Abstract
The decision between survival and death in cells exposed to TNF relies on a highly regulated equilibrium between proapoptotic and antiapoptotic factors. The TNF-activated antiapoptotic response depends on several transcription factors, including NF-κB and its RelA/p65 subunit, that are activated through phosphorylation-mediated degradation of IκB inhibitors, a process controlled by the IκB kinase complex. Genetic studies in mice have identified the IκB kinase-related kinase TANK-binding kinase 1 (TBK1; also called NAK or T2K) as an additional regulatory molecule that promotes survival downstream of TNF, but the mechanism through which TBK1 exerts its survival function has remained elusive. Here we show that TBK1 triggers an antiapoptotic response by controlling a specific RelA/p65 phosphorylation event. TBK1-induced RelA phosphorylation results in inducible expression of plasminogen activator inhibitor-2 (PAI-2), a member of the serpin family with known antiapoptotic activity. PAI-2 limits caspase-3 activation through stabilization of transglutaminase 2 (TG2), which cross-links and inactivates procaspase-3. Importantly, Tg2−/− mice were found to be more susceptible to apoptotic cell death in two models of TNF-dependent acute liver injury. Our results establish PAI-2 and TG2 as downstream mediators in the antiapoptotic response triggered upon TBK1 activation.
Keywords: apoptosis, tumor necrosis factor signaling, transcriptional regulation, posttranslational protein modification
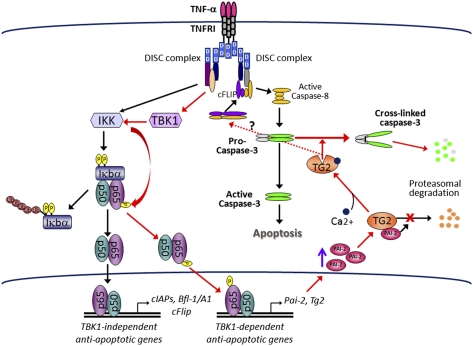
Apoptosis is a highly regulated cell death process that controls cellular homeostasis and prevents survival of injured, damaged, or transformed cells (1). Apoptosis depends on a proteolytic cascade involving intracellular proteases, known as caspases, that are activated in response to cell-intrinsic and -extrinsic insults (2). The proinflammatory cytokine TNF can trigger apoptosis through its main cell surface receptor, TNF receptor (TNFR) type 1 (TNFR1) (3, 4). Engagement of TNFR1 results in assembly of multiprotein signaling complexes around its cytoplasmic death domain, leading to activation of caspase-8, which in turns activates the executioner caspase-3 that ultimately mediates cell death (3, 4). TNF-induced apoptosis is prevented by rapid activation of the IκB kinase (IKK) complex and subsequently NF-κB. NF-κB antagonizes apoptosis and maintains cell survival through induction of antiapoptotic genes encoding factors that tightly control caspase activation, such as members of the cellular inhibitor of apoptosis family (cIAP), the cellular FLICE inhibitory protein, and Bfl-1/A1, a member of the prosurvival Bcl-2 family (5, 6). Besides this intrinsic survival pathway, additional autocrine cascades activated through release of cytokines, such as TGF-α, provide additional prosurvival signals, suggesting the existence of yet unexplored feedback loops controlling cell-fate decisions (7).
TANK-binding kinase 1 (TBK1; also called NAK or T2K) was proposed to serve as an NF-κB activator (8, 9). TBK1 was originally found to interact with the TNFR- associated factor (TRAF) binding protein TANK and form, with TRAF2, a ternary complex able to activate NF-κB in a kinase-dependent manner (8). In addition, TBK1 was found to be recruited to TNFR1 upon TNF binding (10). Mice deficient in TBK1 die during embryonic development from massive liver apoptosis (11, 12), a phenotype also exhibited by mice lacking the NF-κB subunit RelA/p65 or critical components (IKKβ and IKKγ/NEMO) of the IKK complex (13). Embryonic lethality in Tbk1−/− mice was confirmed to be TNF-dependent, as these mice survive upon deletion of TNFRI (11). However, characterization of Tbk1−/− mice failed to explain how TBK1 contributes to NF-κB–dependent gene expression (11), and the mechanism by which TBK1 controls NF-κB activity to exert its antiapoptotic function has remained elusive.
In the present study, we have readdressed the function of TBK1 in TNF-mediated NF-κB activation by using a systematic approach aimed at identifying and characterizing NF-κB–dependent genes activated in a TBK1-dependent manner that encode potential antiapoptotic factors. Defective expression of such genes may account, at least in part, for TNF-induced liver failure in Tbk1−/− embryos. We now show that TBK1 controls the expression of plasminogen activator inhibitor-2 (PAI-2), a member of the ov-serpin family (14). PAI-2 maintains survival of TNF-stimulated cells through the protein modifier transglutaminase 2 (TG2), a pleiotropic enzyme able to cross-link procaspase-3 into inactive dimers. Our data support an unexplored regulatory mechanism in the TNF-activated pathway.
Results
TBK1 Controls TNF-Mediated NF-κB Activation Through RelA Ser534 Phosphorylation.
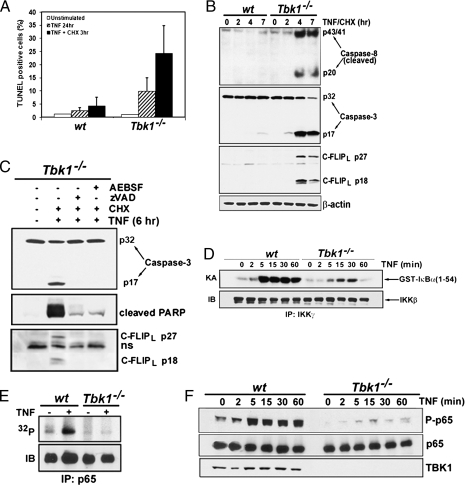
We used a pool of spontaneously immortalized mouse embryonic fibroblasts (MEFs) derived from WT and Tbk1−/− mice to understand how TBK1 prevents apoptosis. Tbk1−/− cells were more sensitive to TNF-induced apoptosis than WT cells when challenged for 24 h, and this was further enhanced in the presence of the protein synthesis inhibitor cycloheximide (CHX) (Fig. 1A). Enhanced TNF-induced apoptosis was associated with elevated caspase-8 and caspase-3 activation in Tbk1−/− MEFs (Fig. 1B). Caspase-dependent cleavage of cFLIPL, the cellular inhibitor of caspase-8, was also observed in Tbk1−/−, and not in WT, MEFs (Fig. 1B), but cFLIP expression, which depends on NF-κB, remained unchanged (Fig. 2C and Fig. S1). TNF-induced apoptosis as determined by cleavage of poly(ADP-ribose) polymerase (PARP), caspase-3, and cFLIPL, was prevented by treatment of Tbk1−/− MEFs with the pan-caspase inhibitor carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (z-VAD-FMK) or the serine protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride, AEBSF (Fig. 1C). Amounts of several other antiapoptotic proteins, including c-IAP1, c-IAP2, Bcl-2, and Bcl-XL, were not substantially reduced in Tbk1−/− MEFs (Fig. S1).
Fig. 1.
