Abstract
An array of photoreceptors including cryptochromes, phototropin, and phytochromes regulates various light responses in plants. Among these photoreceptors, phytochromes perceive red and far-red light by switching between two interconvertible spectral forms (Pr and Pfr). The Pfr form promotes light responses partly by destabilizing negatively acting, phytochrome-interacting basic helix-loop-helix transcription factors (PIFs), thus modulating transcription in the nucleus. The Pfr form is also present in the cytosol. However, the role of phytochromes in the cytosol is not well understood. Here we show that the Pfr form interacts with the cytosolic protein PENTA1 (PNT1) and inhibits the translation of protochlorophyllide reductase (PORA) mRNA. PNT1 possesses five C3H-type zinc finger domains and displays similarity to various RNA binding proteins including Tristetraprolin, which regulates stabilities of mRNAs such as TNF-α mRNA in humans. Consistent with its function as an RNA binding protein, PNT1 directly binds to mRNA of a key chlorophyll biosynthetic gene, protochlorophyllide reductase in vivo and inhibits the translation of PORA mRNA in the presence of phytochromes. The present results demonstrate that phytochromes transmit light signals to regulate not only transcription in the nucleus through PIFs, but also translation in the cytosol through PNT1.
Keywords: light signaling, protein–protein interaction, post transcriptional regulation, translational regulation, nucleocytoplasmic shuttling
Phytochromes are dimeric proteins consisting of an apoprotein and a chromophore. They are synthesized in the cytosol in the Pr form, which can be converted into the Pfr form by red light and back into Pr by far-red light (1). Pfr translocates into the nucleus and inhibits a set of negatively acting phytochrome-interacting basic helix-loop-helix transcription factors (PIFs), partly by destabilizing them (2–7). Among PIFs, four PIFs (PIF1, PIF3, PIF4, and PIF5) play key negative roles to repress light responses in the dark. A pif quadruple mutant (pif1/pif3/pif4/pif5, collectively referred as quadruple mutants pifq) is therefore constitutively photomorphogenic and expresses many light responsive genes, even in the dark (8–10). However, dark-grown quadruple mutant seedlings still have longer hypocotyls than light-grown wild-type seedlings and the correlation coefficient of gene expression is only 0.7 between dark-grown pifq mutant seedlings and red light-grown wild-type seedlings (9, 10). This implies that phytochromes transmit light signals not only through these four PIFs, but also through other phytochrome-interacting proteins such as PIF7 and PHYTOCHROME KINASE SUBSTRATE 1 (PKS1) (8, 11, 12).
Ever since researchers discovered the nuclear translocation of Pfr and identified the nuclear-localized PIFs, the nucleus has been considered the focal site of phytochrome signaling (13). Mutant or transgenic phytochromes that are localized to the cytosol are unable to induce the majority of light responses, including hypocotyl elongation (14–16), supporting the notion that the phytochromes mainly function in the nucleus. The PIFs, which are transcription factors, also function in the nucleus. A genome-wide chromatin immunoprecipitation (ChIP-chip) study coupled with microarray analysis showed that one of the PIFs (PIF1) bound to 748 sites in vivo and regulated the expression of 166 target genes (105 positively and 61 negatively) in imbibed seeds (17). Among PIF1 binding sites, 59% were found to possess G-box elements (CACGTG). Other PIFs have also been shown to bind to individual G-box elements both in vitro and in vivo and to regulate the expression of target genes (18–25). Collectively, these previous results indicate that phytochromes regulate light responses through PIFs in the nucleus.
However, a few lines of evidence suggest that the cytosol should also be considered as a site of phytochrome action and processes occurring in the cytosol as a target of phytochrome signaling. First, cytoplasmic motility is accelerated by red light within a few seconds (26). Second, ion flux is also changed very rapidly by red light (27, 28). Third, hypocotyl negative gravitropism and red-enhanced phototropism are partly regulated by cytoplasmic phytochrome A (phyA) (29). Consistent with the evidence of cytoplasmic phytochrome signaling, phytochromes are present in the cytosol. The Pr form is abundantly present in the cytosol and the presence of the Pfr form of phytochrome is also present in the cytosol, as indicated by the sequestering of the Pfr of phyA in the cytosol and the slow accumulation of phytochrome B (phyB) nuclear speckles after red light irradiation (30–32). In addition, PKS1, which negatively regulates phytochrome signaling, was shown to interact with both phytochrome and phototropin 1 in the cytosol (11, 33). The molecular function of PKS1 has not been fully elucidated.
