Abstract
We investigated the expression of microRNAs (miRNAs) associated with replicative senescence in human primary keratinocytes. A cohort of miRNAs up-regulated in senescence was identified by genome-wide miRNA profiling, and their change in expression was validated in proliferative versus senescent cells. Among these, miRNA (miR)-138, -181a, -181b, and -130b expression increased with serial passages. miR-138, -181a, and -181b, but not miR-130b, overexpression in proliferating cells was sufficient per se to induce senescence, as evaluated by inhibition of BrdU incorporation and quantification of senescence-activated β-galactosidase staining. We identified Sirt1 as a direct target of miR-138, -181a, and -181b, whereas ΔNp63 expression was inhibited by miR-130b. We also found that ΔNp63α inhibits miR-138, -181a, -181b, and -130b expression by binding directly to p63-responsive elements located in close proximity to the genomic loci of these miRNAs in primary keratinocytes. These findings suggest that changes in miRNA expression, by modulating the levels of regulatory proteins such as p63 and Sirt1, strongly contribute to induction of senescence in primary human keratinocytes, thus linking these two proteins. Our data also indicate that suppression of miR-138, -181a, -181b, and -130b expression is part of a growth-promoting strategy of ΔNp63α in epidermal proliferating cells.
Cellular senescence starts as a growth arrest during which cells remain metabolically active but acquire typical morphologic features, appearing enlarged, flattened, and vacuolar, accompanied by changes in their gene expression profiles (1, 2). Senescence may be induced by multiple cellular stress stimuli, including oncogene activation, overproduction of reactive oxygen species, or simply by the end of proliferative potential, that is, replicative senescence (3–6). In tissues with high turnover, such as the epidermis, replicative senescence plays a crucial role in the time-dependent changes occurring in the tissue (7). When normal cells enter a state of replicative senescence, they block progression from the G1 to the S phase by maintaining the retinoblastoma (Rb) protein in a hypophosphorylated state (8, 9). Many regulators of the cell cycle participate in the establishment of replicative senescence, although the most reliable marker to assess the effective entrance to the senescent state is the expression of senescence-associated (SA) β-galactosidase (10). Cellular senescence not only provi des a mechanism that contributes to the natural aging of an organism, but also is a process used to arrest the progression of malignant phenomena, thereby protecting the organism from tumors (4).
The epidermis is a continuously renewing epithelium that requires tight control of keratinocyte proliferative potential to direct the subsequent differentiation stages that occur within its upper layers. In this context, the transcription factor p63 plays a master role in regulating the turnover of basal keratinocytes in both developing and adult epidermis (11). TP63 expression is controlled by two alternative promoters giving rise to the longer-transactivating isoform TAp63 and one lacking the N-terminal transactivation domain ΔNp63. However, ΔNp63 can transactivate a distinct set of target genes due to a C-terminal transactivation domain. Furthermore, alternative splicing at the 3′ end of the p63 transcript generates three more isoforms—α, β, and γ—that are expressed on both TAp63 and ΔNp63 backbones, resulting in a total of six p63 isoforms (12). p63 KO mice completely lack skin and skin derivatives and die within a few hours after birth due to dehydration (13, 14). The most abundant p63 isoform in the epidermis is ΔNp63, which is highly expressed only in the basal compartment of proliferating keratinocytes (13), and its expression is tightly regulated by miR-203 during keratinocyte differentiation (15, 16). Studies have shown that p63 is involved in senescence pathways, as its absence in conditional knockout mice induces premature aging (17–19). It has been reported that the overexpression of TAp63α in both human and murine fibroblasts induces cellular senescence, whereas ΔNp63, expressed specifically in epithelia, is not involved in fibroblast senescence (18). However, the TAp63-selective KO mouse model indicates a totally opposite role for this transcription factor in which it prevents premature aging and promotes adult stem cell maintenance (19). Nevertheless, all these in vitro and in vivo studies do not exclude a possible, and yet unexplored, role for ΔNp63α in cellular senescence and in the maintenance of keratinocyte stemness (20).
microRNAs (miRNAs) have been shown to be necessary for the correct development of the epidermis as demonstrated by the generation of conditional ablations of Dicer or DGCR8, key components of miRNA biosynthesis complexes, which are sufficient to induce severe developmental and structural defects in mouse skin (21–23).
Here, we investigated the involvement of both miRNAs and ΔNp63α in replicative senescence of primary human keratinocytes. By microarray profiling of miRNAs during keratinocyte replicative senescence, differentiation, and after p63 silencing by siRNA, we show that miRNA (miR)-138, miR-181a, miR-181b, and miR-130b levels are up-regulated during keratinocyte replicative senescence and that their direct targets are Sirt1 and p63 mRNAs. These data are therefore evidence of a complex circuitry driving cellular senescence in human epidermal keratinocytes neonatal.
