Abstract
MicroRNA (miRNA) is a class of small noncoding RNA approximately 22 nt in length. Animal miRNA silences complementary mRNAs via translational inhibition, deadenylation, and mRNA degradation. However, the underlying molecular mechanisms remain unclear. A key question is whether these three outputs are independently induced by miRNA through distinct mechanisms or sequentially induced within a single molecular pathway. Here, we successfully dissected these intricate outputs of miRNA-mediated repression using zebrafish embryos as a model system. Our results indicate that translational inhibition and deadenylation are independent outputs mediated by distinct domains of TNRC6A, which is an effector protein in the miRNA pathway. Translational inhibition by TNRC6A is divided into two mechanisms: PAM2 motif-mediated interference of poly(A)-binding protein (PABP), and inhibition of 5′ cap- and poly(A) tail-independent step(s) by a previously undescribed P-GL motif. Consistent with these observations, we show that, in zebrafish embryos, miRNA inhibits translation of the target mRNA in a deadenylation- and PABP-independent manner at early time points. These results indicate that miRNA exerts multiple posttranscriptional outputs via physically and functionally independent mechanisms and that direct translational inhibition is central to miRNA-mediated repression.
MicroRNA (miRNA) is a class of small noncoding RNA approximately 22 nt in length. Previous studies have shown that miRNA plays a wide variety of regulatory roles in animals and plants (1). Animal miRNA silences partially complementary mRNAs (2). However, the mechanism of its action is under intense debate. Biochemical studies have shown a significant repression of miRNA target genes via translational inhibition (3). In contrast, transcriptome analyses of miRNA target genes have revealed that miRNA promotes mRNA degradation (4, 5). Genome-wide studies that analyzed mRNA stability and translation status in parallel have reached different conclusions concerning the relative contributions of these two outputs (6–8). In addition to these outputs, miRNA induces deadenylation of its target mRNA (5, 9, 10). Given that the translation status, poly(A) tail length and mRNA stability are closely linked to each other, whether these three outputs are independently induced by miRNA through distinct mechanisms or rather sequentially induced within single molecular pathway remains unknown (3, 11).
miRNA forms a complex with protein factors, called miRNA induced silencing complex (miRISC), to induce target mRNA silencing. Argonaute (Ago) protein is an integral component of miRISC and directly interacts with miRNA to support the binding of miRNAs to their target mRNAs (12). In addition to Ago, animal miRNA requires another miRISC component, TNRC6/GW182 (hereafter referred to as TNRC6), for target mRNA silencing (13). TNRC6 associates with the Ago-miRNA complex via multiple Ago-binding motifs in its N-terminal region (14, 15). In contrast, the C-terminal regions of TNRC6 have been identified as a silencing domain because this domain is sufficient to elicit posttranscriptional silencing (16, 17). Recent studies have shown that the silencing domains of TNRC6 bind to poly(A) binding protein (PABP). In the case of mammalian TNRC6, the interaction occurs mainly through direct interaction between the conserved sequence PAM2 (PABP-interacting motif 2), which is located at the anterior portion of the silencing domain, and the C-terminal MLLE domain of PABP (18–20). The TNRC6-PABP interaction accelerates deadenylation by CAF1/CCR4 in Krebs cell extract (18, 19). In human and fly cultured cells, the TNRC6-PABP interaction is required to support maximum repression by miRNA (20). These observations have led to a model proposing that, via TNRC6-PABP interactions, miRISC interferes with translation at the initiation step and/or induces mRNA deadenylation/degradation by increasing the susceptibility of the poly(A) tail to deadenylases (11, 18). However, this model does not address the hierarchy of multiple outputs induced by miRNA. In addition, the contribution of the TNRC6-PABP interaction in the miRNA pathway awaits further validation because poly(A) tail-independent repression by miRNA has been reported in several experimental systems (10, 21).
In the current study, we successfully dissected intricate outputs of miRNA-mediated repression using zebrafish embryos as a model system. Our results indicate that translational inhibition and deadenylation are independent outputs mediated by distinct domains of TNRC6A. Translational inhibition by TNRC6A is divided into two mechanisms: PAM2 motif-mediated interference of poly(A)-binding protein (PABP), and inhibition of 5′ cap- and poly(A) tail-independent step(s) by a previously undescribed P-GL motif. Consistent with these observations, we show that, in zebrafish embryos, miRNA inhibits translation of the target mRNA in a deadenylation- and PABP-independent manner at early time points.
Results
TNRC6A Induces Translational Inhibition and Deadenylation Through the Mid Domain in Zebrafish Embryos.
