Abstract
Polyploidy, or whole genome duplication, has played a major role in the evolution of many eukaryotic lineages. Although the prevalence of polyploidy in plants is well documented, the molecular and cytological consequences are understood largely from newly formed polyploids (neopolyploids) that have been grown experimentally. Classical cytological and molecular cytogenetic studies both have shown that experimental neoallopolyploids often have meiotic irregularities, producing chromosomally variable gametes and progeny; however, little is known about the extent or duration of chromosomal variation in natural neoallopolyploid populations. We report the results of a molecular cytogenetic study on natural populations of a neoallopolyploid, Tragopogon miscellus, which formed multiple times in the past 80 y. Using genomic and fluorescence in situ hybridization, we uncovered massive and repeated patterns of chromosomal variation in all populations. No population was fixed for a particular karyotype; 76% of the individuals showed intergenomic translocations, and 69% were aneuploid for one or more chromosomes. Importantly, 85% of plants exhibiting aneuploidy still had the expected chromosome number, mostly through reciprocal monosomy-trisomy of homeologous chromosomes (1:3 copies) or nullisomy-tetrasomy (0:4 copies). The extensive chromosomal variation still present after ca. 40 generations in this biennial species suggests that substantial and prolonged chromosomal instability might be common in natural populations after whole genome duplication. A protracted period of genome instability in neoallopolyploids may increase opportunities for alterations to genome structure, losses of coding and noncoding DNA, and changes in gene expression.
Keywords: compensated aneuploid, segmental allopolyploid
Polyploidy has played a major role in the evolution of many extant eukaryotic lineages (1). In plants, genomic and transcriptomic data associate some ancient polyploidy events with major radiations, including the emergence of seed plants and angiosperms (2) and some large clades within angiosperms (3). The phylogenetic positioning of these ancient polyploidy events suggests that they might have led to key phenotypic innovations or an increased tolerance to extreme environmental changes (1, 2, 4, 5). Within genera, polyploidy has coincided with an estimated 15% of angiosperm speciation events (6). However, over recent time scales, polyploids diversify more slowly than their diploid relatives and are more likely to go extinct, perhaps due in part to the unstable nature of neopolyploids (7).
Classical cytological studies have shown that many newly formed experimental autopolyploids and allopolyploids produce chromosomally variable gametes and progeny (8–19). One important consequence for allopolyploids is that they might not behave strictly as “constant species-hybrids” (20); that is, disomic inheritance at each parental locus may be upset, and genetic heterozygosity between the parental species may not remain fixed after whole genome duplication (18). In some cases, the regularity of meiosis was found to increase rapidly in experimental neoallopolyploids that were initially chromosomally unstable (21, 22); for example, after just five selfed generations, Nicotiana neoallotetraploids displayed bivalent pairing and >99% stainable pollen (22). Very little is known about the extent or duration of chromosomal variability in natural neoallopolyploid populations. Standard chromosome counts can identify numerical aneuploidy, in which there is a change in the total chromosome number; however, aneuploidy is much harder to detect in a polyploid in which chromosome numbers vary within parental subgenomes but the total chromosome number remains unchanged. Such chromosome substitutions have been shown for a subset of chromosomes in a few experimental neoallopolyploids (21, 22). Only one recent molecular cytogenetic study, on the synthetic allotetraploid Brassica napus, was able to detect aneuploidy across all chromosomes, including where it resulted in substitutions, in the early generations postpolyploidization (23). Standard meiotic (24) and mitotic karyotypic analyses (25), as well as the distribution of centromeric and telomeric markers (26), have suggested that the recently formed allopolyploids Tragopogon miscellus and Tragopogon mirus are chromosomally stable. However, preliminary work using genomic and fluorescent in situ hybridization (GISH and FISH, respectively) revealed aneuploidy and translocations in a few individuals of both allotetraploids (27).
Here we present the results of an in-depth molecular cytogenetic survey of a naturally occurring neoallotetraploid, T. miscellus. This species is ∼80 y old (ca. 40 generations for this biennial), having formed repeatedly in North America after the introduction of its diploid (2n = 12) progenitors, Tragopogon dubius and Tragopogon pratensis, from Europe in the early 1900s (24, 28). As noted earlier, preliminary GISH/FISH analyses revealed deviations from the expected additive karyotype in two of the three T. miscellus individuals analyzed (27), but given this small sample size, the extent of this phenomenon in nature remained unclear. Thus, an important aim of the present study was to examine variation within and between populations across a greater part of its range. Using updated in situ methodology, we obtained complete karyotypes of 68 individuals grown from field-collected seed from six natural populations (Fig. 1), an additional sample comprising a set of 10 siblings (Materials and Methods), and eight field-collected plants.
