Abstract
p97 is a key regulator of numerous cellular pathways and associates with ubiquitin-binding adaptors to remodel ubiquitin-modified substrate proteins. How adaptor binding to p97 is coordinated and how adaptors contribute to substrate remodeling is unclear. Here we present the 3D electron cryomicroscopy reconstructions of the major Ufd1-Npl4 adaptor in complex with p97. Our reconstructions show that p97-Ufd1-Npl4 is highly dynamic and that Ufd1-Npl4 assumes distinct positions relative to the p97 ring upon addition of nucleotide. Our results suggest a model for substrate remodeling by p97 and also explains how p97-Ufd1-Npl4 could form other complexes in a hierarchical model of p97-cofactor assembly.
Keywords: ATPase, electron microscopy, mechanism, structure, asymmetric complex
The protein p97 (also known as VCP and Cdc48 in yeast) is a ubiquitous type II AAA ATPase essential for cell viability (1). In complex with a large number of alternative adaptors the ATPase participates in numerous cellular activities including cell cycle regulation, membrane traffic, protein quality control, DNA metabolism, signaling, and apoptosis (reviewed in ref. 2). One of the best-studied adaptors is the heterodimer Ufd1-Npl4, the only essential cofactor in yeast (3–6), which directs p97 into cellular processes regulated by ubiquitin-proteasome degradation such as endoplasmic reticulum-associated degradation (ERAD) (7–10), mitotic progression (11, 12), replication (13), and transcription factor activation (14, 15). In ERAD (16), mitosis (17), and nucleus reformation (12), the p97-Ufd1-Npl4 complex has been shown to recognize ubiquitylated substrates associated with different cellular structures and, with the energy obtained from the binding and/or hydrolysis of ATP, to extract them into the cytosol to be recycled after deubiquitylation or degraded by the proteasome (10, 16, 18, 19). However, it remains unclear how the ubiquitin-binding domains in p97-Ufd1-Npl4 are positioned to recognize the substrate and how the conformational changes in p97 are translated into mechanic force acting on substrate molecules.
The monomer of p97 (∼87 kDa) consists of an N-terminal domain, two AAA domains, termed D1 and D2, and a highly flexible C-terminal region (20–22). The N domain interacts with adaptor proteins (23). The D1 domain is proposed to be responsible for oligomerization and the D2 domain for ATP hydrolysis, although both D1 and D2 are competent for binding and hydrolysis of ATP (24–26). Several studies have shown that p97 hexamerizes into a double ring barrel with a central pore and undergoes conformational changes during the hydrolysis cycle (21, 22, 27–30). The role of the pore in the recruitment and extraction of substrates is still contentious. It has been shown that substrates interact with residues in the D2 pore on the distal side of the ring (31, 32), and it has been suggested that they are not threaded down the central pore as proposed for other members of the AAA family (33–36).
Ufd1 (∼34 kDa) and Npl4 (∼67 kDa) form a heterodimer with a 1∶1 stoichiometry (3, 5). Ufd1 comprises an N-terminal domain, structurally similar to the N domain of p97 and contains binding sites for both poly- and mono-ubiquitin (37), and a flexible C-terminal domain. The structure of Npl4 is less well understood although two domains, the N-terminal ubiquitin-like (UBX-like) domain and the C-terminal NZF (zinc finger), have been solved by NMR (6, 25). The interaction between Ufd1-Npl4 and p97 has been described as bipartite with two different p97-interacting sites located at the C-terminal domain of Ufd1 and the UBX-like domain of Npl4 (4), and it has been proposed that these sites interact with two different p97 N domains (6). We have previously shown that one Ufd1-Npl4 heterodimer binds at the periphery of a p97 hexameric ring to form a complex of ∼640 kDa (5). However, little is known about the 3D arrangement of the p97-Ufd1-Npl4 complex and any of its functional conformations.
Here, using a variety of electron microscopy (EM) techniques we have visualized different conformational states of the p97-Ufd1-Npl4 complex that reveal the dynamic behavior of the complex. These conformational snapshots describe a wide range of positions of Ufd1-Npl4 relative to the p97 ring. Furthermore, the different conformations explain how a p97-Ufd1-Npl4 complex could associate with other p97 cofactors in a hierarchical model of p97-cofactor assembly (38).