Translation is one of the cytosolic processes that could be regulated by phytochromes. To investigate this possibility, we sought to identify phytochrome-interacting proteins that might control translation in the cytosol. The translation of most eukaryotic mRNAs is initiated by attachment of the 43S preinitiation complex (comprising the 40S subunit, the eIF2 complex, eIF3, eIF1, and eIF1A) to the capped 5′ end region of the mRNA (34, 35). This attachment is facilitated by the eIF4F complex, which binds to the cap. Once the 43S complex is attached, it scans downward to find the initiation codon, where it is subsequently joined by the 60S subunit to form the elongation-competent 80S ribosome. In animals, translation is extensively controlled by different mechanisms, including protein modification of eIFs and the selection of specific mRNAs by RNA-binding proteins. Translational control also occurs in plants, where it plays important roles in various plant processes, including the responses to environmental stress, pathogens, and hormonal signaling. However, the role of phytochromes in translational control is not yet known (36, 37). Here, we report that the Pfr forms of phyB can interact with a cytosolic zinc finger protein that binds to the 5′-UTR of the protochlorophyllide reductase (PORA) mRNA to light-dependently inhibit translation of the PORA mRNA. Our results demonstrate that phytochromes can control gene expression not only through transcriptional regulation in the nucleus, but also through translational control in the cytosol.
Results
PENTA1 Is a Cytosolic Phytochrome-Binding Protein That Promotes Far-Red (FR) Block of Greening.
In the present study, a putative cytosolic phytochrome-interacting protein was identified through yeast two-hybrid screening and named PENTA1 (PNT1) on the basis of its structure, which includes five C3H-type zinc finger motifs (Fig. 1A). The five CX8CX5CX2H motifs are present in two clusters that consist of three N-terminal zinc finger motif clusters and two C-terminal zinc finger motif clusters. Proteins with multiple C3H-type zinc finger motifs are found in various eukaryotic proteins ranging from yeast and rice to human proteins, and some of which have been shown to be RNA binding proteins (38, 39). A database search indicated that Arabidopsis contains 10 proteins with similar multiple zinc finger motifs and rice contains 7 proteins. (Fig. S1). One of them, known as ENHANCER OF AG-4 1 (HUA1), was shown to bind an intron of AGAMOUS mRNA to regulate the splicing (40).
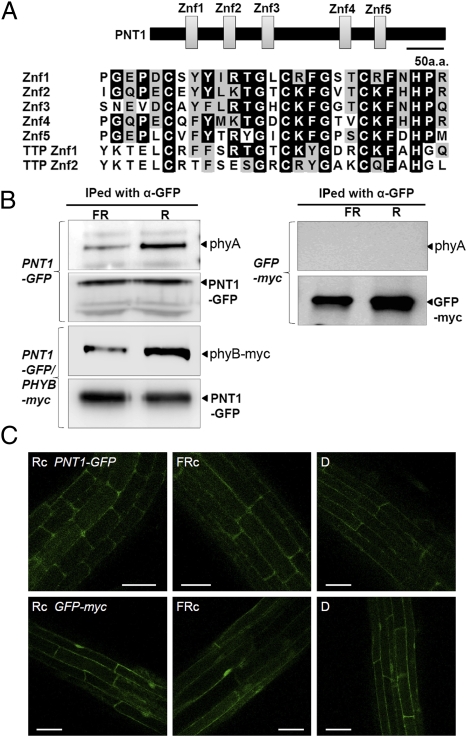
Fig. 1.