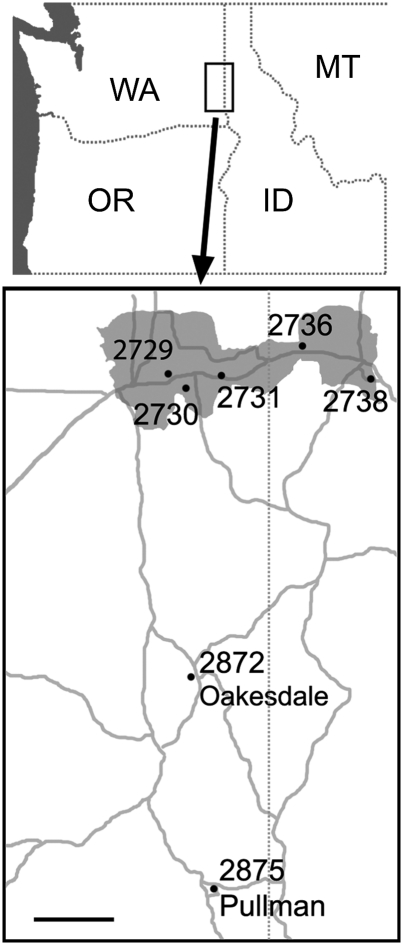
Fig. 1.
Maps showing T. miscellus collection sites in relation to the northwestern United States. (Upper) The Pacific Ocean (gray) and states of Washington (WA), Oregon (OR), Idaho (ID), and Montana (MT) are indicated; a rectangle shows the area in the enlarged map. (Lower) Locations of collection sites, with major roads shown in gray. The greater Spokane area (shaded gray) includes the city of Spokane (collections 2729 and 2730), and the towns of Veradale (2731), Post Falls (2736) and Coeur d'Alene (2738). Two additional collection sites are indicated: Oakesdale (2872) and Pullman (2785/2875-B). The Washington–Idaho state line is indicated by the dotted line. (Scale bar: 10 mi.)
Results and Discussion
GISH and FISH Karyotyping of T. miscellus.
GISH karyotypes readily distinguished chromosomes derived from the diploid progenitors, T. pratensis and T. dubius (referred to as P- and D-subgenomes, respectively), which together constitute the T. miscellus genome (Fig. 2). GISH signals at several positions, including some known to comprise repetitive DNA (Fig. 2), permitted identification of all chromosomes within each subgenome (Fig. 2). The effectiveness of GISH in discriminating DNA of the progenitors was confirmed using a pair of dispersed repeats, each of which is highly represented in only one diploid parent (Fig. 3). These repeats and additional FISH probes confirmed chromosome designations based on GISH.
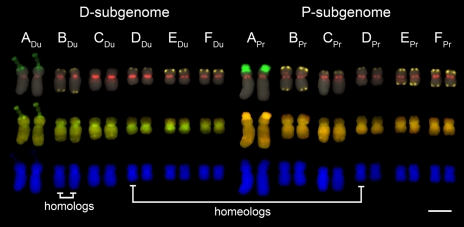
Fig. 2.
Mitotic karyotype of a T. miscellus plant showing an additive chromosome complement. Metaphase chromosomes (from plant 2875–1-1) were first subjected to FISH (top row) using probes for 35S rDNA (green), a centromeric repeat (TPRMBO; red), and a subtelomeric repeat (TGP7; yellow). The same spread was then reprobed with total genomic DNA (GISH; middle row) of T. dubius (green) and T. pratensis (red); chromosomes were counterstained with DAPI (gray). The lower row shows the same chromosomes with only DAPI staining (blue). Each chromosome is present in two copies (disomic). Examples of chromosomes that are homologs and homeologs are indicated. (Scale bar: 5 μm.)
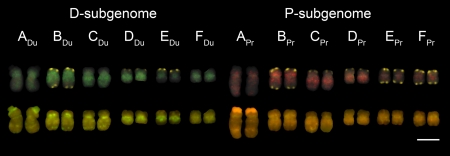
Fig. 3.
Mitotic karyotype of an additive T. miscellus plant probed with dispersed repetitive DNA. Metaphase chromosomes (from plant 2875-B-5) were first subjected to FISH (upper row) using a mixture of two probes for DNA repeats abundant in only one of the diploid parental genomes, T. pratensis (pra001; red) and T. dubius (dub005; green). In addition, a subtelomeric repeat (TGP7; yellow) present in both subgenomes was included, and DNA was counterstained with DAPI (gray; visible where the probe signal is less intense). The same chromosome spread was then reprobed with total genomic DNA (lower row) of T. dubius (green) and T. pratensis (red). (Scale bar: 5 μm.)
Extensive Karyotypic Variation Is Present in All Analyzed Populations.