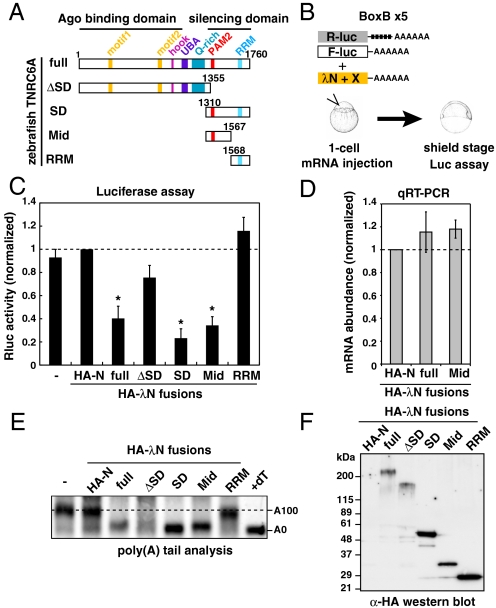
To elucidate the mechanisms of miRNA-mediated repression, we investigated the repression activities of TNRC6 using zebrafish as an in vivo model system. As in other vertebrates, the genome of zebrafish encodes all three TNRC6 paralogues (TNRC6A, B and C) and additional TNRC6B and TNRC6C genes due to genome duplication. We focused on a unique zebrafish orthologue of TNRC6A in this study because all TNRC6 paralogues have a repressive activity in humans (17, 22). Zebrafish TNRC6A contains all of the conserved motifs/domains that were found in other TNRC6 orthologues (13) (Fig. 1A). To identify the functional domain(s) in zebrafish TNRC6A that were responsible for silencing effects, we generated a series of deletion constructs and tested their effects in a λN-BoxB tethering assay in zebrafish embryos (Fig. 1 A and B). Tethering of full-length TNRC6A to Rluc-BoxB-poly(A) mRNA repressed Rluc activity to approximately 40% compared to the control construct encoding the HA-tagged N-peptide (HA-N) (Fig. 1C, full). As reported with other TNRC6 orthologues (16, 17), the silencing domain (SD) of zebrafish TNRC6A was sufficient to induce repression (Fig. 1C, SD). Furthermore, we found that the anterior half of the silencing domain (before the RRM, hereafter referred to as the Mid domain) induced repression (Fig. 1C, Mid). On the other hand, no significant repression was observed with TNRC6A fragments lacking the Mid domain despite detectable expression of effector proteins (Fig. 1F). The repression was mediated via specific binding of N-peptide fusions to BoxB sequences (Fig. S1). qRT-PCR showed that mRNA stability was not changed by tethered TNRC6A proteins during the assay (Fig. 1D). RNaseH-mediated poly(A) tail analysis revealed that the full-length TNRC6A, the silencing domain fragment and the Mid domain fragment induced deadenylation (Fig. 1E). These results reveal that the Mid domain of zebrafish TNRC6A is essential and sufficient to induce translational inhibition and deadenylation.
Fig. 1.
The Mid domain of TNRC6A is sufficient to induce translational repression and deadenylation. (A) Schematic structures of zebrafish TNRC6A and its deletion mutants. (B) Schematic representation of the λN tethering assay in zebrafish embryos. (C) Results of the tethering assay with TNRC6A fragments. The bar graph shows Rluc activity that was normalized to Fluc activity. The normalized Rluc activity with the HA-λN empty construct (HA-N) was set to one. The data show averages of three independent experiments. Error bars show SD. Asterisks indicate p < 0.01 compared to experiments with HA-N. (D) The qRT-PCR analysis of reporter mRNA stability. The normalized Rluc mRNA values [normalized to those of the HA-λN empty construct (HA-N)] were set to one. The data show averages of three independent experiments. Error bars show SD. (E) The poly(A) tail analysis of the Rluc-BoxB-pA reporter mRNA by RNaseH digestion and Northern blot. The lane +dT shows completely deadenylated fragments, which correspond to A0. (F) Western blotting detecting HA-tagged effecter proteins.
A Previously Undescribed P-GL Motif in TNRC6A Contributes to Translational Inhibition in Concert with the PAM2 Motif.