Results
Induction of Replicative Senescence in Human Epidermal Keratinocytes.
To generate a model with which to study the onset of replicative senescence, proliferating primary HEKn cells were monitored through serial passaging. The proliferating or senescent state of the cells was assessed morphologically (Fig. S1). A growth curve that calculated the population doublings at each passage was generated (Fig. S1A). After three passages, the proliferation rate slows down, eventually leading to an almost complete block after five passages. Between p1 and p4 we observed a significant decrease in BrdU incorporation (from 34 to 11%, Fig. S1B) and in relative telomerase activity (Fig. S1C). Finally, p4 senescent cells stained positive for SA β-galactosidase compared with the proliferating p1 population (Fig. S1D). Both promyelocytic leukemia (PML) and p16, which are senescence markers, are induced upon serial passages, whereas Rb shows a transition from an inactive hyperphosphorylated state to an active hypophosphorylated state, thereby indicating a cell cycle block (Fig. S1E). We excluded the possibility that the down-regulation of the rate of keratinocyte proliferation was due to an unwanted differentiation process by verifying that cytokeratin 10 (K10), an early marker of keratinocyte differentiation, was undetectable throughout the experiment (Fig. S1E). We also explored the mRNA and protein expression levels of p53, known to be involved in senescence (24, 25). We found that p53, in contrast to p16 and PML, does not increase during serial passaging of neonatal primary keratinocytes (Fig. S1 E and F), suggesting that it is not involved in this specific type of senescence. As an added control, we also used real-time RT-quantitative PCR (qPCR) to evaluate the expression levels of the other p53 family members: TAp63, ΔNp63, TAp73, and ΔNp73 (Fig. S1F). None of these proteins (and particularly TAp63 and TAp73, which have been linked to senescence) exhibited increased expression levels with serial passaging, suggesting that they are not involved in replicative senescence of keratinocytes.
miRNA Differential Profiling Between Proliferating and Senescent HEKn.
To investigate miRNAs involved in replicative senescence, we used a microarray approach, comparing miRNA expression profiles of proliferating HEKn at passage 1 (p1), with those of senescent HEKn at passage 4 (p4). Several miRNAs were significantly up-regulated during the senescing process (Table S1). After real-time RT-qPCR validation of 20 miRNAs that changed expression in the array (Fig. S2), we selected four miRNAs among the most consistently up-regulated miRNAs, taking into account the ones that have never been associated with cellular senescence. These miRNAs are miR-138 (fold change 2.76 ± 0.06), miR-181a (fold change 1.3 ± 0.05), miR-181b (fold change 1.38 ± 0.03), and miR-130b (fold change 1.38 ± 0.05); P value < 0.032. To discriminate miRNAs directly involved in the senescence process from general antiproliferative miRNAs, we next decided to compare miRNA expression profiles of senescent versus differentiated HEKn. Using the same microarray approach, we demonstrated that miR-138, -181a, -181b, and -130b were up-regulated due to senescence (p4) and not as a result of in vitro differentiation (day 9 of CaCl2 treatment). This conclusion was confirmed by validation with RT-qPCR (Fig. S3).
Sirt1 Is a Direct Target of miR-138, miR-181a, and miR-181b.
To understand the role of the four selected miRNAs, we carried out a detailed analysis of their molecular targets. In silico prediction by TargetScan 5.1 software of miR-138, -181a, and -181b targets showed that the Sirt1 3′-UTR harbors a putative target sequence for these miRNAs that is highly conserved among vertebrates (Fig. S4 A and B). Western blot analysis showed a decrease of Sirt1 protein levels in keratinocytes from p1 to p4 (Fig. S1E), thereby strengthening the hypothesis of its direct regulation by miR-138, -181a, and -181b.
Transfection of premiR-138, -181a, and -181b, but not a scrambled control sequence, reduced the activity of a human Sirt1 3′-UTR–luciferase reporter (about 60% with miR-138 and 40% with miR-181a and b; Fig. 1A), similar to the effect of miR-217, used as a positive control. Site-directed deletion of the single miRNA target sites in the Sirt1 3′-UTR abolished the effect of miR-138, miR-181a, and -181b on luciferase activity under the same conditions (Fig. 1A). As expected, the activity of miR-217 was unaffected by the deletion.