TBK1 protects MEFs from TNF-induced apoptosis and modulates IKK-dependent phosphorylation of RelA/p65. (A) Wt and Tbk1−/− MEFs were treated for 24 h with TNF (25 ng/mL) alone or for 3 h with TNF in the presence of CHX (10 μg/mL). Apoptotic cells were detected by TUNEL assays. The bars represent averages ± SD of three different experiments. Approximately 1,200 cells were counted in each experiment. (B) MEFs treated with TNF and CHX for the indicated times were analyzed for caspase-8 and caspase-3 activation by immunodetection of their cleaved forms. (C) Cell extracts prepared from Tbk1−/− MEFs treated for 6 h with TNF and CHX in the presence of z-VAD-FMK (20 μM), AEBSF (0.5 mM), or DMSO (vehicle) were analyzed by immunoblotting for caspase-3 activation and PARP cleavage. (D) IKK activity in TNF-treated MEFs was measured by immunocomplex kinase assay using GST-IκBα(1-54) as a substrate (19). (E) RelA phosphorylation was examined after its immunoprecipitation from [32P]orthophosphate-labeled and TNF-stimulated MEFs. Phospho-RelA was detected by autoradiography and the total amount of RelA by immunoblotting (IB). (F) Phosphorylation of RelA was examined by immunoblotting using a phospho-specific huRelA(Ser536) antibody.
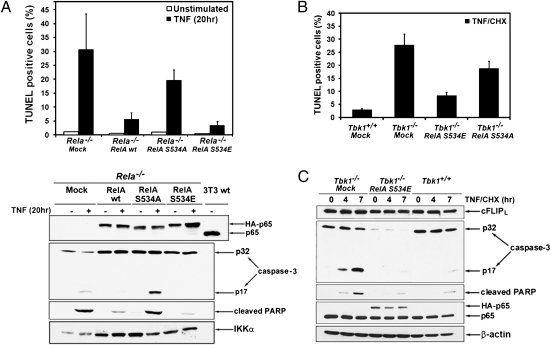
Fig. 2.
Phospho-RelA protects MEFs from TNF-induced apoptosis. (A) Rela−/− MEFs infected with an empty retrovirus (mock) or retroviruses expressing HA-tagged wt RelA, RelA(S534A), or RelA(S534E) were left unstimulated or were treated with TNF (20 h). Apoptotic cells were detected by TUNEL staining. Represented are averages ± SD of three separate experiments. Cell lysates were analyzed for caspase-3 activation, PARP cleavage, and RelA expression by immunoblotting. (B) Tbk1−/− MEFs expressing RelA(S534A) or RelA(S534E) were treated for 3 h with TNF in the presence of CHX. Apoptotic cells were detected by TUNEL assay and quantified as described earlier. (C) Lysates were prepared from cells in B that were stimulated with TNF plus CHX for the indicated times. Caspase-3 activation and protein expression were determined by immunoblotting as in A.
In addition to NF-κB and caspases, TNF activates MAPKs c-Jun NH2-terminal kinase (JNK), p38, and ERK (15). Instead of being strongly enhanced as in RelA- or IKKβ-deficient cells (16), JNK and p38 activities were somewhat reduced in Tbk1−/− MEFs (Fig. S2A). However, activation of the JNK-regulated transcription factors c-Jun and ATF-2 through phosphorylation and nuclear translocation was not affected by TBK1 ablation (Fig. S2B). Phosphorylation of nuclear CREB, a target for p38, was also normal. Interestingly, IKK activation was reduced in TNF-stimulated Tbk1−/− MEFs compared with WT cells (Fig. 1D). This is consistent with our previous observation that TBK1 could activate IKK in vitro (9). However, in agreement with a previous report (11), IκBα and IκBβ were phosphorylated and degraded normally in TNF-stimulated Tbk1−/− MEFs (Fig. S3A), but delayed IκB resynthesis was observed, suggesting a possible defect in NF-κB–directed transcription. Nuclear accumulation of RelA, p50, and c-Rel appeared to be normal in Tbk1−/− MEFs (Fig. S3 B and C), and so was the binding of RelA to its target gene promoters (Fig. S3D).
TBK1 can phosphorylate RelA in vitro (17, 18). Therefore, we examined the role of TBK1 in RelA phosphorylation in living cells. Endogenous RelA, immunoprecipitated from untreated and TNF-stimulated WT MEFs, was labeled with [32P]orthophosphate. TNF stimulated phosphorylation of RelA in WT cells (Fig. 1E and Fig. S4A), but both basal and induced RelA phosphorylations were diminished in Tbk1−/− MEFs (Fig. 1E). Experiments performed in MEFs defective for individual IKK subunits indicated that a functional IKK complex was also required for TNF-induced RelA phosphorylation (Fig. S5A). These results are consistent with our early suggestion that TBK1 stimulates NF-κB activity through the canonical IKK complex (9). By using an established peptide mapping strategy (19), the TNF-inducible phosphorylation sites were localized to a 4.6-kDa CNBr peptide located in the C-terminal transactivation domain of RelA (Fig. S4 B and C). The major TNF-inducible phosphorylation site was identified as Ser534 of mouse RelA (corresponding to Ser536 of human RelA; Fig. S4D). Reduced Ser534 phosphorylation in Tbk1−/− MEFs was confirmed by immunoblotting performed with phospho-specific RelA antibody (Fig. 1F). RelA Ser534 phosphorylation was also impaired in cells defective in both IKK catalytic subunits (Fig. S5B). Ser534 phosphorylation took place in the cytosol, as it was dramatically reduced in IκBα−/− MEFs, in which most of RelA is constitutively nuclear (Fig. S5C).
Ser534 Phosphorylation Regulates RelA Antiapoptotic Function.
To examine whether phosphorylation of RelA at Ser534 was required to protect MEFs from TNF-induced apoptosis, a retroviral vector was used to stably express WT RelA, RelA(S534A), or a phospho-mimic RelA(S534E) mutant in Rela−/− MEFs. Expression levels were similar to that of endogenous RelA in WT MEFs (Fig. 2A), and all the expressed proteins underwent nuclear translocation. Whereas WT RelA and RelA(S534E) protected Rela−/− MEFs from TNF-induced apoptosis, expression of RelA(S534A) provided little protection (Fig. 2A). Similar results were obtained when caspase-3 activation and cleavage of PARP were examined: WT RelA and RelA(S534E) prevented caspase-3 activation, whereas RelA(S534A) did not (Fig. 2A). Expression of RelA(S534E) but not RelA(S534A) also prevented apoptosis (Fig. 2B), PARP cleavage, and caspase-3 activation (Fig. 2C) in Tbk1−/− MEFs. Surprisingly, the amount of procaspase-3 was dramatically reduced in Tbk1−/− MEFs expressing RelA(S534E) (Fig. 2C). This reduction in procaspase-3 protein amount appeared to be specific, as other proteins such as cFLIPL (Fig. 2C) and XIAP (Fig. 3G) were not affected.
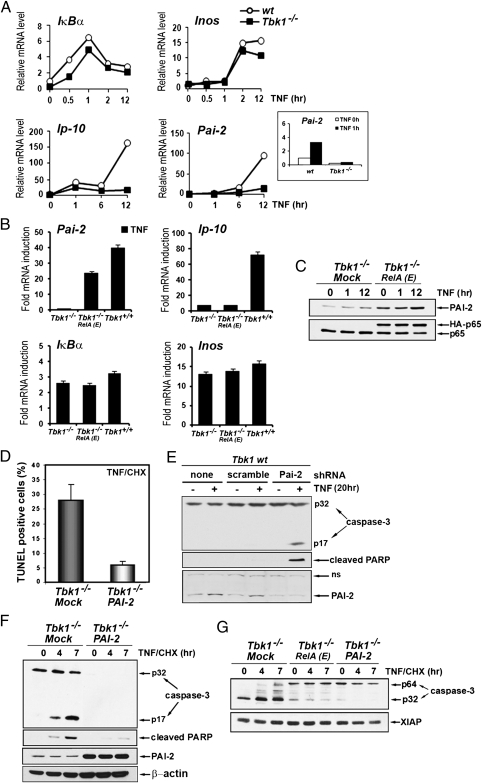
Fig. 3.