From the six populations studied (Spokane-2, Veradale, Post Falls, Coeur d'Alene, Oakesdale, and Pullman), only 31% of T. miscellus plants (n = 18) had additive (euploid) karyotypes (Fig. 4 and Figs. S1–S5), which contain the expected two copies of each parental chromosome (e.g., Fig. 2). The remaining 69% of plants had one or more aneuploid chromosomes (which were not present in two copies); 10% were either 2n = 23 (n = 2) or 2n = 25 (n = 4), and 59% were 2n = 24 (n = 34). The sample of 10 sibling plants from Spokane-1 was also highly variable (Fig. S6 and Table S1) and showed a similar frequency of euploids (n = 2) and aneuploids that were either 2n = 25 (n = 1) or 2n = 24 (n = 7) (Table 1). Such extensive variation in chromosomal copy number is unprecedented within natural populations, but has been reported for synthetic allotetraploid B. napus (25) and, to a lesser extent, for synthetic allohexaploid wheat (29).
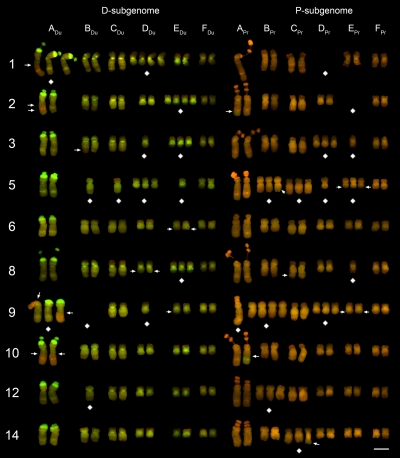
Fig. 4.
Mitotic karyotypes of 10 T. miscellus individuals from Oakesdale, WA. GISH was carried out with total genomic DNA probes of T. dubius (green) and T. pratensis (red). Arrows indicate the positions of translocation breakpoints. Diamond symbols are below aneuploid chromosomes (i.e., those that are not disomic). (Scale bar: 5 μm.)
Table 1.
Summary of T. miscellus karyotypes
| Population | Euploid | Compensated aneuploid | Numerical aneuploid |
| Spokane-1 (sibs) | 2 | 7 | 1 |
| Spokane-2 | 5 | 5 | 0 |
| Veradale | 2 | 5 | 2 |
| Post Falls | 1 | 6 | 2 |
| Coeur d'Alene | 4 | 6 | 0 |
| Oakesdale | 2 | 7 | 1 |
| Pullman | 4 | 5 | 1 |
| Pullman-B* | 6 | 2 | 0 |
*Pullman-B is a sample of eight adult plants collected from Pullman, WA.
Aneuploidy Frequently Results in Chromosome Substitutions.
Plants from all seven natural populations of T. miscellus were pooled for analyses of aneuploidy. Of the 48 aneuploid plants analyzed, 41 (34 from the six sampled natural populations plus 7 of the 10 sibling plants from Spokane-1) represent “compensated aneuploids” (22) because they had the “euploid” number, 2n = 24 (Fig. 4). In these plants, the numbers of chromosomes deviating above and below two copies are equal, giving 24 chromosomes. This is the first report of such extensive compensated aneuploidy in nature, although it has been found in experimental neoallopolyploids (23, 29), and such chromosome substitutions have been used extensively in cereal breeding (30–32). Compensated aneuploidy in natural populations of T. miscellus provides a powerful mechanism influencing the inheritance and fixation of alleles in early allopolyploid evolution.
For all but one of the 41 compensated aneuploid plants, aneuploidy was reciprocal between putative homeologs (e.g., ADu and APr), occurring as either monosomy (one copy) for one homeolog and trisomy (three copies) for the other homeolog, or, less often, nullisomy (zero copies) and tetrasomy (four copies) (Fig. 4). Thus, the sum of each homeologous chromosome group (A–F) was consistently four in 40 of the 41 plants. The single 2n = 24 plant that did not follow this pattern (Coeur d'Alene plant 2738–2-1) was monosomic for the E chromosome of T. dubius origin (EDu) and trisomic for the F chromosome of T. pratensis origin (FPr) (Table 1). This resulted in a total of three group E chromosomes and five group F chromosomes.
Of the 40 plants of T. miscellus with 2n = 24 and aneuploidy between homeologous chromosomes, 29 had only one case of mono-trisomy or nulli-tetrasomy (i.e., a single monosomic or disomic substitution, respectively). The remaining plants showed multiple substitutions involving two (n = 9), three (n = 1), or four chromosome groups (n = 1) (Fig. 4), but in all cases the total number of chromosome copies was four for each of the six groups.