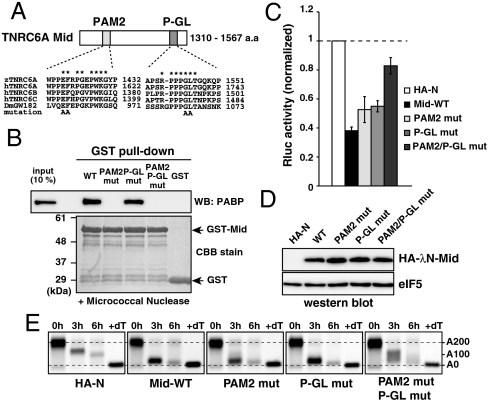
The PAM2 motif was conserved in the Mid domain of zebrafish TNRC6A, with critical residues for the interaction with PABP remaining invariant (Fig. 2A) (18–20). To determine whether the silencing activities of the Mid domain required PAM2–PABP interaction, we introduced a mutation into the PAM2 motif of the TNRC6A Mid domain (Fig. 2A, E1421 and F1422 to A; PAM2 mutation). The PAM2 mutation completely abolished the interaction with PABP, which was assessed using the GST-pulldown assay with zebrafish embryo lysates (Fig. 2B). In the tethering assay, the PAM2 mutation slightly reduced repression activity (Fig. 2 C and D, p < 0.05). Deadenylation of the Rluc-BoxB-poly(A) mRNA was still induced by the PAM2 mutant in this context (Fig. 2E). These results indicate that TNRC6 silencing activity is mediated by the PAM2 motif as suggested in the previous studies (18, 20), yet the contribution might not be predominant in zebrafish embryos.
Fig. 2.
The Mid domain of TNRC6A represses translation via two motifs. (A) Schematic representation of the Mid domain of zebrafish TNRC6A. The two conserved motifs (PAM2 and P-GL) are shown. Sequence alignments of each motif comparing zebrafish TNRC6A, human TNRC6 proteins, and fly GW182 are shown. Conserved residues are marked with asterisks. Alanine substitutions introduced in the current study are shown on the bottom. (B) GST-pulldown assay detecting interaction between the GST-Mid domain and zebrafish PABP. A total of 10% of embryonic lysate was loaded as an input. PABP was detected using Western blotting (Upper). GST fusion proteins were visualized using CBB stain (Lower). (C) The results of the tethering assay with TNRC6A Mid domain mutants. The data were collected and are shown as described in Fig. 1C. (D) Western blot detecting HA-λN-tagged Mid domain proteins. The membrane was probed with anti-eIF5 antibody as a control. (E) The poly(A) tail analysis of the injected Rluc-BoxB-pA reporter mRNA using RNaseH digestion and northern blot at 0, 3 and 6 hours. The lane +dT shows a completely deadenylated fragment (A0).
To characterize repression activities that remained in the PAM2 mutant, we focused on the residues PPPGLT, which are located at the C terminus of the Mid domain and are highly conserved in the TNRC6 family proteins (hereafter referred to as the P-GL motif, Fig. 2A). A mutation introduced into this motif (G1544 and L1545 to A; P-GL mutation) did not affect PABP interaction (Fig. 2B). On the contrary, the P-GL mutation partially reduced the repression activity of the Mid domain (p < 0.01) with no obvious effect on deadenylation (Fig. 2 C and E, P-GL mut). Repression activity of the Mid domain was further impaired when the PAM2 and P-GL motifs were simultaneously mutated (Fig. 2C, PAM2/P-GL mut). Concomitantly, the deadenylation activity was reduced in the double mutant (Fig. 2E). These results suggested that a previously undescribed P-GL motif in the Mid domain contributes to the translational inhibition in concert with the PAM2 motif.
To test the contributions of PAM2 and P-GL motifs in the context of full-length TNRC6A function, we performed two experiments. First, we introduced the mutations into the full-length TNRC6A and tested their effects in the tethering assay (Fig. S2 A–C). The simultaneous loss of both PAM2 and P-GL motifs, but not the loss of one of the two motifs, strongly impaired the repression activity of TNRC6A at translation level. Second, to test the activity of TNRC6A variants in the context of miRNA-mediated repression, we asked if exogenous TNRC6A variants enhance miR-430-mediated repression of the GFP sensor transgene (Fig. S2D; see Fig. 5 for detailed information of the GFP-miR-430 sensor). Overexpression of wild-type, PAM2 mutant, and P-GL mutant versions of TNRC6A promoted miR-430-mediated repression of the GFP-miR-430 sensor. In contrast, the double mutant failed to promote repression by miR-430 (Fig. S2 E and F). These two experiments indicated that our motif analysis with the Mid domain fragment well captured the actions of the full-length TNRC6A.
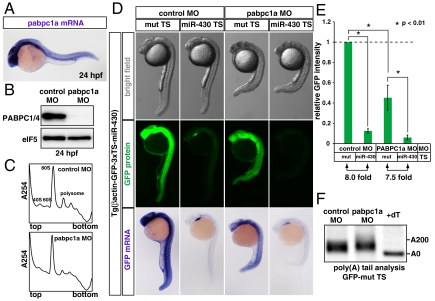
Fig. 5.
