Abstract
RNA sequencing (RNA-Seq) is a powerful tool for transcriptome profiling, but is hampered by sequence-dependent bias and inaccuracy at low copy numbers intrinsic to exponential PCR amplification. We developed a simple strategy for mitigating these complications, allowing truly digital RNA-Seq. Following reverse transcription, a large set of barcode sequences is added in excess, and nearly every cDNA molecule is uniquely labeled by random attachment of barcode sequences to both ends. After PCR, we applied paired-end deep sequencing to read the two barcodes and cDNA sequences. Rather than counting the number of reads, RNA abundance is measured based on the number of unique barcode sequences observed for a given cDNA sequence. We optimized the barcodes to be unambiguously identifiable, even in the presence of multiple sequencing errors. This method allows counting with single-copy resolution despite sequence-dependent bias and PCR-amplification noise, and is analogous to digital PCR but amendable to quantifying a whole transcriptome. We demonstrated transcriptome profiling of Escherichia coli with more accurate and reproducible quantification than conventional RNA-Seq.
Keywords: systems biology, digital counting, next generation sequencing, gene expression, genomics
The central goal of transcriptome profiling is to accurately quantify the abundance of RNA transcripts in a sample. Although hybridization-based approaches like DNA microarrays can provide only a relative, analog measure of transcript abundance, sequencing-based approaches, such as RNA sequencing (RNA-Seq), have the advantage of removing hybridization bias among genes (1, 2) and offer the promise of true digital quantification.
The interpretation of conventional RNA-Seq is complicated by sequence-dependent bias and amplification noise from reverse transcription, adapter ligation, library amplification by PCR, solid-phase clonal amplification, and sequencing (3–5). NanoString technology mitigates these complications by eliminating enzymatic reactions and hybridizing color-coded probes directly to RNA for single-molecule detection (6), although it requires many specific probes. Other methods reduce bias in RNA-Seq by eliminating PCR and directly sequencing single molecules of RNA (7), or sequencing single molecules (8) or clonal populations (9) of cDNA. However, library amplification is desirable for sequencing small samples or single cells (10).
Conventional library amplification is based on PCR, but the exponential amplification afforded by PCR introduces noise, especially at low copy numbers (11). Digital PCR was introduced to circumvent this problem by distributing DNA molecules into many containers, each receiving zero or one molecules, which are amplified and detected by PCR (12). This technique has been successfully applied to RNA counting (13); however, it requires specific primers for each gene, which hinders high-throughput measurements.
Here we report a system-wide method for bias and noise reduction in RNA-Seq that allows the use of PCR to amplify a cDNA library before sequencing, providing accurate digital quantification of the transcriptome. In our approach, each cDNA molecule is attached to a unique barcode sequence from a large pool of barcodes before amplification (Fig. 1A) (14). Deep sequencing then allows quantification of the number of cDNA molecules in the original sample by counting the number of unique barcode sequences associated with a given cDNA sequence. This concept has been applied recently for studying protein-RNA interactions (15), to improve the sensitivity of DNA mutation detection (16, 17) and accuracy of DNA copy-number measurements for individual genes by threshold detection (18), and to perform karyotyping and mRNA profiling (19). However, barcode identification in these studies was not immune to errors incurred during library preparation, amplification, and sequencing, which can convert one barcode into another. Hence, a substantial fraction of reads contained misidentified barcodes (16–19), which in some cases were discarded using an artificial threshold (17–19). To avoid this complication, we designed optimized barcodes that can be ligated and amplified with minimal bias and distinguished from one another despite the accumulation of PCR mutations and sequencing errors.
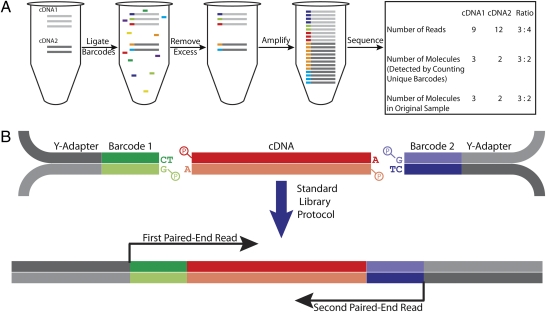
Fig. 1.