TBK-1 controls NF-κB–dependent expression of the survival factor PAI-2. (A) Induction of NF-κB target genes was determined by qRT-PCR amplification of mRNAs prepared from WT and Tbk1−/− MEFs that were stimulated with TNF for the indicated times. Inset: Short time course (0 and 1 h) of Pai-2 mRNA induction in both cell types. (B) Relative mRNA induction was analyzed by qRT-PCR amplification of RNAs prepared from WT (Tbk1+/+), Tbk1−/−, and Tbk1−/− MEFs expressing RelA(S534E), depicted as RelA(E) that were stimulated with TNF for 2 h (IκBα), 6 h (Pai-2 and Inos), or 14 h (Ip10). (C) Expression of endogenous PAI-2 and RelA was examined by immunoblotting in extracts prepared from cells that were stimulated with TNF for the indicated times. (D) Apoptotic cell death, determined by TUNEL assay as described earlier, was quantified in Tbk1−/− MEFs and in cells stably expressing PAI-2 that were stimulated for 3 h with TNF in the presence of CHX. (E) WT MEFs expressing a Pai-2 shRNA or a scrambled shRNA were treated with TNF for 20 h. Caspase-3 activation, PARP cleavage, and protein expression were determined by immunoblotting. (F and G) Caspase-3 activation, PARP cleavage, and protein expression were determined by immunoblotting in extracts prepared from the indicated cells that were stimulated with TNF in the presence of CHX for the indicated times.
TBK1 Controls TNF-Induced Expression of Survival Gene Pai-2/serpinB2.
Genes that mediate the survival function of TBK1 were identified by microarray analysis of RNAs isolated from resting and TNF-stimulated WT and Tbk1−/− MEFs. Selected data were validated by quantitative real-time PCR (qRT-PCR). Induction of most previously described (20) NF-κB–dependent genes was not affected by TBK1 deficiency (Fig. S6). As expected, induction of genes that require activation of IFN regulatory factors in addition to NF-κB, such as IP-10, Rantes, or Irf9 was impaired in TNF-stimulated Tbk1−/− MEFs (Fig. 3A and Fig. S6). Among well established antiapoptotic genes, induction of c-Iap-1, c-Iap-2, Xiap, and BclXL was observed in Tbk1−/− MEFs, whereas c-Flip expression was constitutive and not TNF-inducible in MEFs (Fig. S6). Further examination of less studied NF-κB target genes with a documented survival function identified Pai-2/serpinb2, a gene encoding PAI-2, a serine protease inhibitor previously shown to inhibit TNF-induced apoptosis in cancer cells (21, 22), as a potential candidate (Fig. 3A). Both basal (Fig. 3A, Inset) and TNF-induced expressions of Pai-2 were dramatically reduced in Tbk1−/− MEFs (Fig. 3A). However, Pai-2 expression was restored in Tbk1−/− MEFs upon expression of RelA(S534E), whereas IP-10 induction remained defective (Fig. 3B). Induction of other NF-κB–dependent genes such as IκBα and iNos was not affected (Fig. 3B). Reconstitution of Tbk1−/− MEFs with RelA(S534E) also increased PAI-2 protein expression (Fig. 3C).
The antiapoptotic function of ectopic PAI-2 reintroduced into Tbk1−/− MEFs was examined by retroviral transduction. Reconstituted cells were protected from TNF-induced apoptosis (Fig. 3D) or activation of caspase-3 (Fig. 3F). By contrast, down-regulation of Pai-2 expression by stably expressing a Pai-2-specific shRNA, but not a scrambled shRNA, potentiated TNF-induced caspase-3 activation and apoptosis in WT MEFs (Fig. 3E). Similarly to what was observed in Tbk1−/− MEFs expressing RelA(S534E), the amount of procaspase-3 was dramatically reduced in Tbk1−/− MEFs expressing PAI-2, and a slow-migrating form of procaspase-3 with an apparent molecular weight of 64 kDa (depicted as p64) was observed in Tbk1−/− MEFs expressing either RelA(S534E) or PAI-2 (Fig. 3G). A similar slow-migrating caspase-3 isoform was previously described as a cross-linked procaspase-3 in thapsigargin-treated HCT116 cancer cells and in tumor cells exposed to hypoxia (23, 24). In both cases, caspase-3 cross-linking into nonfunctional dimers or multimers is mediated by TG2, a multifunctional enzyme that induces posttranslational protein modifications by transamidation (25). TG2 was reported to have antiapoptotic function not only in vitro (23, 24) but also in vivo, as TG2-deficient mice show increased sensitivity to apoptosis induced by activation of the CD95/Fas receptor (26), a molecule related to TNFR1.
TG2 Is a TBK1-Regulated Antiapoptotic Factor.
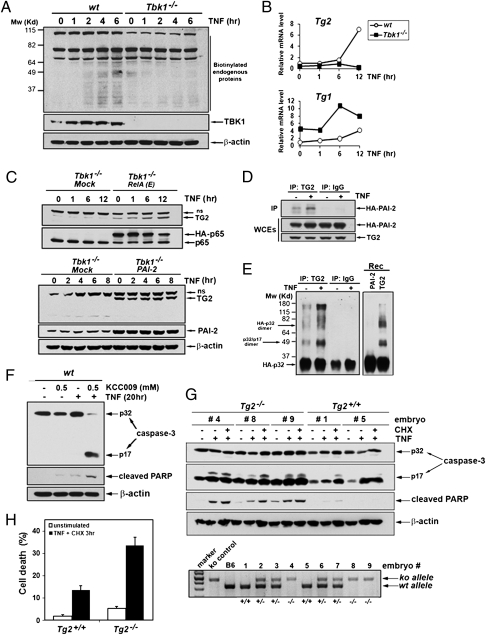
Endogenous TG2 activity was analyzed in WT and Tbk1−/− MEFs stimulated with TNF by examining incorporation of biotinylated pentylamines into cellular proteins (27). TNF induced transamidation in WT MEFs but not in Tbk1−/− MEFs (Fig. 4A). TNF also failed to stimulate Tg2 gene expression in Tbk1−/− MEFs, whereas expression of Tg1 was somehow increased in these cells (Fig. 4B). TG2 protein could hardly be detected in MEFs because of the poor quality of available antibodies combined with its very low level of expression. However, we could observe that basal and TNF-induced endogenous TG2 protein amounts were increased in Tbk1−/− MEFs expressing RelA(S534E) (Fig. 4C, Upper). Interestingly, basal TG2 expression was also increased in Tbk1−/− MEFs constitutively expressing PAI-2, suggesting that PAI-2 might stabilize TG2 (Fig. 4C, Lower). Similarly, MEFs treated with the proteasome inhibitor MG132 contained more TG2 protein (Fig. S7A), suggesting the existence of a tight posttranslational control that down-regulates TG2 protein expression. Consistent with PAI-2's involvement in stabilization of TG2, an interaction between PAI-2 and TG2 was observed by coimmunoprecipitation in MEFs expressing HA-PAI-2 and human TG2 (Fig. 4D). We next examined whether the activity responsible for cross-linking of procaspase-3 was associated with PAI-2. An in vitro transamidation assay was performed with PAI-2 immunoprecipitates from Tbk1−/− MEFs stably expressing untagged PAI-2 that were left untreated or were stimulated with TNF, and recombinant HA-procaspase-3 generated by in vitro translation was used as a substrate. Caspase-3 cross-linking activity was present in PAI-2 immunoprecipitates but not in control immunoprecipitates, and it was strongly elevated in TNF stimulated cells (Fig. S7B). Similarly, TG2 immunoprecipitated from MEFs expressing TG2 and PAI-2 showed inducible transamidation activity toward procaspase-3 (Fig. 4E). Assays performed by using recombinant PAI-2 or TG2 further confirmed that the transamidating activity was carried out by TG2 and not by PAI-2 (Fig. 4E, Right).