miR-430 represses its target mRNA in the absence of PABP during zebrafish embryogenesis. (A) In situ hybridization detecting pabpc1a mRNA in a 24 hpf zebrafish embryo (purple). (B) Western blot detecting PABP protein in 24 hpf embryos injected with control MO or pabpc1a MO. The membrane was probed with anti-eIF5 antibody as a control. (C) The polysome profiles of control MO-injected (Upper) and pabpc1 MO-injected (Lower) zebrafish embryos at 24 hpf. (D) Analysis of miRNA-mediated target mRNA repression in the presence or absence of PABP. Bright field view (Upper panels), GFP fluorescence (Middle panels; green) and GFP mRNA (Bottom panels; purple) of 24 hpf zebrafish embryos expressing the GFP transgene with three copies of the imperfect target site for the ubiquitously expressed miRNA, miR-430 (miR-430 TS) or with mutated target sites (mut TS). Control MO (left columns) or pabpc1a MO (right columns) was injected as indicated. (E) Quantification of GFP expression levels in Fig. 5D. GFP intensity of the embryos with the mut TS and the control MO was set to one. Error bars show SD. Asterisks indicate p < 0.01 compared to the experiment with the mut TS and control MO. (F) The poly(A) tail analysis of the GFP mut TS mRNA with the control MO or pabpc1a MO. The lane +dT shows a completely deadenylated fragment (A0).
TNRC6A Mediates Translational Inhibition Independent of Deadenylation.
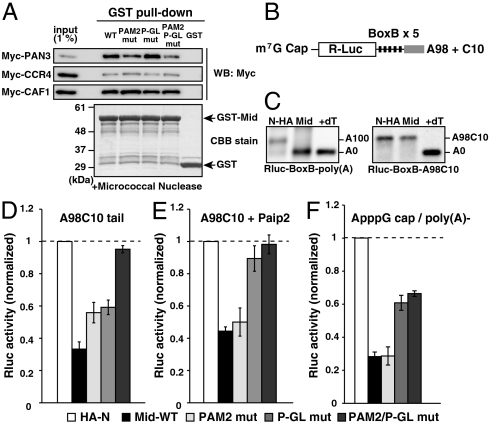
Given the predominant contribution of the PAM2 and the P-GL motifs to translational inhibition with the moderate effect on deadenylation, we next asked if the translational inhibition induced by these two motifs was independent of deadenylation. To this end, we performed two experiments. First, we analyzed the physical interactions between the Mid domain and deadenylase components that have been implicated in miRNA-mediated deadenylation (18, 21, 23, 24). Consistent with its deadenylation activity in the tethering assay, the GST-Mid domain interacted with in vitro translated Myc-tagged PAN3 and CCR4/CAF1 but not with eIF4E (Fig. 3A and Fig. S3 A and B). Notably, the PAM2/P-GL double mutant still interacted with these factors with reduced affinity to PAN3. Second, we asked whether the Mid domain induced translational repression in the absence of deadenylation. To this end, we generated a synthetic poly(A) tail that contained a 98 nt poly(A) sequence followed by a 10 nt poly(C) sequence (A98C10, Fig. 3B). The A98C10 tail was not deadenylated by the Mid domain during the assay (Fig. 3C). On the other hand, the A98C10 tail enhanced Rluc expression similar to the normal poly(A) tail (Fig. S4A). Tethering experiments revealed that the Mid domain strongly silenced tethered mRNA with the A98C10 tail. Furthermore, the coordinated contribution of the PAM2 and P-GL motifs was observed with the A98C10 tail (Fig. 3D). These experiments showed that the Mid domain of TNRC6A inhibited translation via the PAM2 and P-GL motifs in a deadenylation-independent manner.
Fig. 3.
The Mid domain of TNRC6A represses translation via deadenylation-independent mechanisms. (A) The GST-pulldown assay detecting interactions between the GST-Mid domain and deadenylase components translated in rabbit reticulocyte lysate. Total of 1% of in vitro translation reaction was loaded as an input. Myc-tagged proteins were detected using Western blotting (Upper). GST fusion proteins were visualized using CBB stain (Lower). (B) Rluc reporter mRNA containing 5 copies of BoxB sites followed by the A98C10 tail. (C) The poly(A) tail analysis of the Rluc-BoxB reporter mRNAs at six hours, in the presence of control HA-λN peptide (HA-N) or the HA-λN tagged Mid domain (Mid). Left: The reporter mRNA with a normal poly(A) tail [Rluc-BoxB-poly(A)]. Right: The reporter mRNA with an A98C10 tail (Rluc-BoxB-A98C10). The lane +dT shows a completely deadenylated fragment (A0). (D) Tethering assay of the TNRC6A Mid domain with reporter mRNA containing the A98C10 tail in the presence of Myc-GFP. (E) Tethering assay of the TNRC6A Mid domain with reporter mRNA containing the A98C10 tail in the presence of Myc-Paip2. (F) Tethering assay of the TNRC6A Mid domain constructs with a reporter mRNA containing the 5′ ApppG cap without a poly(A) tail. Graphs in D, E, and F show the averages of three independent experiments. Error bars show SD.