Results
The 3D Reconstruction Places Ufd1-Npl4 on top of the p97 Ring.
Previous structural studies have noted the heterogeneous nature of samples containing p97 (reviewed in ref. 39). To visualize the different conformations in p97-Ufd1-Npl4 samples, we initially performed random conical tilt (RCT) experiments using negatively stained electron microscopy (40). During these preliminary studies we clearly observed dissociation of Ufd1-Npl4 from p97, even though it forms a stable complex as assessed by gel filtration (Fig. S1 A and B). In order to further stabilize the complex, we carried out cross-linking using glutaraldehyde in the absence of added nucleotide followed by the removal of unbound p97 by affinity chromatography (see Materials and Methods). Fractions containing cross-linked p97-Ufd1-Npl4 were imaged using negative-stain EM techniques. Approximately 1,000 images were collected and 20,000 particles were selected for analysis. Approximately 15,000 particles displayed the features and size expected for p97-Ufd1-Npl4. The remaining 5,000 particles were significantly smaller and did not correspond to the expected views of p97 or p97-Ufd1-Npl4. It became apparent that these particles were unbound Ufd1-Npl4, confirming dissociation of Ufd1-Npl4 from p97 (Fig. S2). The Ufd1-Npl4 and the p97-Ufd1-Npl4 particles were extracted separately and submitted to single-particle analysis independently.
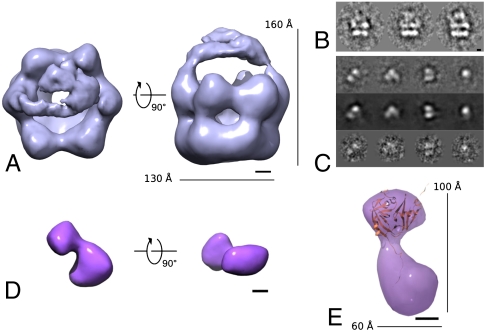
The negative stained dataset of cross-linked p97-Ufd1-Npl4 showed many projections of side-view orientation, where the two layers corresponding to the p97 D1 and D2 rings can be identified as bright parallel streaks (Fig. 1B). These side-view images clearly exhibited extra density on top of the p97 ring, suggesting the presence of bound Ufd1-Npl4. After submitting the particles to a protocol of single-particle analysis without symmetrization, a 3D reconstruction was obtained at ∼28 Å resolution as estimated by Fourier shell correlation (FSC) and 1/2-bit criteria (41) (Fig. 1A, Fig. S1C). The 3D reconstruction measures ∼160 Å by ∼130 Å and shows the hexameric features characteristic of the p97 ring. Additional density is observed above the central pore of the ring, consistent with the density observed in some of the RCT reconstructions of p97-Ufd1-Npl4 (Fig. S1B). The extra density has two contact points with the hexameric ring structure and is therefore likely to correspond to a single Ufd1-Npl4 heterodimer connected to two p97 protomers. These data suggest that in the absence of nucleotide the adaptor binds via two sites on top of the p97 ring.
Fig. 1.
Initial negative-stain 3D reconstructions of cross-linked p97-Ufd1-Npl4 and Ufd1-Npl4 incubated with glutaraldehyde. (A) 3D reconstruction of p97-Ufd1-Npl4 shows additional density placed on top of the ring spanning the central pore of p97 and connected to the ring by two densities. (B) Side-view class averages displaying additional density on top of the p97 double ring structure. (C) Class averages (Top), reprojections (Middle), and raw particles (Bottom) of the Ufd1-Npl4 structure. (D) The 3D reconstruction of cross-linked Ufd1-Npl4. The model displays a bilobed shape of two densities of different sizes. (E) Fitting of the N-terminal domain of Ufd1 [PDB accession code: 1ZC1 (37)] shows an overall agreement in size and shape between the atomic coordinates and the smaller of the two lobes of the EM map. (Scale bars: 20 Å.) (See also Fig. S1.)
The 3D Reconstruction of Ufd1-Npl4 Reveals a Bilobed Complex.