Aneuploidy is expected to result in lowered metabolic efficiency due to disturbances in the normal protein stoichiometry for affected dosage-sensitive genes (33–36). One possible explanation for the repeated occurrence of homeologous substitutions is that the tetrasomic dosage might be maintained for genes present on both homeologs (37). This inference of homeology in T. miscellus is underscored for chromosome groups B–E, which were found in nulli-tetrasomic combinations in T. miscellus. In other allopolyploids, substantial synteny is required for compensation of complete chromosome loss (nullisomy) by chromosomes from another progenitor or related species (23, 32, 38–40). The data that we report suggest that chromosomes A–F of T. dubius and T. pratensis origin correspond to six groups of largely homologous (homeologous) chromosomes. The compensatory pattern may result from mispairing at meiosis (e.g., in multivalents), leading to the missegregation of homeologous chromosomes (41–43).
Aneuploidy Does Not Show a Consistent Parental Bias in T. miscellus.
A combined analysis of the aneuploidy data from the six natural populations did not demonstrate a significant parental bias in terms of either chromosome gains (trisomy or tetrasomy; P = 0.50, one-sample proportions test) or chromosome losses (monosomy or nullisomy; P = 0.20, one-sample proportions test) (Fig. 5). Opposing trends were apparent among populations, however. Only plants from Coeur d'Alene showed a trend toward a gain of P-subgenome chromosomes and a loss of D-subgenome chromosomes. This population was also unusual in that only E and F chromosomes were aneuploid (Table S1 and Fig. S4). When the other five populations (Spokane-2, Veradale, Post Falls, Oakesdale, and Pullman) were analyzed together, D-subgenome chromosomes were gained more often (P = 0.05, one-sample proportions test), and P-subgenome chromosomes were lost more often (P = 0.01, one-sample proportions test). This bias appears irrespective of parentage; plants in Pullman have T. dubius as the maternal parent, whereas all others have T. pratensis as the maternal parent (44).
Fig. 5.
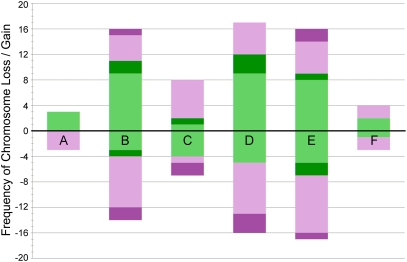
Stacked bar chart showing the number of chromosome losses and gains from GISH karyotypes of seed-grown plants. On the y-axis are the numbers of aneuploid chromosomes observed in the 48 plants grown from seed, which were not chromosomally additive of the parents. Each bar on the x-axis represents one of the six homeologous chromosome groups, A–F, of T. pratensis (magenta) and T. dubius origin (green). Cases of chromosome loss (either monosomy or nullisomy) and gain (either trisomy or tetrasomy) are shown below and above the origin, respectively. The severity of the aneuploidy is indicated by color intensity, with monosomy or trisomy shown by lighter colors and nullisomy or tetrasomy indicated by darker colors.
The bias against chromosomes of T. pratensis origin contrasts with genomic investigations using assays based on single nucleotide polymorphisms, which have shown that gene copies of T. dubius origin are lost more often than those of T. pratensis origin (44–48). Thus, chromosome gains and losses alone cannot explain the gene copy number bias, pointing to other mechanisms. Small-scale nonreciprocal exchanges or deletions, arising via translocations and/or homeologous recombination as observed in other allopolyploids (49–51), also may be occurring in T. miscellus.
Intergenomic Translocations Occur Predominantly Between Homeologous Chromosomes.
Rearrangements between subgenomes were common in T. miscellus, detected on at least one chromosome in 76% of the individuals (44 of 58), not including the 10 sibling progeny from Spokane-1. The group A chromosomes showed the highest frequency of apparent homeologous exchanges with at least five different translocation breakpoints in individuals from six populations (Table S1). Similarly, translocations on the long arms of BDu and BPr were observed in plants from all seven populations (Table S1), suggesting that this region might have a propensity toward intergenomic recombination.
Despite the extensive karyotypic variation seen overall, some populations exhibited distinctive karyotypic signatures. Eight of the 10 plants from Spokane-2 shared what appear to be the same homozygous reciprocal translocations on ADu/APr (Fig. S1). Only individuals from Post Falls had an apparent nonreciprocal translocation on the short arm of DDu (Fig. S3). On careful visual inspection of intergenomic translocations, all appeared to be either reciprocal or nonreciprocal exchanges between homeologous chromosomes.