The PAM2 and P-GL Motifs Contribute to Translational Silencing Through Distinct Mechanisms.
Next, we determined whether the PAM2 and P-GL motifs required PABP for their function in translation repression. To this end, we utilized Paip2 (PABP-interacting protein 2). Paip2 binds to PABP via its PAM1 and PAM2 motifs, and sequesters PABP by displacing it from the poly(A) tail and eIF4G (25). Hence, we predicted that the repression by the Mid domain would be diminished by Paip2 if PABP is the only molecular target for the Mid domain. First we confirmed that the A98C10 tail failed to stimulate translation in the presence of excess Paip2 (Fig. S4B). In the presence of excess Paip2, the Mid domain repressed translation of Rluc-BoxB-A98C10 mRNA (Fig. 3E). This result suggests that the Mid domain inhibits translation via a PABP-independent mechanism. Consistent with this idea, the Mid domain with the PAM2 mutation inhibited translation of Rluc-BoxB-A98C10 mRNA similar to the wild-type Mid domain (p > 0.38). In contrast, the P-GL mutation diminished most of the repressive effect caused by the Mid domain in the presence of Paip2. Hence, the PAM2 motif required PABP for translational inhibition, while the P-GL motif did not. As an alternative approach, we determined whether the m7G cap and the poly(A) tail, which are two important constituents for translation initiation, are required for translational inhibition by the Mid domain. Although the translation efficiency was significantly reduced in the absence of the m7G cap and the poly(A) tail (Fig. S4A), the tethering assay revealed that the Mid domain repressed bound mRNA even in the absence of the m7G cap and poly(A) tail (Fig. 3F). Analysis with Mid domain mutants showed that the P-GL motif, but not the PAM2 motif, contributed to repression of this reporter mRNA. The contribution of the P-GL motif was not attributed to the change in the RNA stability (Fig. S4C), indicating that the P-GL motif inhibited translation. These results strongly suggested that the Mid domain of TNRC6A inhibits translation through two distinct mechanisms.
Direct Translational Inhibition Is a Major Output of miRNA-Mediated Repression During Zebrafish Embryogenesis.
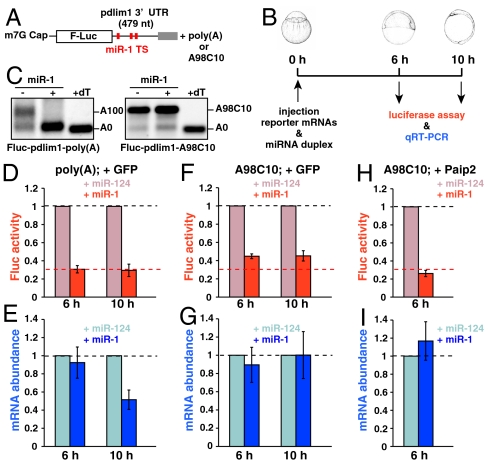
To validate our findings obtained in the tethering assay, we designed a miRNA reporter system in zebrafish embryos using a firefly luciferase (Fluc) mRNA containing the 3′UTR of an endogenous miR-1 target gene, pdlim1 (26) (Fig. 4A). The Fluc-pdlim1 mRNA and a nontargeted mRNA encoding Renila luciferase (Rluc) were microinjected into fertilized eggs together with the miR-1 duplex or control miR-124 duplex. Six or 10 h after the injection, a luciferase assay and quantitative RT-PCR (qRT-PCR) were performed to measure protein expression and mRNA stability (Fig. 4B). As observed in the tethering assay, the A98C10 tail resisted deadenylation by miR-1 in this system (Fig. 4C).
Fig. 4.
miR-1 represses target mRNA in a deadenylation- and PABP-independent manner in zebrafish embryos. (A) Fluc reporter mRNA containing zebrafish pdlim1 3′UTR. Red boxes indicate the target site for miR-1. (B) Scheme of the miR-1 repression assay in zebrafish embryos. (C) The poly(A) tail analysis of the Fluc-pdlim1 3′UTR reporter mRNAs at six hours in the absence (−) or presence (+) of the miR-1 duplex. The left panel shows the reporter mRNA with a normal poly(A) tail [Fluc-pdlim1-poly(A)]. The right panel shows the reporter mRNA with the A98C10 tail (Fluc-pdlim1-A98C10). (D and E) The results of the miR-1 repression assay with reporter mRNA containing a normal poly(A) tail in the presence of control Myc-GFP. (F and G) Results of the miR-1 repression assay with reporter mRNA containing the A98C10 tail in the presence of control Myc-GFP. (H and I) Results of the miR-1 repression assay with reporter mRNA containing the A98C10 tail in the presence of Myc-Paip2. D, F, and H show normalized Fluc activity. E, G, and I show normalized Fluc mRNA levels, which were measured using qRT-PCR. The values of the experiments using miR-124 were set to one at each time point. The data shows averages of three independent experiments. Error bars show SD.