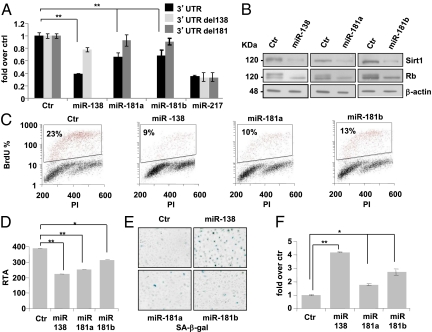
Fig. 1.
miR-138, miR-181a, and miR-181b target Sirt1 mRNA at its 3′-UTR and induce cellular senescence in HEKn cells. (A) Insertion of Sirt1's 3′-UTR in a luciferase reporter vector leads to diminished luciferase activity in the presence of miR-138, miR-181a, and miR-181b in HEK293 cells 24 h after cotransfection (black bars). Deletions of miR-138 (light gray bars) and miR-181a and miR-181b (dark gray bars) target sequences in the Sirt1 3′-UTR abolish the miRNA-mediated effect on luciferase activity. Transfection of all reporter vectors in the presence of miR-217 was performed as a positive control. Histograms show the values resulting as averages ± SD from three independent cotransfections. (B) Western blot analysis of protein extracts from HEKn cells transfected with miR-138, -181a, and -181b versus a scrambled control sequence (Ctr). miR-138, miR-181a, and miR-181b overexpression decreases Sirt1 protein levels and influences the Rb phosphorylation state. β-Actin was used as loading control. (C) Forty-eight hours after transfection of HEKn cells with a scrambled control (Ctr) or miR-138, miR-181a, or miR-181b sequences, cells were subjected to a 4h BrdU pulse, collected, propidium iodide (PI)-stained, and analyzed by flow cytometry. BrdU-positive cells are indicated as S-phase fluorescent populations (red dots) and are assessed by PI staining of DNA content of 2n or 4n (fixed to values of 200 and 400 in the plots). Values reported are averages ± SD of three independent transfections. (D) Relative telomerase activity (RTA) of HEKn 48 h after transfection with a scrambled control or miR-138, miR-181a, or miR-181b sequences. Values reported are averages ± SD of three independent transfections. (E and F) SA β-galactosidase staining and quantification by blue-cell counts/field. Values reported are averages ± SD of three independent stains. *P value <0.01 and **P value <0.005 by Student's t test.
Overexpression of miR-138, -181a, and -181b by transfection in proliferating HEKn led to a strong down-regulation of Sirt1 protein levels (Fig. 1B), thus demonstrating that miR-138, -181a, and -181b were able to repress endogenous Sirt1 expression. miR-138, -181a, and -181b overexpression had additional effects in HEKn cells, including Rb hypophosphorylation by Western blot (Fig. 1B), a decrease of more than 50% in BrdU incorporation in the presence of miR-138 and -181a and about 40% with miR-181b (Fig. 1C), and a significant down-regulation of relative telomerase activity (Fig. 1D). Under identical conditions, we also found an increase in the number of cells positive for SA β-galactosidase staining as shown by the images and blue-cell quantification in Fig. 1 E and F. These results suggest that miR-138, -181a, and -181b overexpression is sufficient per se to induce cellular senescence by down-regulating Sirt1.
p63 Is a Direct Target of miR-130b.
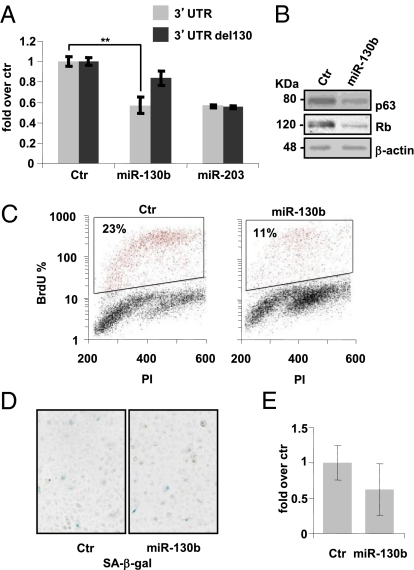
Using TargetScan 5.1 software, we identified the 3′-UTR of p63 as a putative conserved target of miR-130b (Fig. S4 C and D). p63 has already been shown to be involved in the senescence process, in particular its TA isoforms (18, 19). In this study, we observed that the ΔNp63α isoform is down-regulated in HEKn upon serial passaging (Fig. S1E). This inverse correlation with miR-130b expression is consistent with the hypothesis that it might regulate p63. To confirm this, we used the p63 3′-UTR (16) in luciferase assays. Transfection of this reporter construct in the presence of premiR-130b induced about a 50% down-regulation of relative luciferase activity, and this effect was abolished with the mutation of the miR-130 putative target site (Fig. 2A). miR-203 was used as positive control. miR-130b's overexpression in proliferating HEKn down-regulated ΔNp63α levels, as shown in the Western blot in Fig. 2B. The ΔNp63α decrease occurred in parallel with the hypophosphorylation of Rb (Fig. 2B) and the reduction in the percentage of cells incorporating BrdU from 23 to 11% (Fig. 2C), suggesting growth inhibition. Despite this strong effect on cell proliferation, miR-130b alone was not sufficient to increase the number of SA β-galactosidase–positive-staining cells (as shown by the images and histogram in Fig. 2 D and E) or to induce senescence in proliferating keratinocytes.