Selected particles that were thought to correspond to unbound Ufd1-Npl4 were also subjected to single-particle analysis. Such particles were easily detected in negative staining micrographs as a result of the distinct elongated shape of the particles (Fig. 1C). Random Euler angles were initially assigned to the class averages to avoid reference bias. After several rounds of refinement we were able to obtain an asymmetric 3D reconstruction of isolated Ufd1-Npl4 (Fig. 1D). The resolution was estimated to be ∼23 Å by FSC and 1/2-bit criteria (Fig. S1C). The reconstructed model, which measures ∼100 Å by ∼60 Å, exhibits two main lobes of different sizes arranged in a shallow V-shape. Fitting the atomic coordinates of the N-terminal domain of Ufd1 [PDB code: 1ZC1 (37)] shows an overall agreement in size and distinct shape with the smaller lobe with a cross correlation of 0.88 (Fig. 1E). It is notable that the assigned Ufd1-Npl4 density in the reconstruction of p97-Ufd1-Npl4 (Fig. 1A) also has two lobes consistent with the Ufd1-Npl4 reconstruction.
CryoEM Analysis Reveals Different Conformations for p97-Ufd1-Npl4 Complex.
To validate the negative-stain 3D reconstruction of the complex, and to obtain a higher-resolution 3D reconstruction, a cryoEM dataset was collected of p97-Ufd1-Npl4 in the absence of added nucleotide and processed independently (Fig. S3). Approximately 25,000 particles were selected from 100 images. The particles were pretreated and, to retain the characteristic features of p97, aligned to a model of p97 alone generated by back-projecting a top and a side-view extracted from a dataset of p97. Subsequently, class averages were generated from the aligned dataset. In light of the expected heterogeneity, 10 sixfold symmetric initial models were generated from randomly selected class averages corresponding to side-view orientations (see Materials and Methods). The models were subsequently refined without symmetry constraints over several rounds of alignment and projection matching (42), initially using the class averages and subsequently using the entire dataset. The particles distributed approximately in equal numbers (∼5,000 particles in each) among five distinct reconstructions, that were further refined without symmetry over 10 cycles at which point the refinement stabilized.
The final 3D reconstructions (Fig. 2) at resolutions estimated at ∼25 Å (Fig. S4D) exhibit the characteristic ring shape of p97. Three reconstructions (I, II, and III in Fig. 2) measure ∼173 Å in height and ∼140 Å in width and show additional density located above the p97 ring (Fig. S5). Following our negative-stain reconstruction, we attribute the additional density to be Ufd1-Npl4. This extra density generally adopts a bilobed shape and is connected to the p97 ring via density regions protruding from the p97 protomers. The two remaining reconstructions (IV and V in Fig. 2) constitute 40% of the data and do not show any additional density. We assign these reconstructions to p97 with no Ufd1-Npl4 bound, indicating dissociation of Ufd1-Npl4 from p97 as observed in the negative-stain data (Fig. S1B).
Fig. 2.
CryoEM reconstructions of p97-Ufd1-Npl4. Top views (Upper) and side views (Lower) of the five refined 3D reconstructions. All the reconstructions preserve the distinct ring-shape of p97. Among them, reconstructions I, II, and III also exhibit additional density located above the ring in agreement with the negative-stain 3D reconstruction of the complex. Therefore, the first three reconstructions correspond to p97-Ufd1-Npl4 while the last two to p97 alone. (Scale bar: 50 Å.) (See also Fig. S4.)
Correspondence of the p97-Ufd1-Npl4 Reconstructions with the p97 Structure.
To assess the correspondence of the p97-Ufd1-Npl4 reconstructions with the p97 crystal structure, we modeled the structure of p97 [PDB entry code: 1R7R (21)] into the EM density of the two best-defined models (Fig. 2, II and III). Due to the low resolution of these reconstructions, we were unable to carry out detailed fitting. Nonetheless, our fitting shows an overall agreement of size and shape of the p97 ring. However, similar to previous p97 cryoEM reconstructions (43), the D1 ring appears more open than in the p97 crystal structure. Furthermore, no extra density was observed on the side of the D1 ring to account for N domains as positioned in the crystal structure. However, we did observe additional densities protruding from the protomers on the top surface of D1 and we surmised that these densities represent the N domains in an extended conformation (Fig. 3). The position of the N-terminal domains is consistent with the position of Ufd1-Npl4 density, which have been shown to interact biochemically (6). Therefore, each of the N domains were manually fitted independently into the EM maps. In the 3D reconstruction II, two N domains were fitted into the connecting density interacting with the Ufd1-Npl4 heterodimer located on top of the D1 ring (Fig. 3A). In the 3D reconstruction III, we were able to fit only one N domain interacting with Ufd1-Npl4 (Fig. 3B). The fitted models suggest that 3D reconstructions II and III represent different conformational states of Ufd1-Npl4 with either two or one p97 N domain interactions respectively.