T. miscellus Populations Represent Independent Origins.
Microsatellite data obtained for populations sampled in this study, along with previous studies (28, 52), indicate that each of the populations sampled represents an independent polyploid origin (Fig. S7). Microsatellite data suggest that plants of separate origin co-occur in Spokane-2 and Post Falls. In a few cases, genotypic and karyotypic data suggest that crossing might have occurred between plants of different polyploid origins (e.g., Spokane-2: 2730–8-3 and 2730–10-1; Figs. S1 and S7), but this appears to be very limited.
Plants Showing Aneuploidy and Rearrangements Persist in Natural Populations.
The chromosomal variation described above was present in plants derived from field-collected seed; thus, the extent to which this type of variation persists among adults growing in natural populations is unknown. To address this issue, individual rosettes were collected from Pullman, WA. Of eight individuals analyzed, two were compensated aneuploids, showing a single case of monosomy-trisomy between homeologous chromosomes, and six were euploid (Fig. S8); five of the plants also showed translocations. Thus, both aneuploidy and translocations were also observed in plants from natural populations. Of note, two of these eight plants appeared to be fully additive of the progenitor genomes, showing neither aneuploidy nor translocations, whereas only three fully additive plants were found among the 68 plants grown from seed. This finding suggests that in natural populations there may be selection against karyotypes with more extreme aneuploidy and rearrangements; this hypothesis merits further testing.
T. miscellus Remains Chromosomally Variable.
Unlike the diploid progenitors of T. miscellus, T. dubius and T. pratensis, both of which have stable karyotypes (24, 25), none of the ca. 40-generation-old populations of T. miscellus appeared to be chromosomally uniform. No T. miscellus population was fixed for a single karyotype, and few plants had a karyotype that was completely additive of the two parents. Furthermore, the number of aneuploid sibling progeny generated by one plant (the Spokane-1 maternal plant) was comparable to the percentage of aneuploids observed across all of the populations examined here.
The extensive aneuploidy (69%) observed across T. miscellus populations is similar to what has been reported for other synthetic or spontaneous neoallopolyploids, most of which are of younger age than T. miscellus. FISH revealed the presence of a similar percentage of aneuploids (71%) in the fifth generation (S5) of synthetic B. napus; this value continued to rise, reaching 95% by the S10 generation (23). Classical cytological studies revealed that in the first generation of a spontaneous Crepis allotetraploid (21), 78% of the plants were aneuploid (comprising 49% numerical aneuploids and 29% compensated aneuploids) (21), and in the first generation of an induced Cyrtanthus allotetraploid (22), 61% of the plants were aneuploid (comprising 57% numerical and 4% compensated aneuploids). Only a few chromosomal substitutions in Crepis and Cyrtanthus neoallopolyploids were detectable, because not all parental chromosomes were morphologically distinct; thus, these values are likely underestimates (18). Synthetic S2 bread wheat (Triticum aestivum) lines exhibited 0–50% aneuploidy, with variation attributable to progenitor background (29). Instability in wheat neoallopolyploids probably could be increased further in the absence of Ph1, which prevents homeologous pairing (29, 53–55).
Selection for increased fertility should stabilize the genome and reduce the extent of aneuploidy over time (56). However, if a compensated aneuploid plant has little reduction in fitness (i.e., homeologous chromosomes substitute for each other), and if the production of aneuploids is ongoing, then the interplay between generation of and selection against aneuploids is unclear and difficult to predict. Likewise, if new chromosomal combinations arise that are selectively advantageous, then compensated aneuploidy may be maintained. Therefore, chromosomal variation, such as that observed in T. miscellus, perhaps might represent additional variation rather than instability.
Segmental Allopolyploid Behavior.
Stebbins (57) coined the term “segmental allopolyploid” to describe polyploids that do not exhibit strict bivalent formation across all chromosomes or disomic inheritance at all loci. At a time when polyploids were classified as autopolyploids or allopolyploids on the basis of chromosome pairing behavior (quadrivalent vs. bivalent formation, respectively), Stebbins considered the parents of segmental allopolyploids to occupy an intermediate level of chromosomal divergence between those of autopolyploids and allopolyploids (57). He proposed that residual homology between homeologous chromosomes is primarily responsible for inconsistent bivalent formation and nondisomic inheritance. Meiotic irregularities may render segmental allopolyploids unstable (58, 59), with increasingly rearranged karyotypes and shifts to polysomic inheritance (59). Stebbins's definition was based on a concept of structural homology between progenitor chromosomes, but its application was based on patterns of inheritance and/or chromosome behavior, with an underlying assumption of additivity of the parental genomes. However, the same pattern might arise through compensated aneuploidy with pairing between homologs that are, for example, trisomic or tetrasomic. Based on the extensive compensated aneuploidy observed in T. miscellus, it is likely that deviations from strict bivalent pairing and disomic inheritance in other segmental allopolyploids may also reflect postallopolyploidization processes rather than partial chromosomal homology of the parental genomes.