Fig. 2.
miR-130a targets p63 mRNA at its 3′-UTR and decreases HEKn proliferation. (A) Insertion of p63 3′-UTR in a luciferase reporter vector leads to diminished luciferase activity in the presence of miR-130b in HEK293 cells 24 h after cotransfection (gray bars). Deletion of the miR-130b (black bars) target sequence in the p63 3′-UTR abolishes the decrease in miRNA-mediated luciferase activity. Transfection of a reporter vector in the presence of miR-203 was performed as a positive control. Histograms show the values resulting as averages ± SD from three independent cotransfections. (B) Western blot analysis of protein extracts of HEKn transfected with miR-130b versus a scrambled control sequence (Ctr). miR-130b overexpression decreases p63 protein levels and influences Rb phosphorylation. β-Actin was used as a loading control. (C) Forty-eight hours after transfection of HEKn cells with either a scrambled control (Ctr) or pre-miR-130b, cells were subjected to a 4-h BrdU pulse, collected, PI-stained, and analyzed by flow cytometry as described in Fig. 1. Values reported are averages ± SD of three independent transfections. (D and E) SA β-galactosidase staining and quantification by blue-cell counts/field. Values reported are averages ± SD of three independent stains. **P value <0.005 by Student's t test.
Sirt1 or p63 Silencing by siRNA Induces Senescence in Proliferating Primary Keratinocytes.
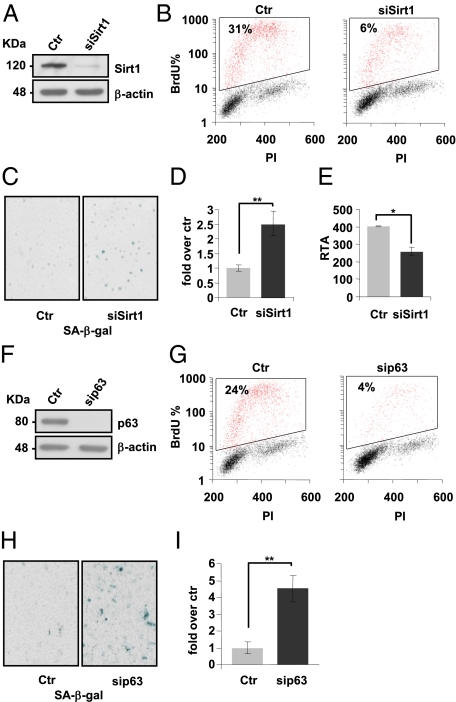
To confirm the effect of senescence-associated miRNAs in HEKn cells, we performed “phenocopy” silencing by siRNA of their direct targets Sirt1 and p63. Sirt1-silenced keratinocytes (Fig. 3A) showed a significant reduction of BrdU incorporation compared with scramble transfected cells (Fig. 3B), an increased staining in SA β-galactosidase activity (Fig. 3 C and D), and a 40% reduction in telomerase activity (Fig. 3E), indicating that silenced cells undergo senescence. This behavior of Sirt1-silenced HEKn is quite similar to Sirt1-targeting miRNA overexpression in the same cells, as can be seen in Fig. 1.
Fig. 3.
Sirt1 and p63 silencing by siRNA induces cellular senescence. (A and F) Western blots of protein extracts of HEKn. Cells were transfected with siSirt1 or sip63 versus a scrambled control sequence (Ctr). β-Actin was used as loading control. (B and G) Forty-eight hours after transfection of HEKn cells with a scrambled control (Ctr), si-Sirt1, or si-p63, cells were subjected to a 4h BrdU pulse, collected, PI-stained, and analyzed by flow cytometry as described in Fig. 1. Values reported are averages ± SD of three independent transfections. (C and D and H and I) SA β-galactosidase staining and quantification by blue-cell counts/field. Values reported are averages ± SD of three independent stains. (E) Relative telomerase activity (RTA) of HEKn 48 h after transfection with a scrambled control or miR-130b sequences. The presented data are averages ± SD. *P value <0.01 and **P value <0.005 by Student's t test.