Fig. 3.
Mapping of the different sequence regions of p97-Ufd1-Npl4 and Ufd1-Npl4 onto the EM reconstructions. (A) Mapping of the different domains in the 3D reconstruction of p97-Ufd1-Npl4 in the conformation where Ufd1-Npl4 interacts with p97 via two binding sites. (B) Mapping of the different domains in the 3D reconstruction of p97-Ufd1-Npl4 in the conformation where Ufd1-Npl4 interacts with p97 via a single binding site. In both cases, the first and second images (top and side view, respectively) shows a fit of the crystal structure of p97 [cartoon representation, PDB entry code: 1R7R (21)] and of the negative-stain reconstruction obtained from cross-linked Ufd1-Npl4 (pink) into the 3D reconstruction of p97-Ufd1-Npl4. The third and fourth images (side view and top view, respectively) are colored by domain according to the sequence representation below. (Scale bar: 20 Å.)
p97-Ufd1-Npl4 Adopts Multiple Conformations upon Nucleotide Addition.
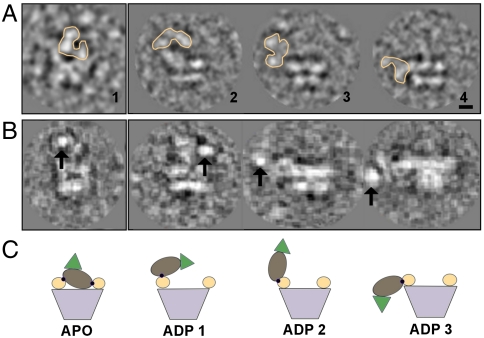
To observe nucleotide-dependent conformational changes, cryoEM datasets were collected for p97-Ufd1-Npl4 incubated with excess of ADP or ATPγS in the presence of Mg2+ (Figs. S6 and S7). Approximately 30,000 and 20,000 particles were collected, respectively. The particles were aligned to a model of p97 alone and those that aligned to references corresponding to side-view orientations were extracted (1,500 and 1,000 side-view particles in the “ADP” and “ATP” states respectively) and submitted to multivariate statistical analysis (MSA) (Fig. S4 A–C). In the case of the ADP dataset, side views with density above the ring and in lateral positions were observed (Fig. 4A), indicating a greater degree of heterogeneity in the presence of this nucleotide. Unlike in the absence of nucleotide, side views analyzed in the ADP and ATP datasets showed that only ∼20% and 10% of particles respectively displayed additional density connected to the p97 ring (Fig. S4). To confirm that the additional density corresponds to Ufd1-Npl4, we conjugated nanogold particles to the histidine-tag at the C terminus of Ufd1. Images showed bright density coincident with the extra density regions observed, but again at variable locations relative to the p97 hexamer (Fig. 4B, Fig. S8). Due to the increased heterogeneity, no 3D reconstructions were attempted. Without a 3D reconstruction it is not possible to definitively identify the different conformations present in the datasets. However, comparing the side views of class averages containing additional density with the reconstruction of p97-Ufd1-Npl4 without added nucleotide, we could observe a significantly wider range of locations of Ufd1-Npl4 relative to p97 (Fig. 4).
Fig. 4.