The analysis using Fluc-pdlim1 mRNA with the normal poly(A) tail revealed strong repression of Fluc activity by miR-1 at 6 h (to approximately 30% compared to the miR-124 duplex), with no obvious change in mRNA abundance (Fig. 4 D and E, 6 h). Substantial mRNA degradation by miR-1 was detected at 10 h. However, miR-1-mediated mRNA reduction (approximately 50% compared to miR-124) does not fully account for the overall repressive effect of miR-1, which was measured by the luciferase activity (approximately 30%) (Fig. 4 D and E, 10 h). These results suggest a major contribution of translational inhibition in the miRNA pathway during zebrafish embryogenesis. To validate the contribution of deadenylation to miRNA-mediated repression, we performed the miR-1 repression assay with the deadenylation-resistant A98C10 tail. This analysis revealed two important findings. First, miR-1 inhibited translation of Fluc-pdlim1 mRNA with the A98C10 tail with a lower efficiency compared to the Fluc-pdlim1 mRNA with the normal poly(A) tail (approximately 45% with the A98C10 tail, versus approximately 30% with the normal poly(A) tail at 6 h; Fig. 4F). Second, the A98C10 tail inhibited miR-1-mediated Fluc mRNA degradation during the assay (Fig. 4G). These analyses confirmed a major contribution of deadenylation-independent translational inhibition during miRNA-mediated repression. Next, we analyzed the involvement of PABP-related mechanism in miRNA-mediated translational inhibition. Inhibition of PABP by Paip2 did not abrogate miR-1-mediated repression. Rather, it promoted miR-1-mediated translational inhibition of the Fluc-pdlim1-A98C10 mRNA to approximately 30% (Fig. 4 H and I). Hence, miR-1 can induce translation repression in parallel to the PABP inhibiton by Paip2. These results reveal distinct contributions of deadenylation, PABP inhibition, and PABP-independent translational inhibition to miRNA-mediated repression in vivo.
A PABP-Independent Mechanism of miRNA-Mediated Repression Operates During Zebrafish Embryogenesis.
To further confirm whether miRNA inhibited its target mRNA via PABP-independent repression pathways in vivo, we depleted PABP from zebrafish embryos and analyzed the effects on miRNA-mediated repression. Among zebrafish orthologues of vertebrate PABP genes, the expression of two PABPC1 paralogues (pabpc1a and pabpc1b) and PABPC4 was detected during zebrafish embryogenesis (Fig. 5A and Fig. S5A). Although pabpc1a mRNA was ubiquitously expressed during development, the expression of pabpc1b and pabpc4 mRNAs was restricted to specific tissues at 24 h post fertilization (hpf). Injection with the translation-blocking morpholino oligo (MO) for pabpc1a mRNA at the one-cell stage reduced total PABP levels to < 1% compared to the injection of embryos with control MO (Fig. 5B and Fig. S5B; note that the antibody we used detected all three PABP proteins described above). Concomitantly, PABP-depleted embryos showed reduced formation of polysomes (Fig. 5C) and strong morphological defects (Fig. 5D, upper panels) at 24 hpf. Therefore, cellular PABP was reduced to nonfunctional levels in pabpc1a MO-injected embryos.
To monitor endogenous miRNA-mediated repression, we generated a transgenic zebrafish that ubiquitously expressed GFP mRNA containing three copies of the miR-430 target site (TS) [Tg(ßactin-GFP-3xTS-miR-430)]. Expression of GFP protein from this reporter gene was strongly repressed throughout the embryo by the ubiquitous miRNA miR-430 in a target-site dependent manner (Fig. 5D, middle panels) (9, 27). Notably, miR-430 repressed the GFP reporter in PABP-depleted embryos. In situ hybridization revealed concomitant degradation of the reporter mRNA (Fig. 5D, lower panels), indicating that the repressive effect we observed in the transgenic embryos might be a combined output of initial translational repression and subsequent mRNA degradation. Similar findings were observed with another GFP transgenic line that visualized repression by muscle specific miRNA miR-1/206 (26) (Fig. S6). The PABP-depletion did not cause shortening of the basal poly(A) tail length as a secondary effect (Fig. 5F). These results with the transgenic lines, together with the results with injected reporter mRNAs in Fig. 4, show that miRNA silences its target mRNA via PABP-independent mechanism(s) during zebrafish embryogenesis.