Under the same conditions, we also silenced p63 by siRNA (Fig. 3F) and observed a marked decrease in BrdU incorporation (5-fold reduction, Fig. 3G) followed by increased staining for SA β-galactosidase activity (4.5-fold increase, Fig. 3H). On the contrary, miR-130b overexpression in proliferating HEKn was not able to significantly augment SA β-galactosidase staining, but did reduce BrdU incorporation (Fig. 2). These partially overlapping results were probably due to the incomplete silencing of p63 in transfected miR-130b cells compared with the silencing of p63 by siRNA (see the Western blots in Fig. 2 and Fig. 3E).
ΔNp63 Controls miR-138, -181a/b, and -130b Expression.
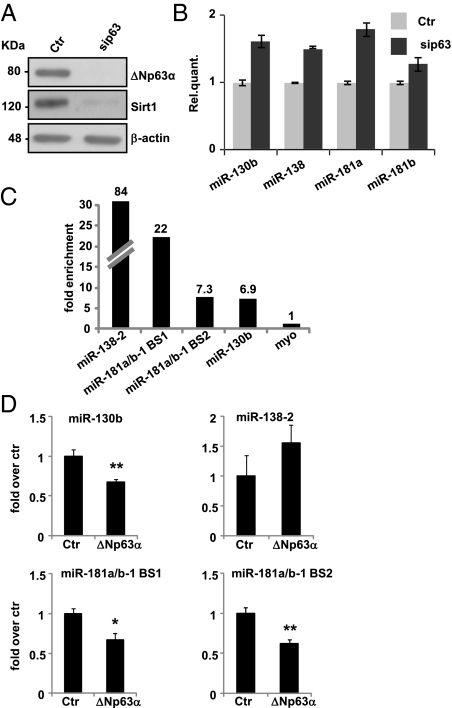
To better investigate the role of ΔNp63 in senescence, we performed a third genome-wide screening comparing the miRNA expression profile of HEKn cells transfected with a scrambled sequence versus those in which p63 has been silenced. Table S2 shows the miRNAs that exhibited the highest up- or down-regulated fold changes upon p63 silencing and their qRT-PCR validation (Fig. S5). The four miRNAs whose expression is increased during senescence are all up-regulated in p63-silenced keratinocytes (Fig. 4B), indicating that p63 inhibits their expression.
Fig. 4.
p63 regulates miR-130b, miR-138, miR-181a, and miR-181b. (A) Western blot of protein extracts of HEKn transfected with si-p63 versus a scrambled control sequence (Ctr). β-Actin was used as loading control. (B) Real-time RT-qPCR showing the up-regulation of selected miRNAs upon p63 silencing in HEKn. Values reported are the average ± SD of three independent transfections. (C) ChIP followed by qPCR analysis was performed in the proliferating human skin keratinocyte HK1 cell line. Binding of p63 at all four binding sites located close to miR-138-2, miR-181a/b-1, and miR-130b were tested and confirmed. The region of myoglobin exon 2 (myo) was used as a non-p63–binding control region. (D) Luciferase assays were performed 24 h after HEK293 cells were cotransfected with pGl3-promoter constructs containing ChIP-positive p63-binding sites as enhancers and an expression vector for ΔNp63α or an empty vector as negative control. Values reported are the average ± SD of three independent cotransfections. *P value <0.05 and **P value <0.01 by Student's t test. BS, binding site.
miR-138 is an intergenic miRNA present in two copies in the human genome: one on chromosome 3 (miR-138–1) and a second on chromosome 16 (miR-138–2). miR-181a and miR-181b are two intronic clustered miRNAs existing in double copies on chromosome 1 (miR-181a/b-1) and on chromosome 9 (miR-181a/b-2). miR-130b is an exonic miRNA located on chromosome 22. To investigate whether the expression of these miRNAs could be directly controlled by p63, we analyzed miR-138–2, miR-181a/b-1, and miR-130b genomic regions to identify p63-responsive elements using a genome-wide p63 DNA-binding profile previously generated in human skin keratinocytes (26, 27). In close proximity (within 25 kb) to these miRNA genes, we found highly reliable p63-binding sites (BS) (Fig. S6). Binding of p63 to these genomic sites was confirmed by independent chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR) (Fig. 4C). Furthermore, these p63 BS colocalized with histone H3 lysine 4 monomethylation and histone H4 lysine 3 di-methylation (Fig. S6), which are considered as markers for distal regulatory elements (28, 29). To demonstrate a direct repressive role of ΔNp63α in controlling the identified BS upstream of these miRNAs, the 200-bp p63-responsive genomic fragments were cloned in reporter constructs as enhancers and used for luciferase assays in the presence of ΔNp63α. Expression of ΔNp63α led to a statistically significant reduction in luciferase activity regulated by miR-130b and the two miR-181a/b-1 p63 BS (more than 30% reduction). The miR-138–2-responsive element was not affected by ΔNp63α within the limits of this specific experiment. These data, together with the observation that p63 silencing affects the down-regulation of Sirt1 protein levels in proliferating keratinocytes (Fig. 4A), indicate that ΔNp63α itself can control Sirt1 expression via the transcriptional regulation of Sirt1-repressing miRNAs.