Our scheme of digital RNA-Seq. (A) General principle of digital RNA-Seq. Assume the original sample contains two cDNA sequences, one with three copies and another with two copies. An overwhelming number of unique barcode sequences are added to the sample in excess, and five are randomly ligated to the cDNA molecules. Ideally, each cDNA molecule in the sample receives a unique barcode sequence. After removing the excess barcodes, the barcoded cDNA molecules are amplified by PCR. Because of intrinsic noise and sequence-dependent bias, the barcoded cDNA molecules are amplified unevenly. Consequently, after the amplicons are sequenced, it appears that there are three copies of cDNA1 for every four copies of cDNA2 based on the relative number of reads for each sequence. However, the ratio in the original sample was 3:2, which is accurately reflected in the relative number of unique barcodes associated with each cDNA sequence. (B) In our implementation of A, we found it advantageous to randomly ligate both ends of each phosphorylated cDNA fragment to a barcoded phosphorylated Illumina Y-shaped adapter. Note that the single T and A overhangs present on the barcodes and cDNA, respectively, are to enhance ligation efficiency. After this step, the sample is amplified by PCR and prepared for sequencing using the standard Illumina library protocol. For each amplicon, both barcode sequences and both strands of the cDNA sequence are read using paired-end deep sequencing.
Results
Barcoding Strategy for Digital RNA-Seq.
Fig. 1A depicts the general concept of digital counting by random labeling of all target nucleic acid molecules in a sample with unique barcode sequences. To achieve unique barcoding of as many target sequences as possible, the set of barcode sequences introduced to the sample must be (i) much larger than the copy number of the most abundant target sequence and (ii) sampled randomly by the target sequences. If these two criteria are satisfied, then digital quantification of the target molecules by this method is limited only by sequencing depth and accuracy. Unlike conventional sequencing-based approaches to nucleic-acid quantification, the digital counting technique is no longer limited by intrinsic amplification noise and bias in downstream sample preparation and sequencing (Fig. 1A).
Implementation of the scheme in Fig. 1A for digital RNA-Seq requires several critical considerations. As noted above, if the barcode sequences are random, then a sequencing error at one position in a barcode will cause that barcode to be misidentified. This error-induced interconversion will occur even if the barcode sequences are nonrandom (18), unless the barcodes are carefully designed so that multiple substitution errors and indels do not obscure their identities (20). Because DNA secondary structure can reduce amplification efficiency, the barcodes should not have significant sequence overlap or complementarity with each other, the adapter and primer sequences used in library preparation and sequencing, or the transcriptome-of-interest. Ideally, the barcode set will not contain sequence motifs that are known to be problematic for sequencing chemistries, such as long homopolymers and regions with high or low GC-content.
We used a computer program to generate a set of 145 barcode sequences 20-bp in length (Dataset S1) that satisfies the above criteria (Materials and Methods and SI Materials and Methods). The barcode sequences can sustain up to four substitution errors and remain unambiguously identifiable. In addition, a barcode that incurs up to nine substitution errors or the combination of one indel and five substitution errors will not take on the sequence of another barcode.
Instead of using a single barcode sequence to identify each target molecule in our sample (15–19), we attached a barcode sequence to both ends of each target molecule (Fig. 1B and SI Materials and Methods). If both ends of a target sequence sample all of the barcodes randomly, the target sequence will have access to 145 × 145 = 21,025 unique labels. The two barcode sequences along with the target molecule sequence were then read out by paired-end sequencing (Fig. 1B). This paired-end strategy dramatically reduces the number of barcodes that must be designed and synthesized, is compatible with conventional paired-end library protocols, and provides long-range sequence information that improves mapping accuracy (21, 22). In addition, attaching barcodes to both ends increases the overall randomness of barcode sampling because the two ends of a target molecule are unlikely to have a similar degree of bias. We tested and characterized this method on a set of quantified DNA spike-in sequences and a cDNA library derived from the transcriptome of Escherichia coli.
Quantification of Spike-in Sequences and Barcode Sampling Bias.
To calibrate our digital RNA-Seq system, we measured the concentrations of five synthetic DNA spike-in sequences using the Fluidigm digital PCR platform and used them as internal standards. The spike-in samples were barcoded, added to the barcoded E. coli cDNA library, and quantified using the sequencing-based digital counting strategy described above. Fig. 2A shows that the number of digital counts (i.e., unique barcodes) observed in deep sequencing is well-correlated with the digital PCR calibration of the spike-in sequences.