Phytochromes interact with PENTA1 (PNT1), a C3H-type zinc finger protein. (A) Diagram showing the five zinc finger motifs in PNT1 and their amino acid sequence alignment. The five zinc finger motifs are indicated by boxes. The amino acid sequences of the five zinc finger motifs (Znf1 to Znf5) are aligned with two zinc finger motifs (TTP Znf1 and TTP Znf2) found in human Tristetraprolin (NP_003398). (B) In vivo coimmunoprecipitation assay showing the preferential interaction between PNT1 and the Pfr form of phyA and phyB. PNT1-GFP or GFP were immunoprecipitated with an anti-GFP antibody. PhyA was detected with an anti-phyA antibody and phyB-myc was detected with anti-myc antibody. The precipitated PNT1-GFP or GFP were detected with an anti-GFP antibody. FR and R indicate plant samples irradiated with far-red (2.4 μmol/m2/s, 15 min) and red light (4.4 μmol/m2/s, 15 min), respectively, before immunoprecipitation. (C) Confocal microscopic images showing the cytosolic localization of PNT1-GFP irrespective of light conditions. D, dark; Rc, continuous red light; FRc, continuous far-red light. GFP alone was used as a control.
The interaction between PNT1 and phytochrome was further probed by in vivo coimmunoprecipitation assay using transgenic plants expressing either GFP-tagged PNT1 for phyA or both GFP-tagged PNT1 and myc-tagged phyB for phyB. PNT1-GFP was functional as its phenotype is similar to transgenic lines expressing PNT1 without any tag (PNT1-OX1). PhyB-myc was also functional (Fig. S2). PNT1-GFP was immunoprecipitated with anti-GFP antibody after either red or far-red light pulse and the precipitated phyA was detected by anti-phyA antibody, whereas the precipitated phyB-myc was detected by anti-myc antibody. The assay showed that PNT1 binds preferentially to the Pfr form of both phyA and phyB (Fig. 1B). A subcellular localization of PNT1 was determined by transgenic plants expressing PNT1-GFP. Similar to PKS1, PNT1 is localized in the cytosol rather than in the nucleus and red or far-red light did not change its subcellular localization (Fig. 1C). Taken together, these results indicate that PNT1 is a cytosolic C3H-type zinc finger protein that interacts preferentially with the Pfr form of both phyA and phyB.
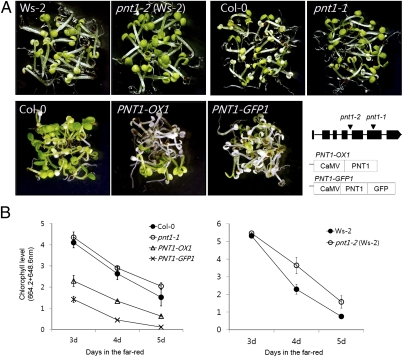
Mutation and overexpression of PNT1 alter a subset of phytochrome-mediated light responses. Among these light responses, FR block of greening was strongly enhanced in PNT1 overexpressing lines, whereas it was mildly suppressed in two pnt1 mutant alleles with different ecotype backgrounds (Col-0 for pnt1-1 and WS-2 for pnt1-2) (Fig. 2A). The FR block of greening response is caused by photobleaching when FR-grown seedlings are transferred to white light (41). The FR block of greening was quantified by measuring the amount of produced chlorophyll after transferring to white light. PNT1-OXs (PNT1-OX1, PNT1-GFP1) accumulated chlorophyll at approximately half the rate of wild type when FR-grown seedlings older than 3 d were transferred to white light, whereas the pnt1 mutants accumulated more chlorophyll than wild-type seedlings (Fig. 2B). A previous study showed that gibberellins (GA) promote photobleaching by repressing the expression of all POR mRNAs (42). GA promoted photobleaching both in wild type and the pnt1 mutant (Fig. S3), suggesting that they function independently. Unlike the FR block of greening, light-dependent hypocotyl elongations were not affected by PNT1 (Fig. S4).
Fig. 2.
PNT1 regulates a subset of phytochrome-mediated light responses. (A) Seedlings displaying more severe (PNT1-OXs) or less severe [pnt1 mutants: pnt1-1 (Col-0) and pnt1-2 (Ws-2)] photobleaching than wild-type plants (Col-0 and Ws-2) are shown. FR (2.4 μmol/m2/s)-grown seedlings were transferred to white light (100 μmol/m2/s) for 4 d. Genomic structure of PNT1 with two T-DNA insertion sites (inverted triangles) and transgenes for two PNT1-Oxs are shown. (B) Quantification of remaining chlorophylls after transferring FR-grown seedlings of various ages to white light for 4 d (SD, n = 3).
