to amino acid changes in GlgB residues that are conserved from Chlamydiales to eukaryotes (Fig S3). (C) Grn mutants display large deposits of insoluble glycogen aggregates (red) within the pathogenic vacuole (“inclusion”) (green) of infected cells, as assessed by Schiff staining after periodic acid treatment. (D) Comparison of glycogen in WT and Grn mutants by transmission electron microscopy. Red arrows denote glycogen deposits.
We next performed a microscopic analysis of HeLa cells infected with Grn mutants and determined that these strains accumulated large precipitates within the lumen of inclusions. These precipitates consisted primarily of glycogen, as assessed by their staining with Schiff reagent after periodic acid treatment (Fig. 2C). Glycogen is prominent in the lumen of inclusions of plasmid bearing Chlamydia strains (10, 11), with extrabacterial glycogen likely arising as a result of spontaneous lysis of bacteria (12). In Grn mutants, extrabacterial glycogen forms large intrainclusion aggregates, as observed by transmission electron microscopy (Fig. 2D). We postulate that these aggregates form as a consequence of the accumulation of insoluble unbranched glycogen (13). By live cell imaging, aggregates emerge suddenly in the inclusion lumen late in infection [>36 h postinfection (hpi)], suggesting that after a critical concentration of unbranched glycogen has been reached, the polymers coalesce into large precipitates (Movie S1).
Generation of Recombinant Chlamydia Strains Permits the Establishment of Causative Associations Between Mutations and Plaque Morphologies.
Despite the strong correlative and functional evidence linking glgB mutations with glycogen aggregates in C. trachomatis, we could not exclude the possibility that additional mutations present in Grn1 through -3 strains contributed to this phenotype. The third component of our strategy (Fig. 1) addresses causative relations between genotypes and phenotypes. To establish a causative link between glgB mutations and the accumulation of insoluble glycogen granules, we took advantage of natural DNA exchange among Chlamydiae upon coinfection of a host cell (14–16) to generate recombinant strains. We coinfected HeLa cells with RifR Grn mutants and spectinomycin-resistant (SpcR) or trimethoprim- resistant (TmpR) Chlamydia variants, harvested EBs, and selected for recombinants that formed plaques on Vero cell monolayers in the presence of the two antibiotics (Rif/Spc or Rif/Tmp). The recombinants derived from these “crosses” were further scored for inheritance of the granular plaque morphology. We then determined by targeted DNA sequencing of mutated regions how individual mutations present in the parental Grn strains segregated among recombinants. Consistent with previous findings (14–16), the genomes of the resulting recombinant strains were mosaics of the parental strains (Fig. 3). Importantly, mutations in glgB were always present in recombinant strains with intrainclusion glycogen deposits but not in strains lacking these aggregates, including recombinant strains where the only contribution from the Grn parent were the mutations in glgB (Fig. 3A). Conversely, we identified a recombinant strain that carried all of the mutations present in the parental Grn mutant except for that in glgB (Fig. S5). As expected, this strain did not accumulate glycogen deposits. Based on these findings, we conclude that loss-of-function mutations in GlgB lead to the accumulation of large glycogen aggregates within inclusions and a granular plaque morphology.
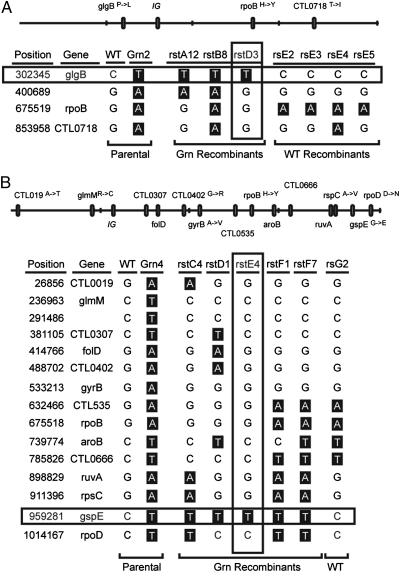
Fig. 3.