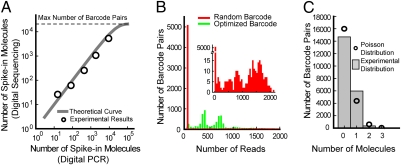
Fig. 2.
Spike-in sequence quantification. (A) Correlation between the number of spike-in molecules for five different spike-in sequences as measured by digital PCR and digital counting of unique barcodes. The theoretical curve, which saturates because of the finite number of barcode pairs (21,025), is calculated based on the Poisson distribution (18). (B) Histograms of the number of reads corresponding to each observed barcode attached to the most abundant spike-in sequence for two experiments. The red histogram corresponds to a spike-in sequence labeled with random barcode sequences, and the green histogram corresponds to a spike-in sequence labeled with our optimized barcodes. Note the left-most bin in the red histogram is >10-times larger than that of the green histogram and contains a large number of unique barcodes with a low number of reads. This discrepancy is caused by various sequencing and PCR amplification errors, which generate new artifactual unique barcodes not present in the original sample and result in a large number of falsely identified unique barcodes (SI Materials and Methods). (Inset) The red histogram in greater detail. (C) Histogram of the number of times a barcode pair was observed with all five spike-in sequences (i.e., the number of spike-in molecules attached to a given barcode pair). Because the spike-in sequences sample the barcode pairs randomly with very little bias, the histogram follows a Poisson distribution.
To evaluate the difference between using random barcode sequences and our optimized barcode sequences, we conducted two experiments. In one experiment, we labeled the spike-in molecules with random barcode sequences (SI Materials and Methods), and in the second experiment we used our optimized, predetermined barcode set. We constructed the histograms of the number of reads for all barcodes observed from the most abundant spike-in sequence (Fig. 2B). When using random barcodes (red histogram in Fig. 2B and SI Materials and Methods), the left-most bin exhibits a large peak because a substantial fraction of barcodes are infrequently read because of sequencing errors; this causes barcodes to interconvert, generating quantification artifacts that were also evident in previous reports (16–19). In stark contrast, the left-most bin, when using optimized barcodes (green histogram in Fig. 2B), has no such peak because our optimized barcode sequences avoid misidentification because of sequencing errors. The effect of sequencing error on both random and optimized barcode counting is clearly shown by simulation (Fig. S1 and SI Materials and Methods).
We note that the green histogram in Fig. 2B is the distribution of the number of reads for the 5,311 uniquely barcoded molecules from a particular spike-in (SI Materials and Methods). Assuming each barcoded spike-in molecule is identical, the green histogram in Fig. 2B is essentially the probability distribution of the number of reads for a single molecule, which spans three orders-of-magnitude. This broad distribution arises primarily from intrinsic PCR amplification noise (11) in sample preparation. Given this broad single-molecule distribution, for low copy molecules in the original sample, counting the total number of reads (conventional RNA-Seq) would be catastrophic. On the other hand, this problem can be circumvented if one counts the number of different barcodes (integrated area of the histogram) using our digital RNA-Seq approach, yielding accurate quantification with single-copy resolution. The two counting schemes give the same results only when the copy number in the original sample is high, assuming there is no sequence-dependent bias.
Random sampling of the barcode sequences by each target sequence is essential for accurate digital counting. Fig. 2C shows that the distribution of observed molecule counts is in excellent agreement with Poisson statistics. Therefore, the five spike-in sequences sample the 21,025 barcode pairs without bias.
Digital Quantification of the E. coli Transcriptome.
We obtained 26–32 million reads from our barcoded cDNA libraries that uniquely mapped to the E. coli genome (Materials and Methods and Dataset S2) in two replicate experiments. Fig. 3A shows the number of conventional and digital counts (unique barcodes) as a function of nucleotide position for the fumA transcription unit. Not surprisingly, the read density is considerably less uniform across the transcription unit than the number of digital counts, presumably because of intrinsic noise and bias in fragment amplification.
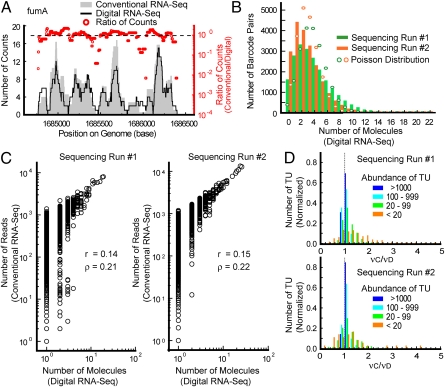
Fig. 3.