Fig. 4.
TG2 is a TBK1-dependent antiapoptotic factor. (A) In vivo transamidation activity was determined in WT and Tbk1−/− MEFs metabolically labeled with BP and stimulated with TNF for the indicated times. Cell extracts were prepared, and biotin-conjugated proteins were detected by immunoblotting using anti–streptavidin-HRP (Pierce). (B) Induction of Tg gene expression was determined by qRT-PCR analysis of mRNAs from WT and Tbk1−/− MEFs stimulated with TNF for the indicated times. (C) Endogenous TG2 protein amounts were determined by immunoblotting in extracts from Tbk1−/− MEFs expressing RelA(E) (Upper), PAI-2 (Lower), or an “empty” retrovirus (mock) and treated with TNF for the indicated times (ns, nonspecific). (D) Coimmunoprecipitation of PAI-2 with TG2 was examined in MEFs stably expressing HA-PAI-2 and human TG2 that were left untreated or were stimulated with TNF for 8 h. Control immunoprecipitations were performed with nonimmune IgGs. (E) In vitro transamidation assay was performed with TG2 or control immunoprecipitates prepared from untreated or TNF-stimulated Tbk1−/− MEFs expressing TG2 and untagged PAI-2 or with recombinant proteins (rec) using HA-procaspase-3 as a substrate. (F) Wt MEFs were treated with TNF (20 h) with or without the transglutaminase-specific inhibitor KCC009 (0.5 mM). Caspase-3 activation, PARP cleavage, and protein expression were determined by immunoblotting. (G) WT and Tg2−/− primary MEFs prepared from littermate embryos were left untreated or treated with TNF alone (20 h) or TNF plus CHX (6 h). Caspase-3 activation and apoptosis were determined as detailed earlier. Genotyping (Lower) was performed by PCR amplification as described (31). (H) The extent of cell death in WT and Tg2−/− MEFs that were stimulated for 3 h with TNF plus CHX was quantified by staining with PI.
We next examined if the protective effect of PAI-2 and TG2 was indeed mediated through inhibition of caspase-3 without affecting alternative (i.e., mitochondrial-mediated) pathways. A slight increase in BID cleavage after stimulation with TNF and CHX was observed in Tbk1−/− MEFs relative to WT cells (Fig. S8A). Expression of PAI-2 in these cells was unable to prevent BID cleavage (Fig. S8B), suggesting that PAI-2 is not a bona fide antiapoptotic factor. We also analyzed TNF-induced apoptosis in cells defective in caspase-3. In agreement with previous studies (28, 29), we found that caspase-3–deficient (Casp3−/−) MEFs were highly resistant to TNF-induced apoptosis (Fig. S8C), highlighting that the absence of caspase-3 does not generate a compensatory cell death pathway (e.g., TNF-induced BID cleavage) in MEFs and that caspase-3 is indeed critical to mediate the TNF response in these cells. Taken together, these experiments clearly indicated that PAI-2 and TG-2 act specifically on TNF-induced, caspase-3–dependent apoptosis.
To examine the antiapoptotic function of TG2, we first used the irreversible TG2 inhibitor KCC009 (30). As shown in Fig. 4F, treatment with KCC009 strongly potentiated TNF-induced caspase-3 activation and apoptosis in WT MEFs. To further confirm the antiapoptotic function of TG2, primary MEFs derived from Tg2+/+ and Tg2−/− littermate embryos (31) were examined for their sensitivity to TNF-induced apoptosis. Apoptosis (PARP cleavage) and caspase-3 activation were detected by immunoblotting (Fig. 4G). Caspase-3 activation was observed in Tg2−/− MEFs treated with TNF without the need for inhibition of de novo protein synthesis, indicating that these cells have an intrinsic defect in the antiapoptotic response to TNF. The extent of cell death was quantified by staining with propidium iodide (PI) (Fig. 4H) and FACS analysis after annexin V and PI staining confirmed that Tg2−/− MEFs showed increased susceptibility to TNF-induced apoptosis (Fig. S9).
TG2 Is Essential for Prevention of TNF-Dependent Liver Injury.
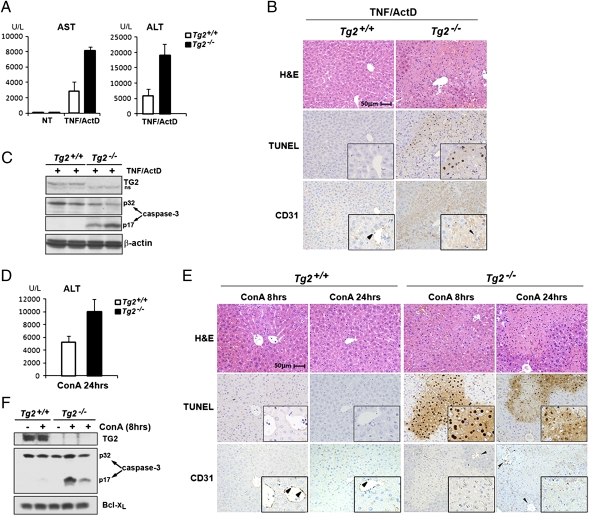
To assess the antiapoptotic function of TG2 in vivo, we used two models of TNF-induced liver injury in mice. WT and Tg2−/− mice were first injected intraperitoneally with TNF and actinomycin D (Act D). Tg2−/− mice showed clear signs of liver failure such as elevated serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels (Fig. 5A), massive infiltration with neutrophils, and presence of numerous apoptotic bodies (Fig. 5B). The presence of a high amount of TUNEL-positive cells at the injury sites indicated that cell death mainly occurred through apoptosis (Fig. 5B). Caspase-3 activation was also observed in liver extracts from Tg2−/− mice injected with TNF and Act D (Fig. 5C). To rule out that apoptosis occurred in endothelial cells, staining with anti-CD31 (PECAM-1), which detects endothelial cells, was performed on sections consecutive to those used for histological evaluation and TUNEL assays. The absence of TUNEL-positive cells in the CD31-positive population, combined with histological evaluation, indicated that hepatocytes were the major cell type undergoing apoptosis in TNF/Act D-challenged Tg2−/− mice (Fig. 5B). Tg2+/+ mice also experienced liver damage, but the response was much weaker than in Tg2−/− mice (Fig. 5A) and no caspase-3 activation could be detected (Fig. 5C). Thus, Tg2−/− mice are more susceptible to TNF-induced liver injury than WT mice. Tg2−/− mice were also highly sensitive to T cell-mediated liver injury induced by i.v. injection of Con A, which is TNF-dependent. Liver damage indicated by ALT release was present in both genotypes but was more severe in Tg2−/− mice (Fig. 5D). Massive hepatocyte cell death detected by TUNEL staining was observed in Tg2−/− liver sections 8 h after Con A injection (Fig. 5E), and caspase-3 activation indicated that cell death in Tg2−/− mice occurred through apoptosis (Fig. 5F). Liver destruction was clearly visible 24 h after Con A injection in Tg2−/− mice, and the apoptotic cell remnants could no longer be detected by TUNEL reaction. Only few apoptotic hepatocytes were found in liver sections from Tg2+/+ mice (Fig. 5E), and caspase-3 activation was not observed (Fig. 5F). These data strongly support the physiological relevance of TG2 as an inhibitor of TNF-induced apoptosis.