Phytochrome and PENTA1 Inhibit the Translation of PORA mRNA Light Dependently.
In previous studies, FR-grown seedlings were shown to photobleach when transferred to white light, partly because phyA represses the expression of PORA mRNA (41), which leads to an imbalance between chlorophyll intermediates and protochlorophyllide reductase (POR) under FR conditions. Irrespective of PNT1 mutation, all plants expressed similar levels of two key chlorophyll biosynthetic genes, HEMA and GUN5 (Fig. 3A). PORA mRNA levels were also either similar in the pnt1 mutant or slightly higher in the PNT1-OXs under far-red light conditions (Fig. 3A). These results suggest that PNT1 does not promote the FR block of greening through transcriptional repression of PORA gene or activation of other chlorophyll biosynthetic genes.
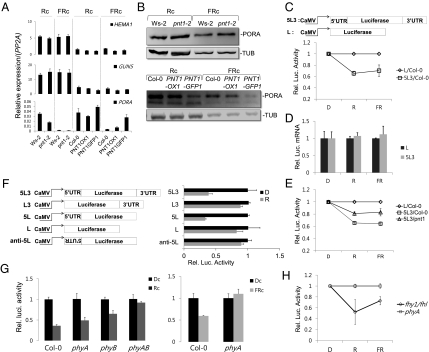
Fig. 3.
Phytochromes inhibit the translation of PORA mRNA through PNT1. (A) Expression levels of chlorophyll biosynthetic gene mRNAs in pnt1 and PNT1-OXs. Results are expressed as relative levels of PP2A mRNA. Four-day-old red light-grown (Rc) or FR-grown seedlings were used for the expression analysis. (B) Altered POR protein levels in pnt1 and PNT1-OX. Tubulin was used as a loading control. (C) Light-dependent decrease of luciferase activity in transgenic Arabidopsis harboring a luciferase reporter gene with 5′- and 3′-UTRs of PORA (5L3) in comparison with a luciferase gene without any UTRs (L). Upper diagrams describe the 5L3 and L constructs. Relative luciferase activities of L samples were taken as one for each light condition. (D) Similar levels of luciferase reporter mRNAs irrespective of UTRs in transgenic Arabidopsis. (E) Reduced light-dependent decrease of luciferase activity in the pnt1 mutant harboring a luciferase reporter gene with 5′- and 3′-UTRs of PORA (5L3). (F) Sufficiency of 5′-UTR of PORA for the translational inhibition of luciferase reporter gene in protoplasts. Various reporter constructs are shown. Relative luciferase activities of dark samples were taken as one for each construct. (G) Abolishment of light-dependent translational inhibition of the luciferase reporter with UTRs of PORA in protoplasts of phytochrome mutants. Relative luciferase activities of dark samples were taken as one for each phytochrome mutant. (H) Light-dependent decrease of luciferase activity in transgenic fhy1/fhl double mutant harboring a luciferase reporter gene with PORA UTRs. Relative luciferase activities of phyA mutant samples were taken as one for each light condition (SD, n = 3).
Instead, PNT1 was found to inhibit the translation of PORA mRNA in the light. Western blot analysis indicated that PORA protein levels were similar or slightly higher in the pnt1 mutant, but much lower in the PNT1-OXs than in the wild type, both under red and far-red light conditions (Fig. 3B). The decrease in PORA protein levels could be caused by the repression of PORA mRNA translation. The effect of PNT1 on the inhibition of the PORA mRNA translation was further investigated by constructing a luciferase reporter gene with or without the 5′- and 3′-UTRs of the PORA gene under the constitutive 35S promoter and generating transgenic Arabidopsis plants. Compared with transgenic plants harboring a luciferase reporter gene without UTRs, transgenic plants harboring the luciferase gene with PORA UTRs showed decreased luciferase activities both under red and far-red light conditions but not in the dark (Fig. 3C). This light-induced decrease of luciferase activity was not due to a decreased stability of luciferase mRNA with UTRs, as levels of luciferase mRNA were similar irrespective of the UTRs (Fig. 3D). The role of PNT1 was also investigated by generating transgenic pnt1 mutants harboring the reporter gene with PORA UTRs. Luciferase activities were decreased less by red and far-red light in the pnt1 mutant than in wild-type plants, indicating that PNT1 is partially responsible for the translational repression in red and far-red light (Fig. 3E). The partial release of the repression in the pnt1 mutant may be caused by the presence of additively acting PNT1 homologs (Fig. S1).