Consequences for Establishment and Evolution of Young Allopolyploids.
Classical cytological studies of synthetic and spontaneous neoallopolyploids indicate that substantial chromosomal instability often follows whole genome duplication. Recent molecular cytogenetic studies of synthetic B. napus (25) and data provided here for the natural allotetraploid T. miscellus also show cytological variability, but reveal that much of the variation involves chromosome substitutions and rearrangements between homeologous regions. These types of changes would have been difficult or impossible to detect using classical approaches. The consequences of such chromosomal instability may be considerable, possibly including gene loss and alterations in gene expression, both of which have been detected in natural populations of T. miscellus (44–47) and in synthetic lines of B. napus (49, 60–62). Even allohexaploid wheat (T. aestivum), which is ca. 10,000 y old (63), still exhibits some chromosomal instability; aneuploids compose ∼1% of intervarietal populations (64) and ∼2–3% of cultivated lines (29).
Chromosomal variation, such as nonreciprocal intergenomic translocations and compensating aneuploidy, will yield segregating genetic variation. Given that all individuals resulting from a single polyploidization event are genetically highly similar or potentially even genetically uniform (following a model of spontaneous hybrid doubling), these chromosomal mechanisms would supply genetic variation in advance of point mutations in the new allopolyploid. Available data suggest that natural populations of neoallopolyploids may undergo a prolonged period of aneuploidy and rearrangements before genomic stabilization, increasing opportunities for alterations in genome structure, losses of coding and noncoding DNA, changes in gene expression, segregating genetic variation and the possibility of genetic and phenotypic novelty.
Materials and Methods
Seed Collections.
Seeds were collected from 60 plants from six populations, plus 10 sibling progeny (sharing at least the maternal parent) collected from a single plant from a seventh population (Spokane; 2729). Details are provided in SI Materials and Methods. Vouchers for each of the populations were deposited at the University of Florida Herbarium.
Plant Collections from the Field.
A sample of 22 plants was collected from Pullman, WA on April 14, 2011, as plants emerged in the spring (just before bolting). These plants had overwintered as basal rosettes. They were shipped to the University of Florida, planted, and grown in the greenhouse, after which roots were obtained for analysis.
Progenitor DNA Repeat Identification/Isolation.
Repetitive sequences were identified from genomic 454 sequences using an approach similar to that described previously (65). Details are provided in SI Materials and Methods.
Chromosome Preparation.
The final 2 cm of growing roots were harvested and pretreated in an aqueous solution of 2 mM 8-hydroxyquinoline (Sigma-Aldrich) for 4.5 h at 4 °C. Metaphase chromosome spreads were prepared as described previously (66). Details are provided in SI Materials and Methods.
GISH and FISH.
Probes of genomic DNA for GISH and repetitive DNA for FISH were fluorescently labeled and then applied to chromosome spreads as described by Birchler et al. (66), with a few minor modifications. Details are provided in SI Materials and Methods.
Nuclear Microsatellite Analysis.
Twelve microsatellite loci for plants from all seven sites were amplified and analyzed as described previously (52) and detailed in SI Materials and Methods.
Statistical Analysis.
One-sample proportion tests were conducted with R version 2.13.0 to identify any significant deviation from a null expectation of equality between subgenomes for chromosome copy number increases (either three or four copies) and for decreases (either zero copies or one copy).
Supplementary Material
Acknowledgments
We thank Chuck Cody (Washington State University) for collecting adult T. miscellus plants, Dr. Robert Harris for assisting in the laboratory, Dr. Patrice Albert for advising on chromosome preparation and in situ hybridization methodology, and Dr. Richard Buggs for commenting on the manuscript. This work was supported by National Science Foundation Grant DEB-0922003. Publication of this article was funded in part by the University of Florida Open-Access Publishing Fund.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN227618 and JN227619).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112041109/-/DCSupplemental.