Establishing a link between EMS-induced mutations and the accumulation of glycogen granules by analysis of recombinant Chlamydia. The mutation responsible for the accumulation of glycogen granules in Grn mutants was determined by analysis of recombinant strains generated after coinfection of HeLa cells with RifR Grn mutants and SpcR C. trachomatis. Recombinants were further crossed to TmpR-resistant C. trachomatis to segregate mutations. The genotypes of wild-type Grn2 and rs or rst recombinants (products of RifR × SpcR and RifR SpcR × TmpR crosses, respectively) are shown. Note the segregation of the Grn phenotype with glgB mutations in Grn2 (A) and gspE mutations in Grn4 (B). A physical map of the parental mutant strains Grn2 and Grn4 is shown (top). IG, intergenic region.
Unlike Grn1 through -3 mutants, Grn4 had 14 EMS-induced transitions but no mutations in glgB. This mutant displayed two phenotypes: glycogen granule accumulation and small plaque morphology. To identify the mutation responsible for glycogen deposits in Grn4, we had to perform sequential crosses to unlink the relative large number of mutations present in this strain. We first crossed Grn4 with a SpcR strain and selected for double drug-resistant recombinants that formed Grn plaques on Vero monolayers. These Grn RifRSpcR recombinants were further crossed with a TmpR strain, followed by selection of recombinant progeny that could form plaques in the presence of Tmp-Spc or Rif-Spc-Tmp. We then determined by targeted DNA sequence how the mutations identified in the parental Grn4 strain segregated among these recombinants. In this manner, we established that a G425E amino acid change in GspE was always present in recombinant strains that accumulated glycogen granules, including one strain (rstE4) where the GspEG425E mutation was the only lesion inherited from the parental mutant strain (Fig. 3B). GspE is homologous to a conserved family of ATPases required for type II secretion (T2S), which exports large folded macromolecules from the periplasm of Gram-negative bacteria (17). The G425 position in GspE is invariant among the Chlamydiales and in more distantly related species (Fig. 4 A and B), suggesting that amino acid changes at this position likely disrupt protein function. By electron microscopy, gspE mutant strains displayed an engorged periplasmic space with marked accumulation of electron-dense material (Fig. 4C), presumably accumulated T2S cargo. We postulate that in these mutants, a key glycogen hydrolase fails to be secreted from bacteria, thus leading to accumulation of glycogen deposits within the inclusion lumen.
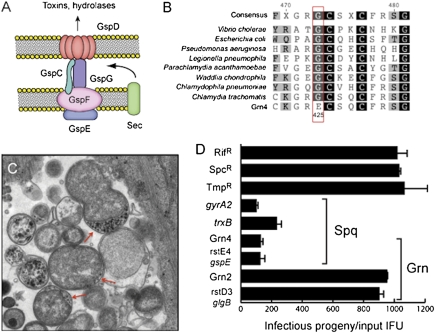
Fig. 4.
Chlamydia mutants with impaired T2S secretion are attenuated for intracellular replication. (A) Schematic representation of a T2S secretion apparatus, a protein translocation system specific for the translocation of la subset of large folded periplasmic proteins (e.g., toxins and hydrolases). Five Gsp proteins of the canonical T2S apparatus are conserved in the Chlamydiae. (B) The ATPase GspE is essential for T2S in eubacteria (18) and the G425 position is invariant among widely diverse bacterial species. Grn4 mutants contain a GspEG425E mutation. (C) C. trachomatis GspEG425E mutants display accumulation of electron dense material in the periplasmic space of reticulate bodies. (D) GspEG425E mutants produce lower yield of infectious units compared with GlgBP606L and wild-type strains used to generate recombinants. Strains rstE4 and rstD3 represent recombinants where the only parental mutation inherited from Grn4 and Grn2 are in gspE and glgB, respectively. The reduction in intracellular replication for GspEG425E mutants is comparable to that observed for small plaque forming mutants GyrA2Q265Stop and TrxBL88F.
A gspE Mutant Is Attenuated for the Production of Infectious Progeny.
All Grn recombinants derived from crosses with Grn4, including the isogenic strain rstE4, generated small plaques. Therefore, the GspEG425E mutation is also responsible for the Spq phenotype. Given the known role of T2S in the secretion of large toxins and hydrolases (18), we inferred that these mutants likely had significant virulence defects. We monitored the generation of infectious progeny at various time points postinfection (hpi) by isogenic glgB and gspE mutant stains and determined that gspE mutants generated approximately five- to sevenfold less infectious progeny compared to wild-type and glgB mutants strains (Fig. 4D). The accumulation of insoluble glycogen granules in gspE mutants was not an indirect consequence of slow growth because other Spq mutants we mapped to DNA gyrase (gyrA2) and thioredoxin (trxB) did not accumulate glycogen aggregates. Because glgB mutants displayed no growth defects in cells, it is unlikely that the growth defect of gspE mutants is the result of abnormal glycogen metabolism. Rather, it is likely that other factors secreted by T2S contribute to bacterial survival within the infected cell. Overall, these findings link T2S defects to decreased Chlamydia proliferation within epithelial cells.