Digital quantification of the E. coli transcriptome. (A) Conventional and digital counting results for the fumA transcription unit (TU) as a function of genome position. The conventional counts were calculated by using a conventional calibration curve that allows regression of the number of reads against the number of input molecules for all spike-in molecules (Fig. 2A). The digital counts were obtained by counting the number of unique barcodes associated with each fragment. The red dots are the ratios of these two numbers for each base. (B) Histograms of the number of times a barcode pair was observed with the E. coli cDNA sequences (i.e., the number of cDNA molecules attached to a given barcode pair) in the two replicates. Barcode sampling is more biased on average for E. coli cDNA fragments, but is still in reasonably good agreement with Poisson statistics. (C) Correlation between the number of reads (conventional counting) and the number of molecules obtained from digital counting of unique barcodes for every mapped fragment in the two replicates. For low copy molecules, the conventional counts are distributed over three orders-of-magnitude; this is because the conventional method counts amplicons, which are subject to intrinsic noise (11), rather than directly counting molecules in the original samples like the digital counting method. We note that higher copy fragments are less affected by intrinsic noise (11), as the number of molecules sequenced is greater; this effectively allows averaging over the read counts of many molecules in conventional RNA-Seq, decreasing the variance of counting in the process. (D) Uniformity of conventional vs. digital counting along the length of each transcription unit as a function of transcription unit abundance across the whole E. coli transcriptome for both replicates. We calculated the variation νD = sD/μD (where μD and sD are the mean and sample SD of the digital counts among 99-base bins in a transcription unit, respectively) associated with digital counting and the variation νC = sC/μC associated with conventional counting within each transcription unit for which at least three bins contained on average at least one read. We then created the histogram of the ratio between conventional and digital counting variation (νC/νD) for transcription units in different abundance ranges for each replicate. Transcription unit abundance is the sum of all digital counts for each fragment in the transcription unit.
It is crucial for transcripts across the E. coli transcriptome to sample all barcodes evenly. Fig. 3B shows this distribution, which is close to Poisson but is somewhat overdispersed. Such biased sampling reduces the effective number of barcode sequences Neff available. However, in our E. coli transcriptome sample, the copy number of the most abundant cDNA ranges from 10 to 40 copies for both counting methods. Based on Poisson statistics, even for the most abundant cDNA fragments in our sample, the required Neff is ∼100–400 for 95% unique labeling of all molecules (18). Because there are 21,025 barcode pairs available, on average the degree of randomness observed in Fig. 3B is sufficient.
The conventional method counts the number of amplicons, a quantity that is subject to bias and intrinsic amplification noise (11), rather than the number of molecules in the original sample. Conversely, in our digital counting scheme, unique barcode sequences distinguish each molecule in the sample, and so the effects of intrinsic noise are minimized. Fig. 3C shows how drastically different digital counting can be from conventional counting at low copy numbers, implying that digital counting of unique barcodes is advantageous, particularly for quantifying low copy fragments. We note that the correlation is stronger for high copy fragments and the same phenomenon is also observed for whole transcription units and genes (Fig. S2).
To demonstrate the superior accuracy of digital counting, we examined the uniformity of our abundance measurements within individual transcripts. Because individual transcription units were, by-and-large, intact RNA molecules following RNA synthesis, the cDNA fragments that map to one region of a given transcription unit should have the same abundance as fragments that map to a different region of the same transcription unit. We histogrammed the ratio between the variation in conventional counting νC and variation in digital counting νD for transcription units in different abundance ranges (Fig. 3D). A variation ratio of νC/νD = 1 indicates that both conventional and digital counting give similarly uniform abundances along the length of a transcription unit. For a transcription unit where νC/νD exceeds one, conventional counting measures abundance less consistently along the transcription unit than digital counting. The mean values of νC/νD in the two replicates are 1.4 (s = 1.5, where s is sample SD) and 1.2 (s = 0.5) for the complete set of analyzed transcription units, indicating that conventional counting is less consistent than digital counting across an average transcription unit. Furthermore, the mean value of νC/νD increases with decreasing copy number and its distribution becomes broader (Fig. 3D). For transcription units in the lowest abundance regime, the mean values of νC/νD are 1.9 (s = 2.4) and 1.3 (s = 0.9) for the two replicates. We conclude that, on average, digital counting outperforms conventional counting in terms of accuracy, and its performance advantage is most pronounced for low abundance transcription units.