Discussion
Due to its intricate outputs, it has been difficult to delineate the direct and primary consequence of miRNA-mediated repression (3, 11). In this study, we addressed a causal relationship between miRNA-mediated translational repression and deadenylation using three different approaches. First, we revealed that the PAM2 and P-GL motifs in the Mid domain of zebrafish TNRC6A induced translational inhibition in a deadenylation-independent manner (Figs. 2C and 3D). Second, we found that the Mid domain carrying the PAM2 and P-GL mutations showed reduced affinity to PAN3 but still interacted with CCR4/CAF1 deadenylases (Fig. 3A and Fig. S3 A and B). Third, miR-1 induced translational repression of its target reporter mRNA even in the absence of deadenylation (Fig. 4F). These observations collectively indicate that miRNA-mediated translational inhibition and deadenylation are independent outputs mediated through distinct molecular actions of TNRC6.
The use of the deadenylation-resistant A98C10 tail allowed us to determine the relative contributions of direct translational inhibition and deadenylation in the miRNA pathway. Although we observed the contribution of deadenylation to miR-1-mediated translational inhibition, we also observed substantial translational inhibition in the absence of deadenylation with miR-1 and the tethered Mid domain (Figs. 3D and 4F). These observations establish that translational inhibition by miRNA is not a mere consequence of deadenylation. Consistent with these findings, mRNAs that were translated in a poly(A) tail-independent manner were repressed by miRNA (10, 21). Hence, we propose that deadenylation plays an auxiliary role in the miRNA-mediated repression by consolidating translational inhibition, possibly through displacement of PABP from mRNA. We also observed that miRNA required deadenylation for target mRNA degradation that occurred at a later time point (Fig. 4E, 10 h). Therefore, we do not exclude the possibility that deadenylation and subsequent mRNA degradation play more active roles in miRNA-mediated silencing after extended periods, which has been suggested by previous genome-wide analyses (6–8). Nevertheless, a rapid and strong translational inhibition in our zebrafish system supported a model in which direct translational inhibition is central to miRNA-mediated gene silencing at early time points.
The mechanisms that mediate direct translational repression by miRNA have not been well characterized. Some of our data are consistent with a model in which miRNA/TNRC6 induces silencing by interacting with PABP via its PAM2 motif (11). In our tethering assay, the PAM2 motif in the Mid domain of zebrafish TNRC6A contributed to translational inhibition when PABP was involved in translation (Fig. 2C). This contribution was deadenylation-independent (Fig. 3D), indicating that the PAM2 motif contributed to translational inhibition by counteracting the function of PABP in translation. These observations are also consistent with previous studies arguing that miRNA targets the m7G cap-dependent translation initiation process (28–31). In addition, our study indicated that the PAM2–PABP interaction was not the only mechanism used by miRISC to inhibit translation. First, the mutation into the PAM2 motif only weakly reduced the repression activity of the Mid domain (Figs. 2 and 3) and the full-length TNRC6A (Fig. S2). Second, miR-1 and the Mid domain induced translational inhibition even in a PABP-independent manner (Figs. 3E and 4F). Third, depletion of PABP from zebrafish embryos allowed endogenous miRNAs to silence their target mRNAs (Fig. 5 and Fig. S6). Hence, zebrafish miRISC is equipped with a mechanism that does not require PABP for target mRNA silencing. In contrast, previous studies in mammalian cultured cells have shown correlations between cellular PABP activity and miRNA-mediated repression (20, 32). It is therefore possible that the contribution of the PAM2–PABP interaction to miRNA-mediated repression is conditional on the general translation status in the cell.
We identified a previously undescribed conserved motif, the P-GL motif, which was within the Mid domain of TNRC6A, as a PABP-independent translation repression mechanism. The action of the P-GL motif was clearly distinct from that of the PAM2 motif in three aspects. First, the P-GL motif was not involved in PABP binding (Fig. 2B). Second, the P-GL motif contributed to translational repression irrespective of the presence of poly(A) tail or the PABP activity (Figs. 3E and F). Third, the P-GL motif contributed to the repression of mRNA with the unnatural 5′ ApppG cap structure (Fig. 3F). Hence, the P-GL motif repressed mRNA translation independent of essential constituents of the canonical translation initiation pathway. Because the use of the ApppG cap structure significantly reduced the basal translation activity (Fig. S4A), the exact contribution and the molecular basis of the observed repression activity need to be interpreted in the context of the m7G capped and polyadenylated mRNA. Nevertheless, it is worth noticing that the cap-independent repression activity observed with the P-GL motif is consistent with post-initiation repression models of miRNA (33–36). Based on these observations, we propose a “double lock model” of miRNA-mediated translational repression, in which miRNA inhibits two distinct translation steps through TNRC6 (Fig. S7).