Discussion
We generated mutants with diverse plaque morphologies, including small plaques, which may underlie defects in infectivity, invasion, or other processes that reflect altered interactions with their hosts. The underlying genetic lesions in these mutants were identified by WGS, which then allowed for strong associations to be made between mutations in a common gene and a common plaque morphotype (Fig. 1). For instance, thee independent mutants that displayed large glycogen precipitates within inclusions had mutations in the glycogen-branching enzyme GlgB (Fig. 2). We then took advantage of natural DNA exchange among Chlamydia to generate recombinant strains and define causal associations between mutations and phenotypes (Fig. 3). In this manner, we confirmed the link between glgB mutations and the formation of glycogen precipitates and defined a role for gspE, encoding a central component of the T2S, in glycogen accumulation and replication in epithelial cells (Fig. 4).
In essence, we have met the basic requirements of genetic analysis, linkage between genotypes and phenotypes, without the aid of conventional tools for insertional inactivation of genes in bacteria. Chemical mutagens provide an additional advantage because these agents generate conditional, hypomorphic, and hypermorphic alleles that expand the spectrum of phenotypes that can be surveyed. This feature is especially useful for obligate pathogens such as Chlamydia, where many genes are likely essential to survival in the host environment. Although we limited our phenotypic scoring to abnormal plaque morphology for proof-of-concept studies, this system can be readily adapted to identify Chlamydia mutants with complex phenotypes such as altered interactions with the host endomembrane system.
Given the forward genetics approach described here, the recent report of stable transformation of C. trachomatis with a shuttle plasmid (19), which will permit extragenic complementation of mutations, and the reverse genetics method recently described by Kari et al. (20), where pools of EMS-generated mutants are screened for specific mutations, it is clear that the term “genetically intractable” no longer applies to Chlamydia. In fact, mutagen-induced gene variation, coupled with robust next-generation genome-sequencing technologies, should enable both reverse and forward genetic approaches in a range of microbes that lack molecular genetic tools. In this manner, we should be able to draw correlations and causal relationships between phenotypes and genotypes for trait discovery in biological systems previously considered intractable to such analysis (21).
Methods
Bacterial Infections.
Strains and Chlamydia infections.
Vero cells (CCL-81; ATCC) were grown in DMEM supplemented with 10% FBS (CellGro; Mediatech). C. trachomatis serovar LGV biovar L2 434/Bu EBs of were purified on Omnipaque (GE Healthcare) density gradients and stored in SPG buffer (0.25 M sucrose/10 mM sodium phosphate/5 nM glutamic acid). LGV-L2 infections were synchronized by centrifugation (2,700 × g for 30 min at 15 °C) of EBs onto prechilled Vero cell monolayers and incubated for the indicated times. To determine the number of inclusion-forming units (IFUs), serial dilutions of EBs were added to Vero cell monolayers seeded on 96-well plates. Cells were fixed and stained with anti-LGV-L2 antibodies at 24 hpi. The plates were analyzed using a Cellomics ArrayScan Vti HCS automated fluorescent imaging system (ThermoFisher) to determine the size and number of inclusions. RifR, SpcR, and TmpR C. trachomatis LGV-L2 variants were generated by repeated passage on cells treated with 200 ng/mL rifampin (Rif), 200 μg/mL spectinomycin (Spc), and 200 μg/mL trimethoprim (Tmp). The genomes of these strains were sequenced to identify the mutations leading to antibiotic resistance: H471Y in CTL0567 (rpoB), G1197 in r01/r02 (16SRNA copies 1 and 2), and G408R in CTL0369 lead to SpcR, SpcR, and TmpR, respectively.
Chemical mutagenesis.
Vero cells were infected with RifR LGV-L2 for 18 h and exposed to 20 mg/mL ethyl methyl sulfonate (EMS) in PBS for 1 h. The cells were washed three times in PBS and incubated in DMEM/10% FBS at 37 °C in a 5% CO2 humidified incubator for 72 h to allow the bacteria to recover and complete their developmental cycle. EBs were harvested by hypotonic lysis of infected cells with sterile water, followed by addition of 5 × SPG to a final concentration of 1 × SPG, titered for IFUs, and stored at −80 °C.