To further determine which UTR is necessary for the translational repression of PORA mRNA, constructs containing the luciferase reporter gene with the 5′-UTR alone or the 3′-UTR alone were generated and a transient expression assay using protoplasts was performed (Fig. 3F). In constructs containing no UTRs or only the 3′-UTR fused to the reporter gene, luciferase activities were not decreased by light. However, when the 5′-UTR or both the 5′-UTR and 3′-UTR were fused, luciferase activities were strongly decreased by light. The correct sequence of the 5′-UTR was required to repress the translation of the reporter mRNA, as insertion of the 5′-UTR in the reverse direction had no effect. These results suggest that PNT1 represses the translation of PORA mRNA through its 5′-UTR.
The role of phytochromes in light signaling-mediated inhibition of the translation of the reporter gene with PORA UTRs was investigated using a transient expression assay. Red light reduced luciferase activity only partially in the phyB mutant (Fig. 3G), suggesting that phyB is not the only phytochrome-mediating red light signaling. A phyA phyB double mutant did not show any inhibition by red light, suggesting that both phyA and phyB additively mediate red light signaling to inhibit the translation of PORA mRNA. Continuous far-red light also reduced luciferase activity in wild type but not in phyA mutants, supporting the role of phyA in mediating far-red light signaling. Consistent with the inhibitory role of phytochromes on the translation of PORA mRNA, the overexpression of phyB caused more severe photobleaching in a PNT1-dependent manner (Fig. S5). Taken together, the present results indicate that phytochromes mediate red and far-red light signaling to inhibit the translation of PORA mRNA in the cytosol.
To demonstrate that cytosolic phytochromes are capable of inhibiting the translation, we introduced the same luciferase reporter gene with the PORA UTRs into phyA mutant and fhy1/fhl double mutant to generate stable transgenic lines. Due to the lack of carrier proteins, phyA protein is localized in the cytosol of the fhy1/fhl double mutant (29, 43). Compared with the phyA mutant, luciferase activities were decreased in the fhy1/fhl double mutant both under red and far-red light conditions but not in the dark (Fig. 3H). Because the inhibition of translation under far-red light condition is solely caused by phyA (Fig. 3G), the results further support that cytosolic phyA can inhibit the translation.
PENTA1 Binds to 5′-UTR of PORA mRNA and Recruits Phytochrome to Inhibit the Translation.
PNT1 possesses a putative RNA binding domain, suggesting that PNT1 binds to PORA mRNA in vivo. RNA immunoprecipitation (RIP) analysis was performed using transgenic plants expressing PNT1-GFP and the luciferase reporter gene sandwiched between the 5′-UTR and 3′-UTR of the PORA gene. Immunoprecipitation of PNT1-GFP by an anti-GFP antibody precipitated high levels of endogenous PORA mRNA compared with levels of PP2A mRNA (Fig. 4A), indicating that PNT1 binds to PORA mRNA in vivo. To further investigate the light dependency of the association between PNT1 and PORA mRNA, transgenic plants harboring a luciferase reporter gene with the PORA UTRs were used. The RNA-IP assay showed that PNT1 precipitated high levels of the luciferase reporter mRNA irrespective of the light conditions, whereas it did not enrich the mRNA of the HPTII transgene (present in the same T-DNA with the luciferase reporter gene) (Fig. 4B). These results indicate that PNT1 is a phytochrome-interacting protein that binds to PORA mRNA through its UTR in vivo irrespective of light conditions.