The multiconformations in the presence of nucleotide. (A) Raw images showing side-view orientations of p97-Ufd1-Npl4 in the absence of nucleotide (inset 1) and in the presence of ADP where Ufd1-Npl4 exhibits a great degree of flexibility (orange contour) (insets 2, 3, and 4). From, the 1,500 side views analyzed, 6% displayed density on the top (similar to conformation shown in APO 1), 9% on the side pointing upwards which equally distributed between conformation ADP 1 and ADP 2, and 5% on the side pointing downwards as shown in ADP 3. (B) Nanogold-labeled particles of the complex in the absence of nucleotide (first inset) and in the presence of ADP (Insets 2, 3, and 4). The nanogold (arrows) indicates the position of Ufd1. (C) Schematic representation of the conformations proposed for p97-Ufd1-Npl4. Black dots represent interaction sites between N domain and Npl4. The color scheme is the same as in Fig. 3 (Scale bar: 20 Å.)
Discussion
Different Conformations of Ufd1-Npl4 Bound to p97.
Ufd1-Npl4 has been shown to bind to p97 either via a single or double binding site (4). Only one binding site is understood in atomic detail: a surface loop on the hydrophobic side of the UBX-like domain of Npl4 (23). A bipartite mechanism of binding has been proposed that involves a secondary binding site in Ufd1 (4). It was speculated that the UBX-like domain in Npl4 is a general p97 binding domain and that the binding site in Ufd1 provides higher affinity and positions the adaptor in the correct orientation with respect to p97 for interaction with ubiquitylated substrates (12). Our reconstructions confirm that both binding modes can occur within the same population of p97-Ufd1-Npl4 complexes, suggesting a high degree of conformational flexibility. Our results also show that in the absence of added nucleotide, Ufd1-Npl4 preferentially binds to p97 on top of the D1 ring above the central pore similar to the adaptor p47 (43). NMR and biochemical experiments show that p47 and Ufd1-Npl4 bind to p97 in a mutually exclusive manner sharing a common binding site on the N domain (6). In the presence of ADP we observe a wider range of locations of Ufd1-Npl4 bound to p97 including locations at the side of the p97 ring similar to our previous 2D negative-stain studies (5).
The Central Pore of p97.
The central pore of p97 appears dilated relative to what is seen in the p97 crystal structures. At the limited resolution of our reconstructions, it is difficult to assign significance to this observation. EM reconstructions at low resolution tend to overestimate the dimensions of cavities, as previously observed for the central pore of p97 (39). The lack of density could be caused by flexible elements of a structure resulting in regions of weak density that are not visible at low resolution. Furthermore crystal structures tend to represent particularly compact conformations that are reinforced by crystal lattice contacts to allow well-ordered diffracting crystals.
Hierarchical Assembly of p97 Adaptor Complexes.
There has been much speculation as to how p97 can interact with so many different adaptor proteins that contain common p97 interaction domains (17, 38, 44). The emerging consensus is that p97 binds a small number of cofactors forming “core” complexes, one of which is Ufd1-Npl4. Under certain cellular conditions additional adaptors either recruit or bind to p97 core complexes (17). The p97-Ufd1-Npl4 structures presented here begin to explain how additional p97 adaptors could bind to a p97-Ufd1-Npl4 core complex. It has been shown (44) that many p97 adaptors comprising UBA-UBX domains bind to a p97-Ufd1-Npl4 complex consistent with previous studies (45). Our cryoEM structures now show that Ufd1-Npl4 binding to p97 would not obstruct the binding of other UBA-UBX adaptors. The adaptors p37 and p47 are exceptions to this paradigm, as they are also thought to form core p97 complexes (17), but exclude the subsequent binding of other adaptor proteins. One possible explanation for this difference could relate to the ability of these adaptors to homooligomerize. p47 binds to p97 as a homotrimer which would prevent other UBA-UBX adaptors from binding p97 due to steric hindrance (6).
Conformational Flexibility may Relate to p97-Ufd1-Npl4 Function.
The function of the p97-Ufd1-Npl4 complex is thought to involve the extraction of ubiquitylated proteins from large multiprotein complexes or from protein channels within the ER membrane. The mechanism of how p97-Ufd1-Npl4 operates is not understood but it involves ATP binding and/or hydrolysis (10). Our data in the presence of nucleotide suggest a large range of conformations of Ufd1-Npl4 in relation to the p97 ring. By inference, the N domain that binds Ufd1-Npl4 can move as well. A number of previous studies have suggested that N domains can adopt different conformations with respect to the D1 ring in a nucleotide-dependent fashion (21, 22, 27, 29, 30, 46) and proposed ways of how this conformational variability could be functionally relevant. Firstly, it has been suggested that conformational changes in the N domain propagate changes to the substrate by altering the adaptor binding arrangement (6). Secondly, refined p97 crystal structures (47) indicated that, upon nucleotide hydrolysis, the D2 domains undergo large conformational changes that are transmitted through the D1-D2 linker to displace N domains. Lastly, a “molecular latch” mechanism was proposed to explain how the locking of previously flexible N domains in the transition state of the hydrolysis cycle could exert the force necessary for protein dislocation (22, 27).