Plaque assays.
Confluent monolayers of Vero cells grown in a 6-well plate were infected with ∼100 IFUs per well. Cells were incubated for 2 h at 37 °C and 5% CO2 to allow bacterial internalization. Infected cells were then overlayed with agarose/DMEM [0.5% SeaKem LE agarose (Lonza), 1 × DMEM, 10% FBS, 25 μg/mL gentamicin, 1 × nonessential amino acids (Gibco), 200 ng/mL cycloheximide] and incubated at 37 °C and 5% CO2 for 7–10 d until plaques became visible. Plaques were plugged using a pipette tip, resuspended in DMEM, and transferred to Vero cell monolayers. After 48 h of growth, infected cells were lysed by hypotonic disruption as described above.
Genetic Analysis.
WGS.
Genomic DNA from gradient purified EBs was prepared with commercial DNA isolation kits (DNeasy; Qiagen) and sheared by ultrasound with an Adaptive Focused Acoustics S220 instrument (Covaris). Sequencing libraries were prepared using genomic library construction kits (Genomic DNA Sample Prep Kits; Illumina) according to the instructions of the manufacturer: DNA fragments of an average size of 400 bp were treated with Klenow, T4 DNA polymerase, and Polynucleotide Kinase for blunting overhangs, adenylated by Exo− Klenow treatment, ligated to oligonucleotide adaptors, and amplified by PCR. Quality and quantification of DNA were assessed by electrophoresis using 2100 Bioanalyzer (Agilent). Samples were sequenced with a Genome Analyzer IIx and a HiSeq2000 (Illumina) sequencing platform. Genome assembly and SNP analysis was performed using reference-guided Geneious Pro v5 software (Biomatters). The C. trachomatis L2 434/Bu (GenBank no. NC_010287) genome was used as the reference genome. Parameters for SNP identification were set to 90% for the minimum variant frequency and 50× for minimum coverage.
Generation of Chlamydia recombinants.
Monolayers of Vero cells grown on 24-well plates were coinfected with two marked strains each at a multiplicity of infection (MOI) of 5. Mutants generated in a RifR background were used in coinfections with SpcR strains and recombinants were selected from plaques in the presence of Spc and Rif. To further segregate mutations, RifR SpcR recombinants were used in coinfections with TmpR strains and selected for Tmp- and Spc-resistant progeny. For all crosses, EBs were collected at 40 hpi by hypotonic lysis, and plaques were isolated after infection of Vero cell monolayers in the presence of the relevant antibiotics (0.2 μg/mL Rif, 200 μg/mL Spc, or 200 μg/mL Tmp).
Genotyping.
Genomic DNA from infected monolayers was extracted by DNazol treatment (Invitrogen) and used as template to amplify by the PCR 300–500-bp regions flanking identified mutations. PCR products were sequenced using dideoxy sequencing (BigDye; Applied Biosystems).
Comparative analysis of GspE sequences.
GenBank accession numbers for GspE were ABJ12335, AAK35046, CCB87516, EGD71373, ZP04416734, ACZ32707.1, YP_001654904.1, YP_003710256.1, ZP_06300623.1. GspE sequences were aligned using ClustalW packages available through Geneious Pro v5 software (Biomatters).
Replication Assays.
Vero cells were infected in duplicate at an MOI of 0.1, with one sample used to assess the exact numbers of infectious particles as described above. The second infected sample was incubated for 48 hpi, and EBs were harvested by disrupting host cells with sterile water and titered. To calculate the yield of infectious progeny generated per bacterium, the number of infectious progeny released from the second sample was divided by the number of infectious particles present in the first sample.
Microscopy.
Immunohistochemistry.
LGV-L2 infections were synchronized by centrifugation (2,700 × g for 30 min at 15 °C) of EBs onto HeLa cell monolayers grown on glass coverslips and incubated for the indicated times. Cells were washed twice in PBS, fixed with 3% formaldehyde/ 0.025% glutaraldehyde in PBS for 30 min, and permeabilized with 0.1% Triton X-100/PBS for 10 min. After blocking with 5% BSA in PBS, cells were incubated with the indicated primary antibodies, followed by fluorophores-conjugated secondary antibodies (Invitrogen) at room temperature. Host and bacterial DNA were stained with Hoechst (Invitrogen).