While this manuscript was under revision, three papers reported additional functional motifs in fly GW182 and mammalian TNRC6 proteins that mediate binding to the CCR4/NOT1 and PAN2/PAN3 deadenylase complexes (37–39). Interestingly, those complexes induced repression not only via deadenylation but also via deadenylation-independent mechanism(s) (37, 39). Moreover, Fukaya and Tomari recently reported that, consistent with our findings in zebrafish, miRNA induces translation repression independent of deadenylation and PABP in the fly in vitro system (40). Although the relative contributions of these pathways need to be clarified, our current study and these recent reports collectively indicate that miRNA system utilizes multiple redundant mechanisms to silence target mRNAs. The characterization of multiple translational inhibition activities of TNRC6 proteins in a wide variety of experimental systems may reconcile contradicting observations on miRNA-mediated translational inhibition (3, 11). It is tempting to speculate that, by having multiple repression mechanisms, miRNA performs robust control of target gene expression under diverse cellular contexts.
Materials and Methods
Tethering Assay in Zebrafish Embryos.
The mRNAs were transcribed using an mMessage mMachine SP6 kit (Ambion). ApppG capped mRNA was synthesized in the presence of an ApppG cap analogue (New England Biolabs) instead of an m7GpppG cap analogue. To add a normal poly(A) tail, the mRNA was polyadenylated in vitro using a poly(A) tailing kit (Ambion). For poly(A)- mRNA, we injected a MO that binds to the end of the mRNA to inhibit polyadenylation (TB-MO). For mRNA injections, Rluc mRNA and Fluc mRNA were diluted to a final concentration of 50 ng/μL each. Effector mRNAs were diluted to obtain solutions with equimolar concentrations of effector mRNA. HA-λN-Mid mRNA was diluted to a final concentration of 100 ng/μL. The mRNA encoding myc-tagged GFP or zebrafish Paip2 was added to a final concentration of 200 ng/μL. Approximately 1,000 pl of solution containing reporter mRNAs and effecter mRNAs was injected into one-cell stage zebrafish embryos. A total of 5–10 embryos were collected at the shield stage (6 hpf) and lysed in Passive lysis buffer (Promega). The luciferase activities were measured using the Dual-Luciferase Reporter assay system and GloMax 20/20 n luminometer (Promega). Rluc activity intensity (IRluc) was normalized to the intensity of Fluc activity (IFluc). The normalized Rluc activity for each experiment with HA-N effector encoding XX was calculated as follows. Fold change = (IFluc + HA-N-XX/IRluc + HA-N-XX)/(IFluc-control/IRluc-control). The values obtained using HA-N empty mRNA were used for controls. Each sample was measured as three replicates. The p value was calculated using Student’s t test.
qRT-PCR.
A total of five embryos were retained after each injection experiment, and total RNA was extracted by ISOGEN (Nippon gene). The cDNA was synthesized using the PrimeScript RT reagent kit (TAKARA). A random hexamer was used for cDNA synthesis to avoid detecting differences in the poly(A) tail length. To assess Fluc mRNA and Rluc mRNA levels, qRT-PCR was performed with SYBR Premix EX Taq II and the Thermal Cycler Dice Real Time System (TAKARA) following a standard protocol. Specific amplification of the PCR products was confirmed by analyzing the dissociation curve, running the products on an agarose gel, and sequencing. Each sample was measured as duplicates, and each experiment was repeated three times.
Additional materials and methods are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Y. Tomari and M. W. Hentze for discussions and helpful comments on our project. We are also grateful to N. Sonenberg, M. R. Fabian, A. J. Giraldez, T. Inada, and M. Wakiyama for discussions; C. B. Chien and K. Kawakami for Tol2 constructs; and A. Kulozik for λN tethering constructs. We thank H. Fukaki and T. Goh for their help in qRT-PCR; K. Fukumura and D. Cifuentes for their critical comments on the manuscript; Y. Takeda for cloning zebrafish PABPC1a and Paip2; R. Kusakabe for sharing miR1/206 MOs; S. Kanamura for fish maintenance; and members of our laboratory for their support. This work was supported by Grants-in-Aid for Scientific Research from Japan Ministry of Education, Culture, Sports, Science and Technology to Y.M (22115510) and K.I (20112003), the Uehara Memorial Foundation and the Senri Life Science Foundation to Y.M, and the Asahi Glass Foundation to K.I. A.F. is a research fellow of the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113350109/-/DCSupplemental.
References
- 1.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 4.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 5.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 9.Mishima Y, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 12.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 13.Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1142. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Till S, et al. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 16.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010;29:4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 26.Mishima Y, et al. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 28.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 29.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathonnet G, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 31.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 32.Walters RW, Bradrick SS, Gromeier M. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010;16:239–250. doi: 10.1261/rna.1795410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc Natl Acad Sci USA. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 36.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 37.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 39.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukaya T, Tomari Y. PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. EMBO J. 2011 doi: 10.1038/emboj.2011.426. 10.1038/emboj.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.