In our current EM studies, we observe that Ufd1-Npl4 is mainly located above the p97 D1 ring in the absence of nucleotide, suggesting that N domains are also located above the D1 ring. In contrast, in the presence of nucleotide, a wide range of locations is observed for Ufd1-Npl4, suggesting altered locations of the N domains. It has been previously shown that p97 alone can form direct interactions with multiubiquitin chains (48). It is therefore possible that the main function of Ufd1/Npl4 is to recruit substrates to p97 and that direct interactions between p97 and target proteins promote unfolding or disassembly through N domain movements upon nucleotide binding and/or hydrolysis similar to those observed here.
Materials and Methods
Protein Purification and Expression.
Mammalian p97 (mouse), Ufd1 (mouse), and Npl4 (rat) were expressed in Escherichia coli, Rosetta strain (DE3) transformed with the following full-length constructs: his6-p97 (21), Ufd1-his6 (3, 49), untagged Npl4 (3, 49), and untagged p97 (43). The complex was formed by colysing the pellets of untagged p97, Ufd1, and Npl4 by sonication in buffer A (25 mM Hepes, 500 mM KCl, 20 mM imidazole, 5% glycerol, pH 8.0). Lysates were clarified and loaded onto a HiTrap chelating column (GE Healthcare) precharged with Ni2+. Unbound proteins were eluted in a wash step with 10% buffer B (25 mM Hepes, 500 mM KCl, 500 mM imidazole, 5% glycerol, pH 8.0). Proteins were then eluted with an imidazole gradient (10% to 80% buffer B). Fractions were assessed by PAGE and further purified by gel filtration (Superdex 200 16/60 prep-grade or Superose 6 analytical grade; GE Healthcare) in buffer C (150 mM KCl, 25 mM Tris and 2.5 mM MgCl2, pH 8.0).
Cross-Linking.
Purified p97-Ufd1-Npl4 at a final concentration of ∼0.05 mg mL-1 was incubated with a final concentration of 0.1% EM-GRADE glutaraldehyde (Fluka) for 1 h at 37 °C in a Hepes buffer (150 mM KCl, 20 mM Hepes, 2.5 mM MgCl2, pH 8.0). The reaction was terminated by the addition of Tris (pH 8.0) to a final concentration of 25 mM. Unbound p97 was then removed by His-tag affinity chromatography, which retained p97-Ufd1-Npl4 thanks to the His-tag on Ufd1.
Nucleotide Incubation.
Incubation of the complex with 100 μM of ADP or ATPγS was carried out for 30 min on ice.
Nanogold Labeling.
Samples containing purified p97-Ufd1-Npl4 were incubated with Ni-NTA nanogold (Nanoprobes Inc.) at final concentration of (5 μM) so that would result in a 1∶1 ratio between the complex and the labelling particle. The incubation was carried out on ice for 30 min. Samples that required ADP or ATPγS, the nanogold labelling incubation was carried out right after the nucleotide incubation.
Electron Microscopy Data Collection and Image Processing.
Images were automatically collected using a Philips CM200 operated at 200 kV at 50 k magnification, using a 4k × 4k CCD camera, resulting in a pixel size of 1.765 Å. Images were collected at nominal defocus values between -1.5 μm and -3 μm. All images were processed using the IMAGIC software (50).
Random Conical Tilt.
Approximately 20 image pairs of negative staining p97-Ufd1-Npl4 were collected at 0° and 45°. The information beyond the first zero in the contrast transfer function (CTF) was discarded by low-pass filtering. Approximately 5,000 particle pairs were extracted from the images to create two datasets. The 0° dataset was pretreated, centered, and submitted to 10 rounds of alignment by classification. After the last round of alignment, the aligned dataset was classified into classes of ∼100 members. For each class, pretreated 45° tilted images were extracted and exact-filter back-projected to generate the 3D models. A similar procedure was applied to a sample containing p97 alone.