Periodic acid Schiff stain.
Formaldehyde/glutaraldehyde fixed infected cells were washed twice in 70% ethanol, followed by incubation in 1% periodic acid (Sigma) at room temperature. Cells were washed with 70% ethanol and stained with 0.5% basic fuchsin (Sigma). After three washes in 70% ethanol, cells were prepared for immunofluorescence staining.
Live cell imaging.
HeLa cells were seeded on glass-bottom six-well plates (In Vitro Scientific) and infected with LGV-L2 without centrifugation at an MOI of 5. Before imaging, the media was removed and replaced with fresh Hepes [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] (Gibco) buffered media. At 30 hpi, cells were placed on an atmospherically controlled stage (37 °C; 5% CO2) and imaged live at 4-min intervals for an additional 20 h on the Inverted Axio Observer microscope (Zeiss). Images were processed using Metamorph software (Molecular Devices).
Transmission election microscopy.
Infected cells were fixed with 2.5% glutaraldehyde/0.05% malachite green (EMS) in 0.1 M sodium cacodylate buffer (pH 6.8) and then postfixed with the following stains: 0.5% osmium tetroxide/0.8% potassium ferricyanide in 0.1 M sodium cacodylate; 1% tannic acid; and 1% uranyl acetate. Samples were dehydrated with graded amounts of ethanol and embedded in Spur resin. Ultrathin sections were processed, poststained with uranyl acetate and lead citrate, and imaged on a Tecnai G2 Twin microscope (FEI).
Supplementary Material
Acknowledgments
We thank J. Coers, J. Heitman, P. Seed, L. Ramakrishnan, and R. Bastidas for comments on the manuscript. This work was supported by funds from a Chancellor's Science Council Pilot Projects award and National Institutes of Health Grant AI085238.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117884109/-/DCSupplemental.
References
- 1.Haggerty CL, et al. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. 2010;201(Suppl 2):S134–S155. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- 2.Donovan P. Confronting a hidden epidemic: The Institute of Medicine's report on sexually transmitted diseases. Fam Plann Perspect. 1997;29:87–89. [PubMed] [Google Scholar]
- 3.Resnikoff S, et al. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 4.Dautry-Varsat A, Subtil A, Hackstadt T. Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 2005;7:1714–1722. doi: 10.1111/j.1462-5822.2005.00627.x. [DOI] [PubMed] [Google Scholar]
- 5.Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci USA. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heuer D, Kneip C, Mäurer AP, Meyer TF. Tackling the intractable - approaching the genetics of Chlamydiales. Int J Med Microbiol. 2007;297:569–576. doi: 10.1016/j.ijmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Binet R, Maurelli AT. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrob Agents Chemother. 2005;49:2865–2873. doi: 10.1128/AAC.49.7.2865-2873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preiss J. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol. 1984;38:419–458. doi: 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- 9.Stephens RS, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 10.Carlson JH, et al. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun. 2008;76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell CM, et al. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect Immun. 2011;79:1044–1056. doi: 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiappino ML, Dawson C, Schachter J, Nichols BA. Cytochemical localization of glycogen in Chlamydia trachomatis inclusions. J Bacteriol. 1995;177:5358–5363. doi: 10.1128/jb.177.18.5358-5363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damotte M, Cattanéo J, Sigal N, Puig J. Mutants of Escherichia coli K 12 altered in their ability to store glycogen. Biochem Biophys Res Commun. 1968;32:916–920. doi: 10.1016/0006-291x(68)90114-9. [DOI] [PubMed] [Google Scholar]
- 14.DeMars R, Weinfurter J. Interstrain gene transfer in Chlamydia trachomatis in vitro: Mechanism and significance. J Bacteriol. 2008;190:1605–1614. doi: 10.1128/JB.01592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demars R, Weinfurter J, Guex E, Lin J, Potucek Y. Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J Bacteriol. 2007;189:991–1003. doi: 10.1128/JB.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother. 2009;53:4604–4611. doi: 10.1128/AAC.00477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandkvist M. Type II secretion and pathogenesis. Infect Immun. 2001;69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, et al. Development of a transformation system for Chlamydia trachomatis: Restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kari L, et al. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci USA. 2011;108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.