Although Fig. 3 demonstrates that digital counting is less noisy and more accurate than conventional counting, Fig. 4 shows that digital counting is also more reproducible. We demonstrate this on the level of a single transcription unit in Fig. 4A, which shows the ratio of counts between the two replicates for both conventional and digital counting along the fumA transcript. This ratio is consistently close to one for digital counting, but fluctuates over three orders-of-magnitude for conventional counting. We analyzed the global reproducibility of the whole transcriptome for quantification of transcription units and genes for both conventional and digital counting in Fig. 4 B and C, respectively. In both cases, the correlation between replicates is noticeably better for digital counting than conventional counting, particularly for low copy transcripts.
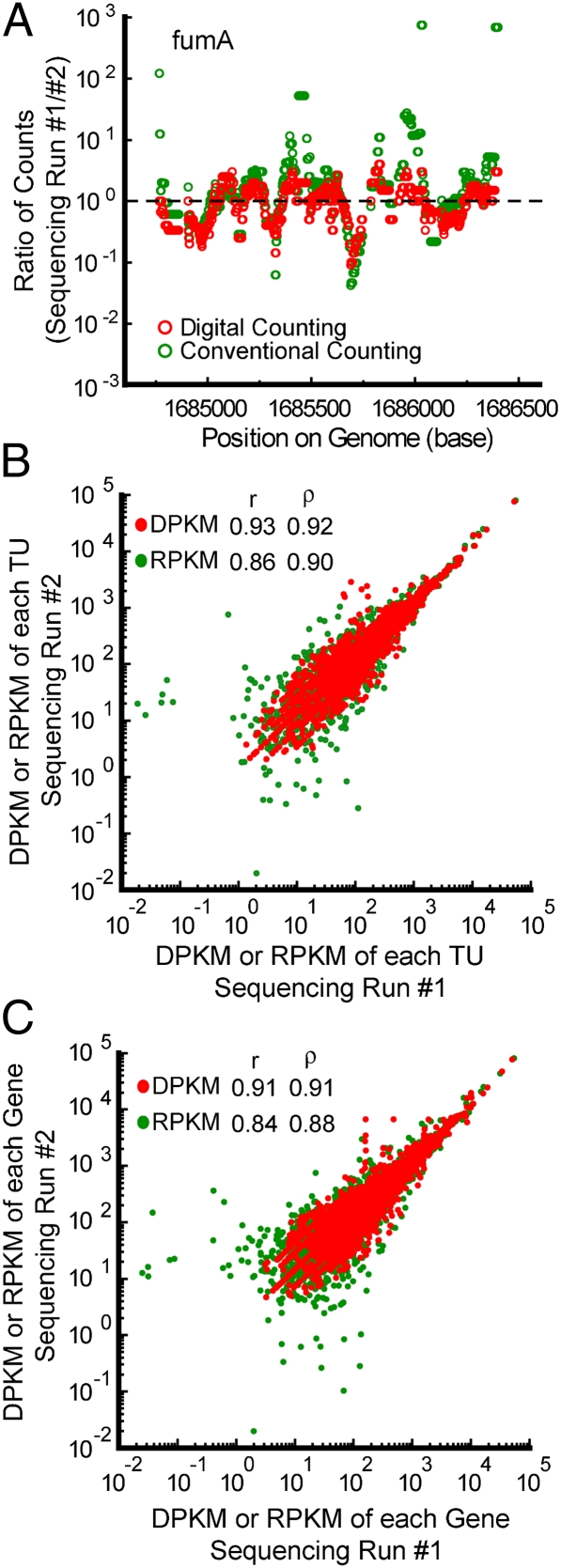
Fig. 4.
Reproducibility of digital and conventional quantification of the E. coli transcriptome. (A) Ratio of counts between two replicate sequencing runs normalized by total uniquely mapped reads for digital counting plotted along with the ratio of counts between the two replicates for conventional counting of the fumA transcription unit. As expected, the ratio fluctuates over a broader range for conventional counting than digital counting along the length of the transcription unit. (B) Correlation between replicate sequencing runs for digital and conventional counting of transcription units. DPKM represents the uniquely mapped digital counts per kilobase per million total uniquely mapped molecules. RPKM represents the uniquely mapped reads per kilobase per million total uniquely mapped reads. (C) Correlation between replicate sequencing runs for digital and conventional counting of genes. Taken together, B and C demonstrate that digital counting is globally more reproducible than conventional counting.