Image Processing and 3D Reconstruction of Negatively Stained Cross-Linked p97-Ufd1-Npl4.
Approximately 1,000 negative-stain CCD images were collected and coarsened by a factor of 2. Particles were picked using PICK-M-ALL using CCF cross-correlation with a disc as a reference and sorted by correlation coefficients. Particles corresponding to p97-Ufd1-Npl4 were extracted into boxes of 128 × 128 pixels and those corresponding to Ufd1-Npl4 were extracted into boxes of 80 × 80 pixels. Boxed particles were filtered using a low frequency cut-off of 100 Å and a high frequency cut-off of 5 Å, and normalized. Following pretreatment, particles were centered and submitted to MSA classification. In the case of p97-Ufd1-Npl4, the initial angular assignment was carried out by alignment and projection matching using a model of p97 alone built from a top and a side view. Angles were refined without symmetry constraints resulting in an initial asymmetric 3D reconstruction. In the case of Ufd1-Npl4, angles were randomly assigned to the raw particles and an initial reconstruction was built resembling a featureless sphere. Both initial 3D reconstructions were further refined over 10 rounds of alignment and projection matching of the individual particles.
Image Processing and 3D Reconstruction of p97-Ufd1-Npl4.
Purified complex was diluted to a concentration of 0.5 mg mL-1 and prepared for cryoEM. Approximately 500 CCD images were collected, CTF-corrected and coarsened by a factor of 2. The images were filtered, normalized, and submitted to the program PICK-M-ALL for particle picking using CCF correlation and a disc as a reference. Particles were cut into a box of 144 × 144 pixels and filtered using 150 Å and 10 Å as parameters. After pretreatment, particles were aligned against references from a model of p97 alone. Such reference model was created by back-projecting with sixfold symmetry a side and a top view extracted from a dataset of p97 alone with no added nucleotide. The aligned dataset was classified by MSA with 10 members per class. Ten random side-view averages were extracted and used to generate 10 initial models by manually assigning Euler angles (α = 0°, β = 90°, γ = 0°) and imposing C6 point-group symmetry during the reconstruction. These were reprojected every 10° in C1 to create the first set of references for angular assignment. The class averages were submitted to competitive projection matching with the set of references generated from the initial models. This procedure resulted in the initial separation of the class averages and the generation of 10 new 3D reconstructions. The new 3Ds were reprojected every 10° in C1 to create a new set of references for another round of projection matching. This procedure was iterated over the class averages first and the particles later until it stabilized.
Fitting and Visualization.
Atomic structures were fitted into the corresponding cryoEM maps using the program Chimera (51), which was also used for the production of molecular graphics images.
Supplementary Material
Acknowledgments.
We thank members of the Freemont/Zhang group and Centre for Biomolecular Electron Microscopy for helpful comments and support, Marin van Heel for advice on EM methods and Tillmann Pape and Raffaela Carzaniga for EM support. This work was supported by a grant to P.S.F. and X.Z. from the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The EM maps have been deposited in the EM data bank under accession codes EMD-2013 (negative stain p97-Ufd1-Npl4), EMD-2014 (negative stain Ufd1-Npl4), EMD-2015 (cryoEM p97-Ufd1-Npl4 II), EMD-2016 (cryoEM p97-Ufd1-Npl4 III).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114341109/-/DCSupplemental.