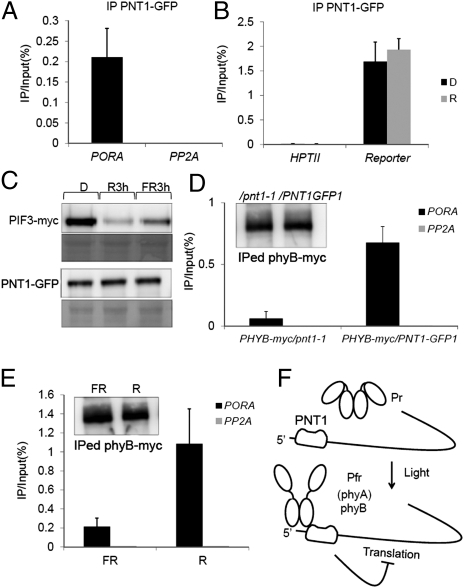
Fig. 4.
PNT1 binds to the UTR of PORA mRNA and recruits phytochromes to inhibit the translation of PORA mRNA. (A) Immunoprecipitation of PORA mRNA by PNT1 using RIP. PP2A mRNA was used as a nonbinding control. (B) Light-independent immunoprecipitation of luciferase reporter mRNA with UTRs of PORA but not HPTII mRNA in transgenic plants. (C) PNT1 was not degraded in response to light. Unlike PNT1, PIF3 is rapidly degraded by light. R3h and FR3h indicate 3 h after the transfer of etiolated seedlings to red and far-red light, respectively. (D) PNT1-dependent immunoprecipiation of PORA mRNA by phyB-myc. phyB-myc/PNT1-GFP1 and phyB-myc/pnt1 indicate transgenic plants expressing phyB-myc either in the PNT1-GFP1 line or in the pnt1-1 mutant. (Inset) Amount of precipitated phyB-myc in different plants. (E) Light-dependent recruitment of phyB to PORA mRNA. R and FR indicate samples treated with either red light or far-red light before precipitation. (Inset) Amount of precipitated phyB-myc under different light conditions. (F) Model showing the translational repression of PORA mRNA by phytochromes and their interacting protein PNT1 (SD, n = 3).
Phytochromes do not regulate the binding of PNT1 to its target RNA as evidenced by the light-independent binding of PNT1 to the reporter gene containing the UTRs of PORA. Phytochromes also do not regulate the protein stability of PNT1, which is in contrast with the degradation of PIF3 under light conditions (Fig. 4C). Instead, phyB was found to be recruited to PORA mRNA through PNT1. The RNA-IP of phyB-myc with an anti-myc antibody enriched the PORA mRNA fraction at much higher levels in PNT1-OX than in the pnt1 mutant (Fig. 4D). Consistent with the preferential binding of the Pfr form to PNT1, the binding of phyB-myc to PORA mRNA was increased by red light (Fig. 4E). Taken together, these results indicate that the Pfr form of phyB inhibits the translation of PORA mRNA after recruited to the 5′-UTR of PORA mRNA through PNT1.
Discussion
The present results uniquely show that phytochrome directly inhibits translation of mRNA in the cytosol. We show that PNT1, possessing multiple C3H-type zinc finger motifs, binds to PORA mRNA in vivo and recruits the Pfr form of phytochrome to the 5′-UTR of PORA mRNA. The light-dependent recruitment of phyB, and presumably phyA too, leads to the translational inhibition of PORA mRNA in the light (Fig. 4F). Our discovery indicates that both nuclear and cytosolic phytochrome signaling events work toward the same goal of reducing PORA protein levels: phytochromes inhibit the transcription of PORA through their effect on nuclear signaling events (10, 41, 44, 45), and they inhibit the translation of PORA mRNA through cytosolic signaling events. This concerted phytochrome-mediated inhibition of both transcription and translation is likely to rapidly reduce the level of PORA protein during the dark-light transition. Finally, it is noteworthy that Pfr is the functional form of phytochromes in both the nucleus and the cytosol.