Discussion
Unlike previously reported methods of eliminating bias and noise from RNA-Seq (7–9), our strategy allows amplification by PCR and uses standard commercial protocols for sample preparation. However, the implementation described above also leaves considerable room for improvement. For example, one could ligate barcoded adapters directly to RNA (23, 24), reducing the bias that occurs during reverse transcription. Alternatively, a recently described protocol for processing mature mRNA from single mammalian cells could be modified to include barcoded primers for reverse transcription and second-strand synthesis before amplification (10), obviating the need for ligation.
One disadvantage of our technique is that it requires higher sequencing coverage than conventional RNA-Seq. This requirement is because both the transcriptome and the barcode set must be evenly sampled for accurate counting. However, the cost-per-base of deep sequencing continues to decrease rapidly. In our experiment, the mean number of reads per fragment was ∼400. However, the spike-in sequencing reads can be randomly down-sampled 10-fold (SI Materials and Methods) without perturbing the correlation between abundance measured by digital PCR and digital barcode counting (Fig. S3), which implies that significantly lower coverage will suffice in many cases.
For applications where many cycles of PCR are required for sensitive detection, bias and noise reduction are crucial for accurate quantification. Although we demonstrated our technique on the E. coli transcriptome, we note that the maximum copy number for polyadenylated mRNA in a single mouse blastomere was found to be ∼2,400 (10). With 155 optimized barcode sequences (10 more than were used in this study), one could uniquely label nearly every identical molecule in this system (with 95% unique labeling for even the most abundant transcript). Hence, we expect this technique will be readily applicable to eukaryotic systems without substantial modification. In addition, we analyze the performance of digital and conventional counting in a simulation of differential expression analysis, a key application of RNA-Seq (Fig. S4). Our simulation, which accounts for experimentally measured copy number, barcode sampling bias and amplification noise distributions, shows that digital counting of unique barcodes outperforms conventional counting for differential expression analysis (SI Materials and Methods). Although it is always more difficult to reject the null hypothesis for low-abundance transcripts, we expect our digital counting scheme to be nonetheless more accurate than conventional counting for differential expression analysis at low copy numbers.
In addition to single-cell applications, we expect this technique to be particularly useful for nascent transcript sequencing by run-on (25) or RNA polymerase capture (26), ribosome profiling (27), and profiling of miRNA and other regulatory RNAs, which typically exist at low copy numbers. Significant recent progress has been made in minimizing bias induced by sample barcodes for multiplexed miRNA-Seq (28), and we expect that this technique could be applied to the introduction of barcodes for digital counting in any RNA-Seq experiment. In addition, one could use our approach to improve DNA sequencing experiments, such as ChIP sequencing (ChIP-Seq) (29), which is procedurally related to RNA-Seq and exposed to similar sources of bias and noise (30).
RNA-Seq holds substantial promise for basic research in biomedicine and may ultimately impact clinical diagnostics (21, 31, 32). However, challenges ranging from bias in sample preparation to limited sensitivity and remain significant. Digital RNA-Seq, along with continued improvements to sequencing technology, will lead to new applications and allow RNA-Seq to reach its full potential.
Materials and Methods
Generation and Optimization of Barcodes.
We generated 2,358 random 20-base barcode candidates using a computer, such that even if a barcode accumulated nine mutations, it would not take the sequence of any other generated barcode sequences [unlike the random barcode case (SI Materials and Methods and Dataset S3)]. Barcode candidates containing homopolymers longer than four bases or GC-content less than 40% or greater than 60% were discarded. Barcode candidates were also discarded if each exceeded a certain degree of complementarity or sequence identity (total matches and maximum consecutive matches) with (i) the Illumina paired-end sequencing primers (33), (ii) the Illumina PCR primers PE 1.0 and 2.0, (iii) the 3′ end of the Illumina PCR primers PE 1.0 and 2.0, (iv) the whole E. coli genome [K-12 MG1655 strain (U00096.2)], and (v) all other generated barcode candidates (SI Materials and Methods). Any barcode candidate for which an indel mutation would place it within five point mutations of another barcode candidate was also discarded. The final population consisted of 150 barcodes, of which 145 were randomly chosen and used (Dataset S1).
E. coli RNA Preparation and cDNA Generation.
The cDNA library of E. coli [K-12 MG1655 strain (U00096.2)] was generated by a standard method (SI Materials and Methods).