References
- 1.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 2.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 3.Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 4.De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood TE, et al. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayrose I, et al. Recently formed polyploid plants diversify at lower rates. Science. 2011;333:1257–1257. doi: 10.1126/science.1207205. [DOI] [PubMed] [Google Scholar]
- 8.Poole CF. Constant species hybrids. Am Nat. 1933;67:188–190. [Google Scholar]
- 9.Newton WCF, Pellew C. Primula kewensis and its derivatives. J Genet. 1929;20:405–467. [Google Scholar]
- 10.Upcott M. The nature of tetraploidy in Primula kewensis. J Genet. 1939;39:79–100. [Google Scholar]
- 11.McNaughton IH. Synthesis and sterility of Raphanobrassica. Euphytica. 1973;22:70–88. [Google Scholar]
- 12.Tokumasu S. The increase of seed fertility of Brassicoraphanus through cytological irregularity. Euphytica. 1976;25:463–470. [Google Scholar]
- 13.Howard HW. The fertility of amphidiploids from the cross Raphanus sativus × Brassica oleracea. J Genet. 1938;36:239–273. [Google Scholar]
- 14.Buxton B, Darlington C. Behaviour of a new species, Digitalis mertonensis. Nature. 1931;127:94–94. [Google Scholar]
- 15.Shkutina FM, Khvostov VV. Cytological investigation of Triticale. Theor Appl Genet. 1971;41:109–119. doi: 10.1007/BF00277752. [DOI] [PubMed] [Google Scholar]
- 16.Marchant CJ. Corrected chromosome numbers for Spartina × townsendii and its parent species. Nature. 1963;199:929. [Google Scholar]
- 17.Gottschalk W. Open problems in polyploidy research. Nucleus. 1978;21:99–112. [Google Scholar]
- 18.Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annu Rev Ecol Syst. 2002;33:589–639. [Google Scholar]
- 19.Davis BM. An amphidiploid in the F1 generation from the cross Oenothera franciscana × Oenothera biennis, and its progeny. Genetics. 1943;28:275–285. doi: 10.1093/genetics/28.4.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winge O. On the origin of constant species-hybrids. Sven Bot Tidskr. 1932;26:107–122. [Google Scholar]
- 21.Poole CF. The interspecific hybrid, Crepis rubra × C. foetida, and some of its derivatives, II: Two selfed generations from an amphidiploid hybrid. Univ Calif Publ Agr Sci. 1932;6:231–255. [Google Scholar]
- 22.Ising G. Cytogenetic studies in Cyrtanthus, I: Segregation in an allotetraploid. Hereditas. 1966;56:27–53. [Google Scholar]
- 23.Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci USA. 2011;108:7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ownbey M. Natural hybridization and amphiploidy in the genus Tragopogon. Am J Bot. 1950;37:487–499. [Google Scholar]
- 25.Ownbey M, McCollum G. The chromosomes of Tragopogon. Rhodora. 1954;56:7–21. [Google Scholar]
- 26.Pires JC, et al. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. Am J Bot. 2004;91:1022–1035. doi: 10.3732/ajb.91.7.1022. [DOI] [PubMed] [Google Scholar]
- 27.Lim KY, et al. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae) PLoS ONE. 2008;3:e3353. doi: 10.1371/journal.pone.0003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soltis DE, et al. Recent and recurrent polyploidy in Tragopogon (Asteraceae): Cytogenetic, genomic and genetic comparisons. Biol J Linn Soc Lond. 2004;82:485–501. [Google Scholar]
- 29.Mestiri I, et al. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 2010;186:86–101. doi: 10.1111/j.1469-8137.2010.03186.x. [DOI] [PubMed] [Google Scholar]
- 30.Unrau J, Person C, Kuspira J. Chromosome substitution in hexaploid wheat. Can J Bot. 1956;34:629–640. [Google Scholar]
- 31.Law CN, Worland AJ. Inter-varietal chromosome substitution lines in wheat, revisited. Euphytica. 1996;89:1–10. [Google Scholar]
- 32.Miller TE. The homoeologous relationship between the chromosomes of rye and wheat: Current status. Can J Genet Cytol. 1984;26:578–589. [Google Scholar]
- 33.Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- 34.Goff SA. A unifying theory for general multigenic heterosis: Energy efficiency, protein metabolism, and implications for molecular breeding. New Phytol. 2011;189:923–937. doi: 10.1111/j.1469-8137.2010.03574.x. [DOI] [PubMed] [Google Scholar]
- 35.Birchler JA, Yao H, Chudalayandi S. Biological consequences of dosage-dependent gene regulatory systems. Biochim Biophys Acta. 2007;1769:422–428. doi: 10.1016/j.bbaexp.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridges CB. The origin of variations in sexual and sex-limited characters. Am Nat. 1922;56:51–63. [Google Scholar]
- 37.Khush GS. Cytogenetics of Aneuploids. New York: Academic; 1973. [Google Scholar]
- 38.Linde-Laursen I, Heslop-Harrison JS, Shepherd KW, Taketa S. The barley genome and its relationship with the wheat genomes: A survey with an internationally agreed recommendation for barley chromosome nomenclature. Hereditas. 1997;126:1–16. [Google Scholar]