Fig. 5.
TG2 protects mice from TNF-dependent liver apoptosis. (A–C) WT and Tg2−/− mice were injected with TNF and ActD and analyzed 16 h later. (A) Serum AST and ALT levels were determined in untreated mice (NT) or mice treated with TNF and Act D. Data are averages ± SD (n = 3). (B) Histological analysis (H&E staining), TUNEL staining, and anti-CD31 immunostaining were performed on sequential liver sections of WT and Tg2−/− mice 16 h after TNF and Act D injection. Insets: Higher magnification of a selected area. Arrowheads show vascular endothelial cells (CD31-positive), which are TUNEL-negative. CD31 immunostaining in Tg2−/− liver sections displayed some nonspecific background staining as a result of the presence of cell debris. (C) Caspase-3 activation was determined by immunoblot analysis of liver protein extracts prepared 16 h after TNF and Act D injection. (D–F) The same analyses were performed on WT and Tg2−/− at 8 h (E and F) or 24 h (D and E) after injection of Con A (35 mg/kg). (D) Serum AST and ALT levels measured after Con A injection. Data are averages ± SD (n = 2). (E) Histological analysis, TUNEL assays, and anti-CD31 staining were performed on sequential liver sections of WT and Tg2−/− mice at 8 h or 24 h after injection of Con A. Insets: Higher magnification of a selected area. TUNEL-positive cells (nuclear staining) in Tg2−/− liver sections are CD31-negative and represent apoptotic hepatocytes. Twenty-four hours of treatment with Con A induced massive liver degeneration in Tg2−/− mice, and dead cells were no longer detectable by TUNEL staining. Arrowheads point to vascular endothelial cells (CD31-positive cells), which are TUNEL-negative. (F) Liver extracts prepared from WT and Tg2−/− mice challenged with Con A (8 h) or PBS solution were analyzed for caspase-3 activation.
Discussion
The role of TBK1 in mediating the innate immune response to viruses and dsDNA through induction of type I IFN is well documented. By contrast, the role of TBK1 in IKK–NF-κB signaling has been debated, because of the observation that, although TBK1 affects NF-κB target gene expression, it is not required for activation of NF-κB DNA binding (11). Early studies found TBK1 to form a complex with the adaptor molecules TRAF2 and TANK (8), which mediate assembly of signaling complexes at the intracellular tails of cytokine receptors (32). Indeed, TBK1 is recruited to TNFR1 upon TNF binding (10) and interacts with IKKγ/Nemo, the regulatory subunit of IKK (33, 34). We reported that TBK1 activates NF-κB through phosphorylation-mediated activation of the IKK complex (9). Consistent with a role for TBK1 in NF-κB signaling, Tbk1−/− mice die during embryonic development from massive liver apoptosis and, like Rela−/−, Ikkβ−/−, and Ikkγ−/− mice, they can be rescued by ablation of TNFR1 (11, 13). Cells derived from Tbk1−/− mice were reported to exhibit normal NF-κB activation without increased sensitivity to TNF-induced apoptosis (11). However, the present results show that TBK1 can directly phosphorylate RelA and thereby control the expression of a subset of NF-κB target genes that inhibit TNF-induced apoptosis. Our findings that Tbk1−/− MEFs are highly susceptible to TNF-induced apoptosis are consistent with the phenotype of Tbk1−/− fetuses, which exhibit all the characteristics of a defective TNF-induced and NF-κB–dependent antiapoptotic response (13). The divergence between our results and those reported earlier may result from different experimental conditions and detection methods. For instance, Bonnard et al. (11), examined cell viability by PI staining after stimulation of MEFs with a low dose of TNF (10 ng/mL), whereas we used a higher dose of TNF (25 ng/mL) without or with CHX and evaluated the apoptotic response by TUNEL assays and direct analysis of caspase-3 cleavage.
Molecular analysis of signaling events downstream of TNFR1 revealed that Tbk1−/− cells exhibit impaired IKK-mediated RelA phosphorylation at Ser534, confirming the earlier suggestion that TBK1 modulates IKK activity (9). Importantly, expression of a phospho-mimic RelA(S534E) variant in Tbk1−/− MEFs restored the antiapoptotic response, demonstrating a specific biological function associated with RelA Ser534 phosphorylation, although this phosphorylation event is not critical for activation of most NF-κB target genes. These findings are consistent with previous reports that TNF-induced NF-κB activation is largely normal in Tbk1−/− cells (11). Phosphorylation of RelA at Ser534 does not affect most RelA functions, including inhibition by IκBs, nuclear translocation, and binding to target gene promoters (Fig. S3). Instead, it modulates its ability to transactivate a specific subset of NF-κB target genes. Among these, we have identified Pai-2 as a critical antiapoptotic gene and showed that reexpression of PAI-2 in Tbk1−/− MEFs inhibited the TNF-induced apoptotic response. Although NF-κB activation through induced IκB degradation is a well established signaling mechanism, it has long been suspected that expression of individual NF-κB target genes is further modulated through specific posttranslational modifications of NF-κB subunits (35, 36). We now show that one such mechanism involves RelA Ser534 phosphorylation, which depends on TBK1 and IKK activity. However, it is not yet clear how TBK1 directs IKK to specifically phosphorylate RelA Ser534 while having no effect on other IKK-dependent functions.
We also do not understand what causes the initial activation of caspase-8 in Tbk1−/− cells and how PAI-2 affects caspase-8 activation. We showed that cFLIPL is cleaved in a caspase-dependent manner when Tbk1−/− MEFs are stimulated with TNF in the presence of CHX (Fig. S1), and this may prompt initiation of the proapoptotic cascade. This is consistent with previous studies reporting that TNF promotes caspase-8 activation by elimination of cFLIPL when NF-κB–mediated cFlip induction is defective (5), de novo protein synthesis is blocked (37), or cFLIP degradation is enhanced (38). Furthermore, it was recently proposed that, in response to death receptor ligation, cFLIP and caspase-8 could form catalytically inactive heterodimers, which prevent the initiation of apoptosis (39).
Our results suggest that PAI-2 exerts its antiapoptotic function through interaction with and stabilization of TG2, which prevents caspase-3 activation by cross-linking of procaspase-3. Although it remains to be determined whether TG2 can also cross-link other caspases, including caspase-8, this observation provides a missing molecular link that explains the observation made more than a decade ago that the C-D interhelical domain or C-D loop of PAI-2, a conserved protein binding domain among ov-serpin family members (14), was essential for the antiapoptotic function of PAI-2 in TNF-stimulated cells (22). It was postulated that an interaction between the C-D loop and unknown proteins was likely to be important for resistance to apoptosis. We now show that PAI-2 interacts with TG2 in a TNF-dependent manner. This interaction presumably protects TG2 from proteolysis allowing TG2 to cross-link procaspase-3 and promote cell survival. Interestingly, another ubiquitously expressed ov-serpin family member, serpinB10 (PI10), was also found to provide protection against TNF-induced apoptosis (40). The authors observed the formation of high molecular weight SDS-stable PI10-containing complexes in cells treated with TNF in the presence of CHX (40). These complexes were suggested to contain a serine protease that is activated during this process, but this activity was never identified. Nevertheless, these observations raise the hypothesis that some ov-serpins may share a common function in controlling cell death, and that their specific physiological roles may have been obscured as a result of redundancy (14, 41). Although it remains to be further investigated, our observations that PAI-2 and TG2 interact in cells, and mediate a common antiapoptotic response, suggest that these unconventional antiapoptotic factors participate in a posttranslational mechanism that controls caspase-3 activation. Identification of the mechanism that controls TG2 turnover will provide additional insights into this antiapoptotic process.