Different explanations may account for how phytochromes and PNT1 inhibit the translation of the PORA mRNA. The recruitment of phytochromes to the 5′-UTR of the PORA mRNA may inhibit the activity of one or more of the eIFs required for loading of the 43S preinitiation complex onto the 5′ cap and the subsequent formation of the 48S initiation complex (34, 35). In mammals, the phosphorylation of various eIFs (e.g., eIF1, eIF2, eIF3, eIF4E, and eIF4G) by mammalian kinases can affect the activities of these eIFs. Because phytochromes were shown to have protein kinase activity in vitro (46), the recruited phytochrome may phosphorylate some of the eIFs, thereby inhibiting their activities. Alternatively, the phy-PNT1 complex on the 5′-UTR may inhibit the loading of the 43S preinitiation complex or other proteins onto the 5′-UTR by steric hindrance. A well-known example of this process is the interaction between the iron response element (IRE) in the 5′-UTR of ferritin mRNA and the iron response protein (IRP) (47). When the concentration of iron is low, IRP binds to the IRE and blocks loading of the 43S preinitiation complex onto the ferritin mRNA (48), thereby inhibiting translation of the ferritin mRNA. Analogous to IRP, the phy-PNT1 complex could inhibit translation by blocking the loading of the 43S preinitiation complex or other protein components. However, future work will be required to unravel the detailed molecular mechanism(s) of translational inhibition by PNT1 and phytochromes.
Our results further suggest that there may be additional cytosolic phytochrome-mediated signaling events. The translational regulation of the PORA mRNA does not seem to account for the classical cytoplasmic light responses (e.g., rapid changes in cytoplasmic motility and ion fluxes) (26–28), indicating that there is likely to be cytosolic phytochrome-mediated signaling other than the PNT1-mediated translational control described herein. In addition, although PNT1 was identified as a PORA-binding protein, PNT1 may also bind and regulate other mRNAs. PNT1 itself belongs to a small protein family that has 10 members in Arabidopsis and 7 members in rice (Fig. S1). It is somewhat analogous to the PIFs, which belong to a subgroup of the bHLH transcription factors having 15 members in Arabidopsis and at least 6 members in rice (49, 50). As many PIF family members play key roles in nuclear phytochrome signaling, it would be interesting to determine the potential roles of other PNT1 family members in cytosolic phytochrome signaling.
Methods
In Vivo Pull Down Assay.
In vivo pull down assay was performed with 4-d-old dark-grown seedlings. Total proteins were solubilized in extraction buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, 1× complete mini protease inhibitor, 100 μM MG132) and filtered through a QIAshredder. The filter through was divided into two and anti-GFP antibody was added. Each sample was irradiated with either red (4.4 μmol/m2/s) or far-red (2.4 μmol/m2/s) light for 15 min and incubated in the dark for 1 h with gentle rotation at 4 °C. Antibody-bound protein complexes were precipitated by protein A agarose. Beads were recovered and washed three times with 500 μL binding buffer in spin columns (Pierce). Proteins were eluted and analyzed by Western blot using anti-phyA (Agrisera) and anti-myc (Santa Cruz) antibody. All procedures were performed in the dark or under safety green light.
RIP.
All procedures were basically performed as described previously, which was reported in the mammalian cell (51). To optimize RIP in Arabidopsis we introduced minor changes. In brief, 2 g of long-day grown PNT1-GFP1 plants were dark adapted for 24 h before sampling and grinding in liquid nitrogen. Alternatively, 2 g of 4- to 6-d-old dark grown seedlings were used in the experiments. Total proteins were solubilized in the same volume of polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes pH 7.0, 0.5% Nonidet P-40, 1 mM DTT, 100 units ml−1 RNase inhibitor, 400 μM VRC, 1× complete mini protease inhibitor). In the case of phyB-myc RIP, we added 100 μM MG132 to prevent the degradation of phyB-myc during the procedure. The lysate was filtered through two layers of miracloth and diluted two-fold with NT2 buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM MgCl 2, 0.05% Nonidet P-40). A total of 15 μg of GFP antibody (Ab frontier) and 100 μL of protein A agarose/salmon sperm DNA slurry (Millipore) were added and incubated 4 h in 4 °C with gentle rotation. Beads were collected and washed thoroughly five to six times with NT2 buffer. RNAs were released by adding 10 μL proteinase K. DNase1 was treated to remove genomic DNA contamination. The RNAs were purified by TRIzol. Reverse transcription was performed with total eluted RNA, random hexamer and oligo dT. Transcript levels were detected by real-time PCR with specific primers for each gene. Amplified products were reconfirmed by gel electrophoresis. For the light treatment experiments, all procedures were performed in the dark or under safety green light.