Sirt1, p63, and Senescence-Associated miRNA Expression Levels Change During Skin Aging in Vivo.
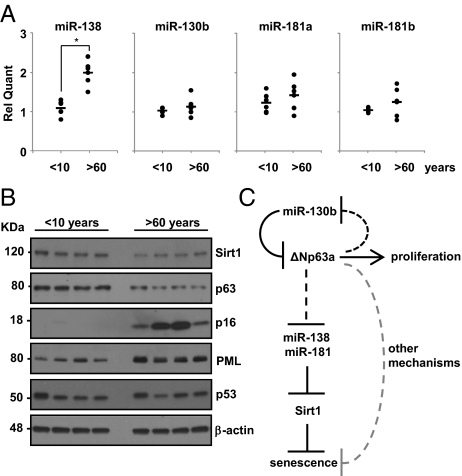
We analyzed senescence-associated miRNAs, Sirt1 and ΔNp63 expression levels in total RNA, and protein extracts from primary keratinocytes cultured from skin biopsies of young (<10 y old, n = 6) and aged (>60 y old, n = 6) healthy subjects. We observed that miR-138 expression is significantly increased in keratinocytes of aged skin (P value < 0.01) and that there is a positive trend for miR-130b, miR-181a, and miR-181b, albeit not statistically significant (Fig. 5A). Sirt1 and ΔNp63α protein levels are consistently reduced during aging as shown by Western blot analysis (Fig. 5B). As a positive control, we included the typical senescence markers p16 and PML. We also showed, by Western blotting, that expression of p53 does not change in the keratinocytes of young and old subjects (Fig. 5B).
Fig. 5.
miR-138, miR-181a, and miR-181b are up-regulated in aged skin. (A) Relative quantification by RT-qPCR of miR-138, miR-181a, miR-181b, and miR-130b expression levels in primary keratinocytes cultured from young (<10 y) and aged (>60 y) healthy skin biopsies. *P value <0.01 by Student's t test. (B) Western blot of primary keratinocyte protein extracts cultured as in A showing Sirt1, p63, p53, p16, and PML levels. β-Actin was used as a loading control. (C) Model of the feedback circuitry during HEKn replicative senescence involving ΔNp63α, Sirt1, and miRNAs.
Discussion
Our data indicate that miR-138, miR-181a, miR-181b, and miR-130b levels are up-regulated during keratinocyte senescence, leading to growth inhibition and induction of SA β-galactosidase activity. We demonstrate that Sirt1 and p63 mRNAs are directly regulated by these miRNAs, linking these two factors to replicative senescence in keratinocytes. Furthermore, ΔNp63 inhibits miR-138, miR-181a, and miR-181b expression and consequently permits Sirt1 expression, thus counteracting replicative cellular senescence (Fig. 5C). We also showed that, in human healthy subjects, miR-138 is significantly up-regulated in aged skin, whereas miR-181a, miR-181b, and miR-130b show a positive trend in our patient cohort, although this failed to reach significance, possibly because of the small numbers of samples analyzed. In parallel, we showed that their identified targets, p63 and Sirt1, were down-regulated in the skin of aged subjects whereas p16 and PML increased consistently in all of the patients analyzed.
miR-138 and miR-181 are strongly associated with senescence because their expression is sufficient per se to induce premature senescence in proliferating keratinocytes, whereas overexpression of miR-130b acts mainly on proliferation. Sirt1 is a member of the NAD+-dependent deacetylase family, which has both histone and nonhistone substrates (30, 31). It can protect against age-related diseases by deacetylating proteins that regulate different cellular processes, including stress responses, replicative senescence, inflammation, and metabolism (31). Sirt1 has already been linked to senescence but not in the context of skin where it has been reported to protect keratinocytes from different cellular stresses (32) and to modulate differentiation (33). Our previous studies showed that Sirt1 levels are modulated by miR-217 in replicative endothelial cell senescence (34), and a recent report indicated that miRNAs can also regulate Sirt1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues (35). p63 deficiency causes cellular senescence in proliferating keratinocytes, leading to accelerated aging in vivo, as shown in p63 heterozygous mice (17). However, the contribution of the different isoforms (TAp63 and ΔNp63) has yet to be fully investigated. Because senescence is considered a tumor-suppressive mechanism, the role of p63 has been studied mainly in oncogene-induced senescence. In this context, TAp63γ is up-regulated in mouse embryonic fibroblasts (MEFs) during Ras-induced oncogene senescence (18), and, in general, TAp63 isoforms are considered mediators of senescence (18, 19). In skin-derived progenitor cells of the hair follicle, which are distinct from both keratinocytes and MEFs, the selective loss of TAp63 isoform expression induces hyperproliferation that culminates in senescence. The sarcomas developed in TAp63-selective KO mice express high levels of senescence markers (19). These findings suggest that both p63 isoforms are involved in cellular senescence even though it is not clear what their specific contributions are to the different types of senescence in different cellular systems. The results presented here clearly indicate that ΔNp63 counteracts replicative senescence in human keratinocytes by inhibiting miR138, miR-181a, miR181b, and miR-130b.