The tumor suppressor retinoblastoma protein (Rb) is another intracellular target of PAI-2, which protects Rb from cleavage by calpains and thereby contributes to tumor cell survival (42). It was also shown that TG2 could protect Rb from caspase-7–mediated cleavage in fibroblasts (43). In light of our present results, it is tempting to speculate that PAI-2 and TG2 or a related enzyme may also control Rb-dependent cell survival. Notably, PAI-2 was also found to protect macrophages from pathogen-induced cell death (44), acting as a modulator of the innate immune response (45).
The antiapoptotic function of TG2 has been widely debated (25, 46), primarily because no spontaneous cell death was observed in Tg2−/− mice (31, 47, 48). However, TG2 protects cancer cells from thapsigargin- or hypoxia-induced death (23, 24), as well as staurosporine-induced apoptosis (49). Initial evidence for the antiapoptotic function of TG2 in vivo came from the work of Sarang et al. (26), who showed that Tg2−/− mice were more susceptible to Fas-mediated cell death. We now show in two different models of liver injury triggered by injection of TNF to Act d-sensitized mice or by Con A administration that Tg2−/− mice display increased hepatocyte apoptosis relative to WT counterparts (Fig. 5). These data strongly support a role for TG2 in inhibition of cell death mediated by members of the TNF/TNFR1 superfamily. However, an important question that needs to be answered is why Pai-2−/− and Tg2−/− mice are viable whereas genetic ablation of Tbk1 leads to embryonic lethality. It could be argued that the newly identified PAI-2 and TG2-dependent pathway is not the only way by which TBK1 inhibits cell death. In addition, PAI-2 may act redundantly with PI10, as mentioned earlier. Furthermore, Pai-2 is only one of several NF-κB–regulated antiapoptotic genes, such as cIaps, cFlip, Bcl-XL, and others (5, 20), whose expression is needed to suppress TNF-induced liver destruction. Although the antiapoptotic function of these genes has never been disputed, genetic ablation of each one of them in isolation does not result in embryonic lethality as a result of liver failure. The current challenge is to identify the minimal set of antiapoptotic genes that needs to be expressed at a given time and in a particular environmental context during liver development and adult life to suppress TNF-driven hepatocyte death. It will also be interesting to test whether the combined ablation of both Pai-2 and Tg2 will result in a more severe phenotype that approaches that of Tbk1−/− mice. The identification of additional antiapoptotic factors whose expression or activity depends on TBK1 will shed further light on this question. Such factors may act independently of or in conjunction with the PAI-2–TG2 pathway.
Materials and Methods
Reagents.
CHX, puromycin, z-VAD-FMK, MG-132, and protease inhibitors were from Calbiochem, and Act D, Con A, AEBSF, and Polybrene were from Sigma-Aldrich. Recombinant human PAI-2 was from Peprotech. Antibodies against RelA/p65 (S536) (no. 3031), phospho-IκBα (no. 9241), phospho-MAPKs (no. 9910), phospho-cJun (no. 9261), ERK, p38 (no. 9212), TBK1 (no. 3012), Bcl-XL (no. 2764), c-IAP1 (no. 4952), mBID (no. 2003), caspase-3 (no. 9662), cleaved caspase-3 (no. 9661), and cleaved PARP (no. 9544) were from Cell Signaling; antibodies to IKKγ (no. 557383), JNK1 (no. 551196), XIAP (no. 610716), CD95/Fas receptor (554254), caspase-8 (no. 559932), and procaspase-3 (no. 65906E) were from BD Transduction Laboratories; antibodies to IKKα (no. IMG-136A), IKKβ (no. IMG-129A), TBK1 (IMG-139A), and IκBα (no. IMG-127A) were from Imgenex; antibodies against RelA (sc-372 and sc-372-G), RelB, cRel, IκBβ, HSP60, and PAI-2 (sc-25746) were from Santa Cruz Biotechnology; anti-p65/RelA (CT), anti–phospho-ATF-2 (no. 05–891), anti-TG2 (no. 06–471), and anti-caspase-1 (no. 06–503) were from Upstate Biotechnology; anti–CREB-1 (AB3006) was from Millipore; anti-FLIPα (CT; no. 1161) was from ProSci, anti-TG2 (AB-4) was from Neomarkers (Lab Vision), anti-HA (clone 3F10) was from Roche, anti-cAIP2 was from R&D Systems, and anti-CD31 was from Abcam.
Immunoblotting.
Whole-cell extracts were obtained by lysing cells in a buffer containing 50 mM Tris-HCl, pH 7.6, 250 mM NaCl, 1% Triton X-100, 0.5% Nonidet P-40, 3 mM EDTA, 3 mM EGTA, 10% glycerol, 2 mM DTT, 1 mM PMSF, 1 mM sodium orthovanadate, and a protease inhibitor mixture (Calbiochem). Nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce). Proteins were separated by SDS/PAGE and blotted onto PVDF membranes (Millipore). The membranes were probed with the appropriate antibodies and the antigen–antibody complexes detected by SuperSignal Western Pico Luminol/Enhancer solution (Pierce).
Kinase Assays.
The IKK complex was immunoprecipitated from cell extracts using an anti-IKKγ antibody and the IKK activity was measured by in vitro kinase assay as described (19, 50) by using GST-IκBα(1-54) or GST-p65(354-551) as substrates.
Phosphorus-32 Metabolic Labeling and Phospho-Peptide Mapping.
Cells were labeled with [32P]orthophosphate as previously described (19, 50). Briefly, cells incubated with [32P]orthophosphate (2 mCi/mL) for 5 h were stimulated with TNF (25 ng/mL) for 15 min, washed with PBS solution, and harvested in RIPA buffer. RelA was immunoprecipitated from precleared lysates by using an anti-RelA/p65 antibody (C-20; Santa Cruz Biotechnology). Immune complexes were washed in RIPA buffer. The proteins were resolved by SDS/PAGE and transferred to PVDF membranes (Millipore) for autoradiography and immunoblot analysis. Phospho-peptide mapping of phospho-labeled RelA/p65 was performed as described (19, 50).
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed using the Quick Exchange Mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Retroviral Transductions.
cDNAs were subcloned into the pLPCX retroviral vector (Stratagene). Recombinant retroviruses were produced by cotransfection with pCL-Eco into Phoenix packaging cells using Lipofectamine Plus reagent (Invitrogen). Supernatants containing recombinant retroviruses were collected 2 d after transfection, filtered to remove cell debris, and used directly for infection. MEFs at subconfluence were infected with viral stocks in the presence of Polybrene at 8 μg/mL, and two or three consecutive infections were performed over a period of 24 h. One day after the last infection, the infected cells were selected in medium supplemented with 2 μg/mL puromycin. Experiments were performed with pools of stable cells.
Gene Silencing Using shRNAs.
Retroviral vectors (pRS plasmids) encoding shRNAs specific to mPAI-2 or a scrambled shRNA were purchased from Origene. Production of retroviruses, infection of cells and selection of stable cells were performed as described earlier. The most efficient Pai-2–specific shRNA construct among four tested was used.
Apoptosis Assays.
Cells gown on Permanox Lab-Tek chamber slides were left untreated or stimulated with TNF (25 ng/mL) in the presence or absence of CHX (10 μg/mL) for the indicated times. Cells were fixed with 4% paraformaldehyde and permeabilized for 5 min with 0.1% Triton X-100 in 0.1% sodium citrate at 4 °C. Apoptotic cells were detected by TUNEL staining (Roche or Promega). DAPI staining was performed for total cell counting.