Sample-Adapter Ligation, Sequencing Sample Preparation, and Sequencing.
The cDNA library was ligated to the barcode adapter mixture, and the sequencing sample was prepared by the standard Illumina protocol with some modifications along with an internal standard (SI Materials and Methods and Dataset S4).
E. coli Transcriptome Analysis.
From the raw sequencing data, we isolated reads that contained barcode sequences that corresponded to our original list of 145 barcodes in both forward and reverse reads for each sequencing cluster that had at most one mismatch. We then aligned the first 28 bases (26 bases for the second sequencing run) of the targeted sequence of both the forward and reverse reads of each cluster to the E. coli genome and kept the sequences that uniquely align fewer than three mismatches and where the two reads did not map to the same sense or antisense strand of the genome. The remaining sequences were mapped to transcription units (34) and sorted by starting and ending position, as well as forward and reverse barcodes (unique tag). Mapped sequence fragments with a length of at least 1,000 bases were discarded. All sequences within the same transcription unit that had the same unique tag were analyzed further. We determined that more than one sequence with the same unique tag were identical if the distance between their center positions was less than 4 bp and if the difference in length was less than 9 bp (Figs. S5 and S6). Thus, the read counts for sequences deemed identical were summed and the sequence with more read counts was deemed as the actual correct sequence. Then, for each unique sequence, we counted the number of unique barcode tags that appeared to determine the copy number of each sequence. The genome-wide expression profile by digital counting and conventional counting are visualized by Integrated Genome Browser (SI Materials and Methods; see Datasets S5, S6, and S7 for specific values for datapoints shown in Fig. S2).
Supplementary Material
Acknowledgments
Illumina sequencing was performed at the Harvard Faculty of Arts and Sciences Center for Systems Biology, with the help of Christian Daly, and at the Tufts University School of Medicine Genomics Core Facility, with the help of Kip Bodi and James Schiemer; digital PCR was performed by the Fluidigm Genetic Analysis Facility at the Molecular Genetics Core Facility of the Children's Hospital Boston Intellectual and Developmental Disabilities Research Center, with the help of Hal Schneider and Ta-Wei Lin; and base-calling analysis was performed at the Harvard Faculty of Arts and Sciences Research Computing Group by Jiangwen Zhang. This work was supported by National Institutes of Health (NIH) National Human Genome Research Institute Grant HG005097-01 (to X.S.X.) and NIH National Human Genome Research Institute Recovery Act Grand Opportunities Grant 1RC2HG005613-01 (to X.S.X.); and K.S. was supported by a Postdoctoral Fellowship for Research Abroad from the Japanese Society for the Promotion of Science and a fellowship from The Uehara Memorial Foundation.
Footnotes
Conflict of interest statement: Harvard University has filed a provisional patent application based on this work.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34449).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118018109/-/DCSupplemental.
References
- 1.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W, Chung LM, Zhao H. Bias detection and correction in RNA-Sequencing data. BMC Bioinformatics. 2011;12:290. doi: 10.1186/1471-2105-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 7.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 8.Lipson D, et al. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 9.Mamanova L, et al. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nat Methods. 2010;7:130–132. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 11.Peccoud J, Jacob C. Theoretical uncertainty of measurements using quantitative polymerase chain reaction. Biophys J. 1996;71:101–108. doi: 10.1016/S0006-3495(96)79205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 14.Hug H, Schuler R. Measurement of the number of molecules of a single mRNA species in a complex mRNA preparation. J Theor Biol. 2003;221:615–624. doi: 10.1006/jtbi.2003.3211. [DOI] [PubMed] [Google Scholar]
- 15.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casbon JA, Osborne RJ, Brenner S, Lichtenstein CP. A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res. 2011;39:e81. doi: 10.1093/nar/gkr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu GK, Hu J, Wang PH, Fodor SP. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc Natl Acad Sci USA. 2011;108:9026–9031. doi: 10.1073/pnas.1017621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kivioja T, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2011 doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 20.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Au KF, Jiang H, Lin L, Xing Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38:4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 24.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 25.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alon S, et al. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011;21:1506–1511. doi: 10.1101/gr.121715.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 30.Nix DA, et al. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozsolak F, et al. Amplification-free digital gene expression profiling from minute cell quantities. Nat Methods. 2010;7:619–621. doi: 10.1038/nmeth.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah SP, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 33.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keseler IM, et al. EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39(Database issue):D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.