Plant materials and other methods including a primer list (Table S1) can be found in SI Methods.
Supplementary Material
Acknowledgments
We thank Mathias Zeidler for fhy1/fhl double mutant seeds and Dr. Woo Yong Lee for confocal microscopic pictures. This work was supported in part by National Research Foundation of Korea Grants 2007-0056940, 2009-0077873, and ABC2011-0031339 and Rural Development Administration Grant SSAC-PJ008120 (to G.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109683109/-/DCSupplemental.
References
- 1.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer D, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park E, et al. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004;45:968–975. doi: 10.1093/pcp/pch125. [DOI] [PubMed] [Google Scholar]
- 4.Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- 5.Oh E, et al. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- 6.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 7.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 8.Leivar P, et al. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin J, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fankhauser C, et al. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- 12.Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- 13.Lorrain S, Genoud T, Fankhauser C. Let there be light in the nucleus! Curr Opin Plant Biol. 2006;9:509–514. doi: 10.1016/j.pbi.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita T, Mochizuki N, Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 15.Toledo-Ortiz G, et al. Subcellular sites of the signal transduction and degradation of phytochrome A. Plant Cell Physiol. 2010;51:1648–1660. doi: 10.1093/pcp/pcq121. [DOI] [PubMed] [Google Scholar]
- 16.Huq E, Al-Sady B, Quail PH. Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 2003;35:660–664. doi: 10.1046/j.1365-313x.2003.01836.x. [DOI] [PubMed] [Google Scholar]
- 17.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huq E, et al. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-García JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 20.Oh E, et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19:1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- 22.Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 25.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EOMB J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takagi S, Kong SG, Mineyuki Y, Furuya M. Regulation of actin-dependent cytoplasmic motility by type II phytochrome occurs within seconds in Vallisneria gigantea epidermal cells. Plant Cell. 2003;15:331–345. doi: 10.1105/tpc.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownlee C, Kendrick RE. Ion fluxes and phytochrome protons in mung bean hypocotyl segments: II. Fluxes of chloride, protons, and orthophosphate in apical and subhook segments. Plant Physiol. 1979;64:211–213. doi: 10.1104/pp.64.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brownlee C, Kendrick RE. Ion fluxes and phytochrome protons in mung bean hypocotyl segments: I. Fluxes of potassium. Plant Physiol. 1979;64:206–210. doi: 10.1104/pp.64.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rösler J, Klein I, Zeidler M. Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci USA. 2007;104:10737–10742. doi: 10.1073/pnas.0703855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie JM, Jr, Coleman RA, Briggs WR, Pratt LH. Reversible redistribution of phytochrome within the cell upon conversion to its physiologically active form. Proc Natl Acad Sci USA. 1975;72:799–803. doi: 10.1073/pnas.72.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kircher S, et al. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lariguet P, et al. PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc Natl Acad Sci USA. 2006;103:10134–10139. doi: 10.1073/pnas.0603799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robaglia C, Caranta C. Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 2006;11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi R, Bailey-Serres J. Regulation of translational initiation in plants. Curr Opin Plant Biol. 2002;5:460–465. doi: 10.1016/s1369-5266(02)00290-x. [DOI] [PubMed] [Google Scholar]
- 38.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE. 2008;3:e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Y, Kato N, Wang W, Li J, Chen X. Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell. 2003;4:53–66. doi: 10.1016/s1534-5807(02)00399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua NH. Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell. 1996;8:601–615. doi: 10.1105/tpc.8.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheminant S, et al. DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell. 2011;23:1849–1860. doi: 10.1105/tpc.111.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiltbrunner A, et al. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47:1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- 44.Hu W, Su YS, Lagarias JC. A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant. 2009;2:166–182. doi: 10.1093/mp/ssn086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leivar P, et al. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh KC, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 48.Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 49.Khanna R, et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura Y, Kato T, Yamashino T, Murakami M, Mizuno T. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci Biotechnol Biochem. 2007;71:1183–1191. doi: 10.1271/bbb.60643. [DOI] [PubMed] [Google Scholar]
- 51.Keene JD, et al. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.