Immunofluorescence.
Cells cultured on Lab-Tek chamber slides were left untreated or stimulated for 30 min with TNF (25 ng/mL). Cells were fixed with 4% paraformaldehyde, and the subcellular localization of RelA/p65 was examined by fluorescent immunostaining by using a polyclonal antibody against p65 (C-20) and Alexa-labeled anti-rabbit purified IgG (Jackson ImmunoResearch).
qRT-PCR and Microarray Analysis.
Total RNA was extracted using TRIzol LS (Invitrogen) and purified on RNeasy Miniprep columns (Qiagen). For qRT-PCR analysis, cDNAs were synthesized by using a SuperScript II cDNA synthesis kit (Invitrogen). Real-time PCR amplifications were performed in 96-well optical reaction plates with Power SYBR Green PCR Master Mix (Applied Biosystems). Primers were designed by using Primer Express software. Primer sequences are available upon request. For microarray analysis, preparation of cDNA probes, hybridization to a mouse 10k oligo DNA microarray, scanning of the microarrays, and data analysis were performed by the Nippon Laser and Electronics Lab (Nagoya, Japan).
ChIP Assays.
ChIP assays were performed as described (51) by using a polyclonal antibody against RelA/p65 (C-20; Santa Cruz Biotechnology). Samples were analyzed by PCR. Sequence information of the promoter-specific primers is available upon request.
In Vivo Transamidation.
Assays were performed essentially as described previously (27). Cells were metabolically labeled with 1 mM pentylamine-biotin (BP; Pierce) added to the culture medium for 1 h before stimulation with TNF (25 ng/mL) for the indicated times. Cells were washed with PBS solution and harvested, and cell extracts were prepared by sonication in urea-containing buffer (50 mM Tris HCl, pH 7.6, 250 mM NaCl, 2 M urea, 0.05% SDS, 40 mM DTT, and a protease inhibitor mixture). Proteins were resolved by SDS/PAGE (10% gel) and transferred onto nitrocellulose membranes (Schleicher and Schuell), and proteins that incorporated BP were detected by using HRP-conjugated streptavidin and chemiluminescence (Pierce).
In Vitro Transamidation Assays.
The mouse procaspase-3 cDNA was HA-tagged and subcloned into pBluescript KS(+) (Stratagene). The protein was expressed by in vitro coupled transcription-translation in reticulocyte lysate (Promega), immunoprecipitated with anti-HA beads (Roche), and eluted with HA-peptide. The protein was further incubated with PAI-2 or TG2 immunoprecipitated from cell extracts or with recombinant proteins in a buffer containing 50 mM Tris HCl, pH 8.5, 150 mM NaCl, 5 mM CaCl2, and a protease inhibitor mixture (Calbiochem). The reactions were carried out for 1 h at 37 °C. HA-tagged procaspase-3 was detected by SDS/PAGE and immunoblotting using anti-HA or anti–procaspase-3 antibodies.
Mice.
Tg2−/− mice were generated by homologous recombination (31) and were back-crossed to C57BL/6 mice for more than eight generations.
Preparation of MEFs.
MEFs were prepared from individual littermate embryos at embryonic day 13.5 by using a standard procedure. Briefly, after removal of the head and the liver, the embryonic tissue was washed twice with PBS solution and treated with trypsin/EDTA for 30 min. The homogenates were transferred into a 150-mm dish containing complete culture medium (DMEM supplemented with 10% FBS, 0.1 mM β-mercaptoethanol, and antibiotics). Cells were passaged every 2 to 3 d. Experiments were performed with cells at passages three and four. The genotype of MEFS was determined and confirmed by PCR using genomic DNA extracted from yolk sac and MEFs.
Liver Injury Models.
All experimental protocols were conducted in accordance with the Korean law on animal protection and approved by the institutional animal care and use committee at the National Cancer Center of Korea. Mice (9–10 wk old) were injected intraperitoneally with 20 μg of Act D and 0.3 μg of mouse TNF (no.575202; Biolegend). Alternatively, Con A was injected through the tail vein at 35 mg/kg. In both cases, PBS solution was injected in control animals. Mice were killed 8 h, 24 h (Con A), or 16 h (TNF and Act D) after challenge. Blood was collected by cardiac puncture, and the livers were surgically removed. Serum ALT and AST levels were determined by using Fuji Dri-Chem Slides AST-/ALT-PIII (FujiFilm) according to the manufacturer's instructions. Liver tissue samples were fixed in 10% buffered formalin and processed for paraffin embedding and histological evaluation. Pieces of liver tissue were snap-frozen and used for preparation of whole-liver protein extracts. Histological analysis was performed on liver sections (2–3 μm thick) after routine H&E staining. In situ TUNEL assays were performed on tissue sections by using an in situ apoptosis detection kit (Takara Bio) according to the manufacturer's instructions. Briefly, deparaffinized sections were treated with proteinase K and washed with PBS solution, and endogenous peroxidase activity was inactivated in 3% H2O2. After terminal-deoxynucleotidyl transferase enzymatic reaction, the signal was detected by using an HRP-labeled anti-FITC antibody and visualized with DAB as substrate.
Immunohistochemistry.
Deparaffinized tissue sections were incubated with anti-CD31 (PECAM-1; Abcam) at a dilution of 1:50. Antibody binding was detected by using a HRP-linked secondary antibody and revealed by conventional immunostaining performed in an autoimmunostaining apparatus (HX System; Ventana) using DAB as substrate.
Supplementary Material
Acknowledgments
We thank D. Rothwarf for critical reading of the manuscript and helpful suggestions; W.-C. Yeh, S. Akira, A. Beg, A. Hoffmann, R. Flavell, and Z.-W. Li for providing Tbk1−/−, RelA/p65−/−, IκBα−/−, caspase-3−/−, and Ikkα/β−/− MEFs, respectively; P. Brouckaert for providing mTNF; M. Hernandez (Applied Biosystems) for advice on quantitative PCR; C. H. Jeon for technical assistance; and D. Ginsburg, M. Montminy, T. Kato, and S. Imajoh-Ohmi for plasmids and antibodies. M.D. was supported by a grant from the Claudia Adams Barr Program in Cancer Research. K.S.K. was supported by the National Institutes of Health (NIH). This work was supported by National Cancer Center (Korea) Research Grants NCC 0510270 and NCC1110011-1 (to H.L. and S-Y.K.), National Research Foundation Grant 2010-0029919 funded by the Korean government (to S-Y.K.), NIH Grant AI043477 (to M.K.), and Grants-in-Aid for Scientific Research on Priority Area and for Scientific Research (B) by the Ministry of Education, Science, Sports, and Culture of Japan (M.N.), M.K. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 1005.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119296109/-/DCSupplemental.