miR-138, miR-181a, miR-181b, and miR-130b have been proposed as tumor-suppressor miRNAs that are down-regulated in different malignancies (36–40). Among the miRNAs up-regulated in keratinocytes during replicative-induced senescence, miR-130b is of particular interest because it was found to be a direct transcriptional target of TAp63 in metastasis suppression (39, 41). In keratinocytes, we implicate an auto-regulatory loop between ΔNp63 and miR-130b, and we detect only weak expression of the TAp63 isoform at the mRNA level and conclude that it is not involved in replicative-induced senescence. Our data also indicate that p53 is not involved in keratinocyte replicative senescence because p53 expression, both at RNA and protein levels, does not change during keratinocyte serial passaging (Fig. S1 E and F). Our finding that miR-130b per se is not sufficient to trigger senescence suggests the existence of several parallel regulatory pathways, including miR-138, miR-181a, and miR-181b, and possibly other miRNAs (24), that need to be activated simultaneously to achieve senescence induction in keratinocytes.
In summary, we have evaluated the molecular pathways of replicative senescence in human keratinocytes, and our results strongly indicate that miRNAs play an important role in this process together with ΔNp63 and Sirt1, targets of the identified miRNAs. These findings may be relevant for young patients with genetic defects leading to precocious senescence, such as xeroderma pigmentosum and progeria, and as an in vitro model for physiological aging of the skin.
Materials and Methods
Cell Culture and Transfection.
HEKn (Cascade, Invitrogen) were cultured and transfected as described in SI Materials and Methods.
microRNA Profiling.
Five micrograms of RNA from each sample was labeled with biotin by reverse transcription using random octamers. Hybridization was carried out on the second version of miRNA-chip (MDACC miRNA Expression Bioarray 19k v5). Details about data analysis are in SI Materials and Methods.
RNA Extraction and Real-Time PCR Analysis.
Total RNA isolation and miRNA quantification by RT-qPCR are described in SI Materials and Methods.
Senescence-Associated β-Galactosidase Staining.
Cells were grown in 12-well culture plates, washed with PBS, and fixed and stained for 24 h at 37° using a standard protocol. The reaction was stopped by replacing the staining solution with 70% glycerol.
Cell Proliferation and Cell Cycle Analysis.
Incorporation of BrdU during DNA synthesis was evaluated with the Click-iT EdU flow cytometry assay kit, following the manufacturer's protocol (Molecular Probes). Cell cycle was analyzed using a FACS Calibur flow cytometer (BD Biosciences). Fifteen thousand events were evaluated using the Cell Quest (BD) software.
Luciferase Assay and Constructs.
The pGL3Control-p63 3′-UTR construct has been described in Lena et al. (16). The miR-130 predicted target site (7 bp, UUGCACU) was deleted by PCR. pGL3Control-Sirt1 3′-UTR has also been described (34). miR-138 (7 bp, CACCAGCA) and miR-181 (7 bp, UGAAUGU) predicted target sites were also deleted by PCR. PCR primers used to obtain them are available on request. Luciferase assays were performed as described in Lena et al. (16). Cells were transfected with 100 ng of pGL3 vectors, 12 pmol of pre-miRNA or a scrambled sequence (Ambion), and 10 ng of Renilla luciferase pRL-CMV vector (Promega). To demonstrate a ΔNp63α-mediated repression on miR-138-2, miR-181a/b-1, and miR130b ChIP-positive p63-binding sites, luciferase assays were performed in HEK293 cells as described before (42).
Western Blotting.