References
- 1.Peters JM, Walsh MJ, Franke WW. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J. 1990;9:1757–1767. doi: 10.1002/j.1460-2075.1990.tb08300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreveny I, et al. p97 and close encounters of every kind: a brief review. Biochem Soc Trans. 2004;32:715–720. doi: 10.1042/BST0320715. [DOI] [PubMed] [Google Scholar]
- 3.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruderer RM, Brasseur C, Meyer HH. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J Biol Chem. 2004;279:49609–49616. doi: 10.1074/jbc.M408695200. [DOI] [PubMed] [Google Scholar]
- 5.Pye VE, et al. Structural insights into the p97-Ufd1-Npl4 complex. Proc Natl Acad Sci USA. 2007;104:467–472. doi: 10.1073/pnas.0603408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaacson RL, et al. Detailed structural insights into the p97-Npl4-Ufd1 interface. J Biol Chem. 2007;282:21361–21369. doi: 10.1074/jbc.M610069200. [DOI] [PubMed] [Google Scholar]
- 7.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarosch E, Geiss-Friedlander R, Meusser B, Walter J, Sommer T. Protein dislocation from the endoplasmic reticulum ‘pulling out the suspect’. Traffic. 2002;3:530–536. doi: 10.1034/j.1600-0854.2002.30803.x. [DOI] [PubMed] [Google Scholar]
- 10.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 11.Hetzer M, et al. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 12.Ramadan K, et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- 13.Mouysset J, et al. Cell cycle progression requires the CDC-48UFD-1/NPL-4 complex for efficient DNA replication. Proc Natl Acad Sci USA. 2008;105:12879–12884. doi: 10.1073/pnas.0805944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoppe T, et al. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 15.Rape M, et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 16.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrynin G, et al. Cdc48/p97-Ufd1-Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells. J Cell Sci. 2011;124:1571–1580. doi: 10.1242/jcs.069500. [DOI] [PubMed] [Google Scholar]
- 18.Jentsch S, Rumpf S. Cdc48 (p97): a ‘molecular gearbox’ in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 21.Huyton T, et al. The crystal structure of murine p97/VCP at 3.6 A. J Struct Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 22.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 23.Yeung HO, et al. Insights into adaptor binding to the AAA protein p97. Biochem Soc Trans. 2008;36:62–67. doi: 10.1042/BST0360062. [DOI] [PubMed] [Google Scholar]
- 24.Song C, Wang Q, Li C-CH. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J Biol Chem. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, et al. Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J Biol Chem. 2003;278:20225–20234. doi: 10.1074/jbc.M300459200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briggs LC, et al. Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J Biol Chem. 2008;283:13745–13752. doi: 10.1074/jbc.M709632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouiller I, et al. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 28.Beuron F, et al. Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantized elastic deformational model. J Mol Biol. 2003;327:619–629. doi: 10.1016/s0022-2836(03)00178-5. [DOI] [PubMed] [Google Scholar]
- 29.Davies JM, Tsuruta H, May AP, Weis WI. Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-Ray scattering. Structure. 2005;13:183–195. doi: 10.1016/j.str.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 30.DeLaBarre B, Brunger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 31.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 32.Rothballer A, Tzvetkov N, Zwickl P. Mutations in p97/VCP induce unfolding activity. FEBS Lett. 2007;581:1197–1201. doi: 10.1016/j.febslet.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 33.Ogura T, et al. From the common molecular basis of the AAA protein to various energy-dependent and -independent activities of AAA proteins. Biochem Soc Trans. 2008;36:68–71. doi: 10.1042/BST0360068. [DOI] [PubMed] [Google Scholar]
- 34.Matsushita-Ishiodori Y, Yamanaka K, Hashimoto H, Esaki M, Ogura T. Conserved aromatic and basic amino acid residues in the pore region of Caenorhabditis elegans spastin play critical roles in microtubule severing. Genes Cells. 2009;14:925–940. doi: 10.1111/j.1365-2443.2009.01320.x. [DOI] [PubMed] [Google Scholar]
- 35.Wendler P, et al. Motor mechanism for protein threading through Hsp104. Mol Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13:995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Hänzelmann P, Buchberger A, Schindelin H. Hierarchical binding of cofactors to the AAA ATPase p97. Structure. 2011;19:833–843. doi: 10.1016/j.str.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Pye VE, et al. Going through the motions: the ATPase cycle of p97. J Struct Biol. 2006;156:12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Micro Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 41.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Harauz G, Ottensmeyer FP. Direct three-dimensional reconstruction for macromolecular complexes from electron-micrographs. Ultramicroscopy. 1984;12:309–320. [Google Scholar]
- 43.Beuron F, et al. Conformational changes in the AAA ATPase p97-p47 adaptor complex. EMBO J. 2006;25:1967–1976. doi: 10.1038/sj.emboj.7601055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexandru G, et al. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 46.Tang WK, et al. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- 49.Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.