- 39.Sears ER. Cytogenetic studies with polyploid species of wheat, II: Additional chromosomal aberrations in Triticum vulgare. Genetics. 1944;29:232–246. doi: 10.1093/genetics/29.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longwell JH, Sears ER. Nullisomics in tetraploid wheat. Am Nat. 1963;97:401–403. [Google Scholar]
- 41.Phillips LL. Segregation in new allopolyploids of Gossypium, IV: Segregation in New World × Asiatic and New World × wild American hexaploids. Am J Bot. 1962;49:51–57. [Google Scholar]
- 42.Phillips LL. Segregation in new allopolyploids of Gossypium. V. Multivalent formation in New World × Asiatic and New World × wild American hexaploids. Am J Bot. 1964;51:324–329. [Google Scholar]
- 43.Kostoff D. Studies on polyploid plants, X: On the so-called “constancy” of the amphidiploid plants. Compt Rend Acad Sci URSS. 1935;1:653–657. [Google Scholar]
- 44.Tate JA, Joshi P, Soltis KA, Soltis PS, Soltis DE. On the road to diploidization? Homoeolog loss in independently formed populations of the allopolyploid Tragopogon miscellus (Asteraceae) BMC Plant Biol. 2009;9:80. doi: 10.1186/1471-2229-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buggs RJA, et al. Gene loss and silencing in Tragopogon miscellus (Asteraceae): Comparison of natural and synthetic allotetraploids. Heredity. 2009;103:73–81. doi: 10.1038/hdy.2009.24. [DOI] [PubMed] [Google Scholar]
- 46.Tate JA, et al. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics. 2006;173:1599–1611. doi: 10.1534/genetics.106.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buggs RJA, et al. Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol Ecol. 2010;19(Suppl 1):132–146. doi: 10.1111/j.1365-294X.2009.04469.x. [DOI] [PubMed] [Google Scholar]
- 48.Buggs RJA, et al. Rapid, repeated and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr Biol. 2012;22:1–5. doi: 10.1016/j.cub.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 49.Lukens LN, et al. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 2006;140:336–348. doi: 10.1104/pp.105.066308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmon A, Flagel L, Ying B, Udall JA, Wendel JF. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 2010;186:123–134. doi: 10.1111/j.1469-8137.2009.03093.x. [DOI] [PubMed] [Google Scholar]
- 51.Chantret N, et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops) Plant Cell. 2005;17:1033–1045. doi: 10.1105/tpc.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Symonds VV, Soltis PS, Soltis DE. Dynamics of polyploid formation in Tragopogon (Asteraceae): Recurrent formation, gene flow, and population structure. Evolution. 2010;64:1984–2003. doi: 10.1111/j.1558-5646.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- 53.Sánchez-Morán E, Benavente E, Orellana J. Analysis of karyotypic stability of homoeologous-pairing (ph) mutants in allopolyploid wheats. Chromosoma. 2001;110:371–377. doi: 10.1007/s004120100156. [DOI] [PubMed] [Google Scholar]
- 54.Riley R, Chapman V. Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature. 1958;182:713–715. [Google Scholar]
- 55.Sears E, Okamoto M. X International Congress of Genetics. Toronto, Canada: University of Toronto Press; 1958. Intergenomic chromosome relationships in hexaploid wheat; pp. 258–259. [Google Scholar]
- 56.Kostoff D. Studies on polyploid plants, XXI: Cytogenetic behaviour of the allopolyploid hybrids Nicotiana glauca Grah. Nicotiana langsdorffii Weinm. and their evolutionary significance. J Genet. 1938;37:129–209. [Google Scholar]
- 57.Stebbins GL., Jr Types of polyploids: Their classification and significance. Adv Genet. 1947;1:403–429. doi: 10.1016/s0065-2660(08)60490-3. [DOI] [PubMed] [Google Scholar]
- 58.Stebbins GL. Variation and Evolution in Plants. London: Oxford Univ Press; 1950. [Google Scholar]
- 59.Sybenga J. Chromosome pairing affinity and quadrivalent formation in polyploids: Do segmental allopolyploids exist? Genome. 1996;39:1176–1184. doi: 10.1139/g96-148. [DOI] [PubMed] [Google Scholar]
- 60.Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szadkowski E, et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 2010;186:102–112. doi: 10.1111/j.1469-8137.2010.03182.x. [DOI] [PubMed] [Google Scholar]
- 62.Song KM, Lu P, Tang KL, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feldman M, Levy AA. Allopolyploidy—a shaping force in the evolution of wheat genomes. Cytogenet Genome Res. 2005;109:250–258. doi: 10.1159/000082407. [DOI] [PubMed] [Google Scholar]
- 64.Riley R, Kimber G. Aneuploids and cytogenetic structure of wheat varietal populations. Heredity. 1961;16:275–290. [Google Scholar]
- 65.Macas J, Neumann P, Navrátilová A. Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Birchler JA, Albert SA, Gao Z. Stability of repeated sequence clusters in hybrids of maize as revealed by FISH. Trop Plant Biol. 2008;1:34–39. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.