References
- 1.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 3.Chen G, Goeddel DV. TNF-R1 signaling: A beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 4.Wajant H, Scheurich P. TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 2011;278:862–876. doi: 10.1111/j.1742-4658.2011.08015.x. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 6.Burstein E, Duckett CS. Dying for NF-kappaB? Control of cell death by transcriptional regulation of the apoptotic machinery. Curr Opin Cell Biol. 2003;15:732–737. doi: 10.1016/j.ceb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Janes KA, et al. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 8.Pomerantz JL, Baltimore D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999;18:6694–6704. doi: 10.1093/emboj/18.23.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tojima Y, et al. NAK is an IkappaB kinase-activating kinase. Nature. 2000;404:778–782. doi: 10.1038/35008109. [DOI] [PubMed] [Google Scholar]
- 10.Kuai J, et al. NAK is recruited to the TNFR1 complex in a TNFalpha-dependent manner and mediates the production of RANTES: identification of endogenous TNFR-interacting proteins by a proteomic approach. J Biol Chem. 2004;279:53266–53271. doi: 10.1074/jbc.M411037200. [DOI] [PubMed] [Google Scholar]
- 11.Bonnard M, et al. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 2000;19:4976–4985. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemmi H, et al. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerondakis S, et al. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 14.Izuhara K, Ohta S, Kanaji S, Shiraishi H, Arima K. Recent progress in understanding the diversity of the human ov-serpin/clade B serpin family. Cell Mol Life Sci. 2008;65:2541–2553. doi: 10.1007/s00018-008-8049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 16.Tang G, et al. Inhibition of JNK activation through NF-kappaB target genes. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 17.Fujita F, et al. Identification of NAP1, a regulatory subunit of IkappaB kinase-related kinases that potentiates NF-kappaB signaling. Mol Cell Biol. 2003;23:7780–7793. doi: 10.1128/MCB.23.21.7780-7793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buss H, et al. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-kappaB at serine 536 is mediated by multiple protein kinases including IkappaB kinase (IKK)-alpha, IKKbeta, IKKepsilon, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J Biol Chem. 2004;279:55633–55643. doi: 10.1074/jbc.M409825200. [DOI] [PubMed] [Google Scholar]
- 19.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 20.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Baglioni C. Protection from tumor necrosis factor-mediated cytolysis by overexpression of plasminogen activator inhibitor type-2. J Biol Chem. 1991;266:20960–20964. [PubMed] [Google Scholar]
- 22.Dickinson JL, Norris BJ, Jensen PH, Antalis TM. The C-D interhelical domain of the serpin plasminogen activator inhibitor-type 2 is required for protection from TNF-alpha induced apoptosis. Cell Death Differ. 1998;5:163–171. doi: 10.1038/sj.cdd.4400324. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi H, Wang HG. Tissue transglutaminase serves as an inhibitor of apoptosis by cross-linking caspase 3 in thapsigargin-treated cells. Mol Cell Biol. 2006;26:569–579. doi: 10.1128/MCB.26.2.569-579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang GY, et al. Transglutaminase 2 suppresses apoptosis by modulating caspase 3 and NF-kappaB activity in hypoxic tumor cells. Oncogene. 2010;29:356–367. doi: 10.1038/onc.2009.342. [DOI] [PubMed] [Google Scholar]
- 25.Lorand L, Graham RM. Transglutaminases: Crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 26.Sarang Z, et al. Tissue transglutaminase (TG2) acting as G protein protects hepatocytes against Fas-mediated cell death in mice. Hepatology. 2005;42:578–587. doi: 10.1002/hep.20812. [DOI] [PubMed] [Google Scholar]
- 27.Shin DM, et al. Cell type-specific activation of intracellular transglutaminase 2 by oxidative stress or ultraviolet irradiation: implications of transglutaminase 2 in age-related cataractogenesis. J Biol Chem. 2004;279:15032–15039. doi: 10.1074/jbc.M308734200. [DOI] [PubMed] [Google Scholar]
- 28.Lakhani SA, et al. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masud A, et al. Endoplasmic reticulum stress-induced death of mouse embryonic fibroblasts requires the intrinsic pathway of apoptosis. J Biol Chem. 2007;282:14132–14139. doi: 10.1074/jbc.M700077200. [DOI] [PubMed] [Google Scholar]
- 30.Siegel M, Khosla C. Transglutaminase 2 inhibitors and their therapeutic role in disease states. Pharmacol Ther. 2007;115:232–245. doi: 10.1016/j.pharmthera.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DS, et al. Transglutaminase 2 gene ablation protects against renal ischemic injury by blocking constant NF-κB activation. Biochem Biophys Res Commun. 2010;403:479–484. doi: 10.1016/j.bbrc.2010.11.063. [DOI] [PubMed] [Google Scholar]
- 32.Napolitano G, Karin M. Sphingolipids: The oil on the TRAFire that promotes inflammation. Sci Signal. 2010;3:pe34. doi: 10.1126/scisignal.3141pe34. [DOI] [PubMed] [Google Scholar]
- 33.Chariot A, et al. Association of the adaptor TANK with the I kappa B kinase (IKK) regulator NEMO connects IKK complexes with IKK epsilon and TBK1 kinases. J Biol Chem. 2002;277:37029–37036. doi: 10.1074/jbc.M205069200. [DOI] [PubMed] [Google Scholar]
- 34.Bouwmeester T, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 36.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 38.Chang L, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 39.Green DR, Oberst A, Dillon CP, Weinlich R, Salvesen GS. RIPK-dependent necrosis and its regulation by caspases: A mystery in five acts. Mol Cell. 2011;44:9–16. doi: 10.1016/j.molcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schleef RR, Chuang TL. Protease inhibitor 10 inhibits tumor necrosis factor alpha -induced cell death. Evidence for the formation of intracellular high M(r) protease inhibitor 10-containing complexes. J Biol Chem. 2000;275:26385–26389. doi: 10.1074/jbc.C000389200. [DOI] [PubMed] [Google Scholar]
- 41.Dougherty KM, et al. The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival. Proc Natl Acad Sci USA. 1999;96:686–691. doi: 10.1073/pnas.96.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tonnetti L, et al. SerpinB2 protection of retinoblastoma protein from calpain enhances tumor cell survival. Cancer Res. 2008;68:5648–5657. doi: 10.1158/0008-5472.CAN-07-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boehm JE, Singh U, Combs C, Antonyak MA, Cerione RA. Tissue transglutaminase protects against apoptosis by modifying the tumor suppressor protein p110 Rb. J Biol Chem. 2002;277:20127–20130. doi: 10.1074/jbc.C200147200. [DOI] [PubMed] [Google Scholar]
- 44.Park JM, et al. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis—CREB and NF-kappaB as key regulators. Immunity. 2005;23:319–329. doi: 10.1016/j.immuni.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Medcalf RL. Plasminogen activator inhibitor type 2: Still an enigmatic serpin but a model for gene regulation. Methods Enzymol. 2011;499:105–134. doi: 10.1016/B978-0-12-386471-0.00006-7. [DOI] [PubMed] [Google Scholar]
- 46.Fesus L, Piacentini M. Transglutaminase 2: An enigmatic enzyme with diverse functions. Trends Biochem Sci. 2002;27:534–539. doi: 10.1016/s0968-0004(02)02182-5. [DOI] [PubMed] [Google Scholar]
- 47.De Laurenzi V, Melino G. Gene disruption of tissue transglutaminase. Mol Cell Biol. 2001;21:148–155. doi: 10.1128/MCB.21.1.148-155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nanda N, et al. Targeted inactivation of Gh/tissue transglutaminase II. J Biol Chem. 2001;276:20673–20678. doi: 10.1074/jbc.M010846200. [DOI] [PubMed] [Google Scholar]
- 49.Rossin F, D'Eletto M, Macdonald D, Farrace MG, Piacentini M. TG2 transamidating activity acts as a reostat controlling the interplay between apoptosis and autophagy. Amino Acids. 2011 doi: 10.1007/s00726-011-0899-x. [DOI] [PubMed] [Google Scholar]
- 50.Delhase M. IkappaB kinase and NF-kappaB signaling in response to pro-inflammatory cytokines. Methods Mol Biol. 2003;225:7–17. doi: 10.1385/1-59259-374-7:7. [DOI] [PubMed] [Google Scholar]
- 51.Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]