Total cell extracts were resolved on an SDS polyacrylamide gel and blotted onto a Hybond P PVDF membrane (G&E Healthcare). The following antibodies were used: anti-p63 (Ab4, Neomarkers; dilution 1:500), anti–β-actin (Sigma; dilution 1:5,000), anti-p16 (Santa Cruz Biotechnology; dilution 1:1,000), anti-Rb (BD Biosciences; dilution 1:1,000), anti-Sirt1 (Abcam; dilution 1:500), anti-p53 (Santa Cruz Biotechnology; dilution 1:300), anti-PML (Santa Cruz Biotechnology; dilution 1:400), and anti-K10 (Covance; dilution 1:1,000).
Relative Telomerase Activity Assay.
Telomerase activity in HEKn was assayed by the TeloTAGGG Telomerase PCR ELISAplus kit (Roche Molecular Biochemicals) following the manufacturer's protocol.
Chromatin Immunoprecipitation Assay.
ChIP-qPCR was performed according to a previously published procedure (26) where 4 μg/sample of the anti-p63 H129 (Santa Cruz Biotechnology) was used. qPCR primers are available on request.
Bioinformatics.
Analysis of microRNA target sites were performed using the TargetScan 5.1 software available at http://www.targetscan.org.
Supplementary Material
Acknowledgments
We thank Drs. Richard Knight, Amanda Formosa, Angelo Peschiaroli, and Giovanna Zambruno for scientific discussions and Massimo Teson for technical support. The work reported in this manuscript has been supported by Theleton Grant GGP09133, Associazione Italiana per la Ricerca sul Cancro Grant 2011-IG11955; Ministero Università Istruzione e Ricerca (MIUR), Ministero Sanità (Italy) Grants RF73, RF57 and ACC12; Odysseus/VIB (Belgium) (to G.M.); by Ministero Università Istruzione e Ricerca (MIUR), Ministero Sanità (Italy) Grants RF73, RF57, and ACC12 (to E.C.). This work was also supported by the Ministry of Education and Science of the Russian Federation Grant 11.G34.31.0069.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112257109/-/DCSupplemental.
References
- 1.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams PD. Healing and hurting: Molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell. 2009;36:2–14. doi: 10.1016/j.molcel.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Narita M, Lowe SW. Senescence comes of age. Nat Med. 2005;11:920–922. doi: 10.1038/nm0905-920. [DOI] [PubMed] [Google Scholar]
- 4.Campisi J. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Cristofalo VJ, Lorenzini A, Allen RG, Torres C, Tresini M. Replicative senescence: A critical review. Mech Ageing Dev. 2004;125:827–848. doi: 10.1016/j.mad.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordisco S, et al. Bmi-1 reduction plays a key role in physiological and premature aging of primary human keratinocytes. J Invest Dermatol. 2010;130:1048–1062. doi: 10.1038/jid.2009.355. [DOI] [PubMed] [Google Scholar]
- 8.Blagosklonny MV. Cell senescence: Hypertrophic arrest beyond the restriction point. J Cell Physiol. 2006;209:592–597. doi: 10.1002/jcp.20750. [DOI] [PubMed] [Google Scholar]
- 9.Chicas A, et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candi E, et al. Differential roles of p63 isoforms in epidermal development: Selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 12.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 13.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 14.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 15.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lena AM, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 17.Keyes WM, et al. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo X, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su X, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 21.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 22.Yi R, Fuchs E. MicroRNA-mediated control in the skin. Cell Death Differ. 2010;17:229–235. doi: 10.1038/cdd.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi R, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin KH, et al. Identification of senescence-inducing microRNAs in normal human keratinocytes. Int J Oncol. 2011;39:1205–1211. doi: 10.3892/ijo.2011.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campisi J, d'Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 26.Kouwenhoven EN, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raney BJ, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39(Database issue):D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 29.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donmez G, Guarente L. Aging and disease: Connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 31.Haigis MC, Sinclair DA. Mammalian sirtuins: Biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao C, et al. SIRT1 confers protection against UVB- and H2O2-induced cell death via modulation of p53 and JNK in cultured skin keratinocytes. J Cell Mol Med. 2009;13(9B):3632–3643. doi: 10.1111/j.1582-4934.2008.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blander G, et al. SIRT1 promotes differentiation of normal human keratinocytes. J Invest Dermatol. 2009;129(1):41–49. doi: 10.1038/jid.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menghini R, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120:1524–1532. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 35.Saunders LR, et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2010;2:415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi L, et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 37.Nicoloso MS, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, et al. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009;286:217–222. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitomo S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adorno M, et al. A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 42.Browne G, et al. Differential altered stability and transcriptional activity of ΔNp63 mutants in distinct ectodermal dysplasias. J Cell Sci. 2011;124:2200–2207. doi: 10.1242/jcs.079327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.