Abstract
Single-cell analysis is gaining popularity in the field of mass spectrometry as a method for analyzing protein and peptide content in cells. The spatial resolution of MALDI mass spectrometry (MS) imaging is by a large extent limited by the laser focal diameter and the displacement of analytes during matrix deposition. Owing to recent advancements in both laser optics and matrix deposition methods, spatial resolution on the order of a single eukaryotic cell is now achievable by MALDI MS imaging. Provided adequate instrument sensitivity, a lateral resolution of ~10 µm is currently attainable with commercial instruments. As a result of these advances, MALDI MS imaging is poised to become a transformative clinical technology. In this article, the crucial steps needed to obtain single-cell resolution are discussed, as well as potential applications to disease research.
Keywords: biomarker, clinical, histopathology, immunohistochemistry, MALDI MS imaging, mass spectrometry, prognosis, single cell
Introduction to MALDI mass spectrometry imaging
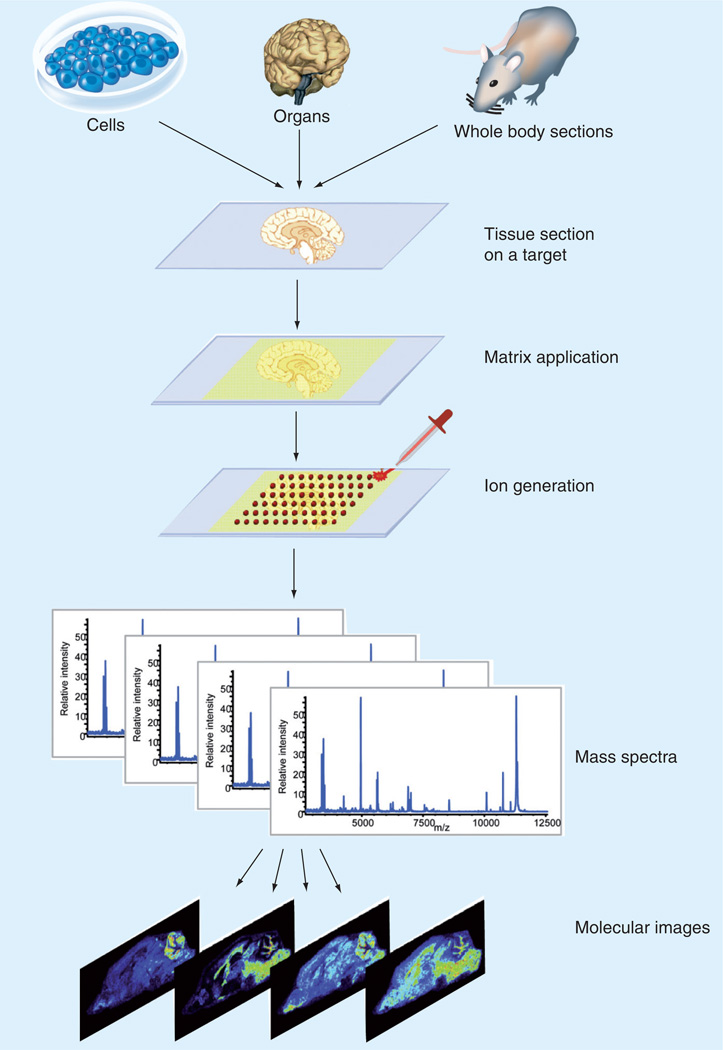
Matrix-assisted laser desorption/ionization mass spectrometry (MS) imaging, also referred to as MALDI imaging MS and MS imaging, is a method that allows for the visualization of the spatial distribution of compounds across tissue sections. This method does not require specific antibodies such as those required by immunohistochemistry, thus allowing for a direct discovery approach to tissue analysis. This is achieved by obtaining an array of mass spectra using a pulsed laser. Once an array of spectra is obtained, it is possible to create an ion image of a particular mass-to-charge (m/z) ratio. This image represents the spatial distribution and relative abundance of that particular ion. An array of MALDI mass spectra (MS image) therefore contains hundreds of ion images, each representative of a different molecule of interest in the same tissue section. MS images can be correlated with various imaging modalities such as fluorescence, histological stains and MRI [1]. An overview of this process is shown in Figure 1.
Figure 1. MALDI mass spectrometry imaging process.
The tissue/cell of interest is placed onto a MALDI target or an indium tin oxide-coated glass slide. A matrix is applied to the tissue/cell and mass spectra are acquired in a raster pattern across the tissue section. The spatial distribution of a single m/z can be represented as a 2D ion density map.
Reprinted with permission from [118].
The MALDI MS imaging experiment described in Figure 1 is a suitable approach for biomarker discovery, and can detect molecules that would not otherwise be detected in tissue homogenates. For example, as a result of the limited dynamic range of MS, a molecule that is confined to a relatively small area of the brain would be detected by MS imaging, but not necessarily in a whole-brain homogenate. The mass lists from MALDI MS images can be used to create classes of compounds to be analyzed by statistical methods such as principal component analysis or hierarchical clustering [2,3]. Principal component analysis reduces high-dimensional data into a working set of principle components that are representative of the highest amount of variance in the data set. These components can be visualized in order to determine how specific data points relate to one another [2]. An a priori approach can also be taken to tissue analysis, such as histology-directed analysis where regions of interest are identified by a histopathology expert.
Another form of analysis that will not be discussed in this article, but merits acknowledgement is secondary-ion MS (SIMS). SIMS boasts higher spatial resolution (~500 nm on tissues) than MALDI MS imaging, allowing for subcellular resolution. SIMS can therefore be applied to the characterization of individual organelles, and most often detects elemental composition or small molecules (such as vitamin E [4]) and metabolites, but generally does not detect peptides, proteins and most lipids [4,5]. SIMS should therefore be regarded as a technique that is complementary to MALDI MS imaging. Ambient MS imaging methods, such as desorption electrospray ionization [6,7] and laser ablation electrospray ionization [8,9], have a significant advantage of minimal sample preparation, but their discussion lies outside the scope of this article.
MALDI MS imaging & histochemistry
Hematoxylin and eosin (H&E) staining of tissue is a method of widespread use in the preparation of clinical tissue specimens, and allows the visualization of cells with bright field microscopy through the respective labeling of the nucleus and cytoplasm of cells [10,11]. The microscopic review of H&E-stained tissue sections by the trained eye of pathologists is then used to establish diagnosis based upon criteria such as cellular composition. Malignancies are also graded according to characteristics such as proliferation, cellular and nuclear morphology, vascularization and the visual presence of biomarkers.
Methods have been developed that allow for the co-registration of histological features observed by various histological stains with that of the MALDI MS ion image. There are two methods that are widely used to correlate histology results with that of the MALDI MS image. The first approach is to apply the histological stain of choice to a serial section of tissue from that which is being analyzed by MALDI MS imaging. By staining a serial section, the researcher need not concern themselves with the compatibility of the stain with the mass spectrometer [12,13]. The second method is to stain the same tissue section from which the MALDI MS image was obtained. This can be carried out either before, by employing a MS-compatible stain [14], or after acquiring the ion image, by washing off the matrix and staining the tissue section post-imaging [15,16].
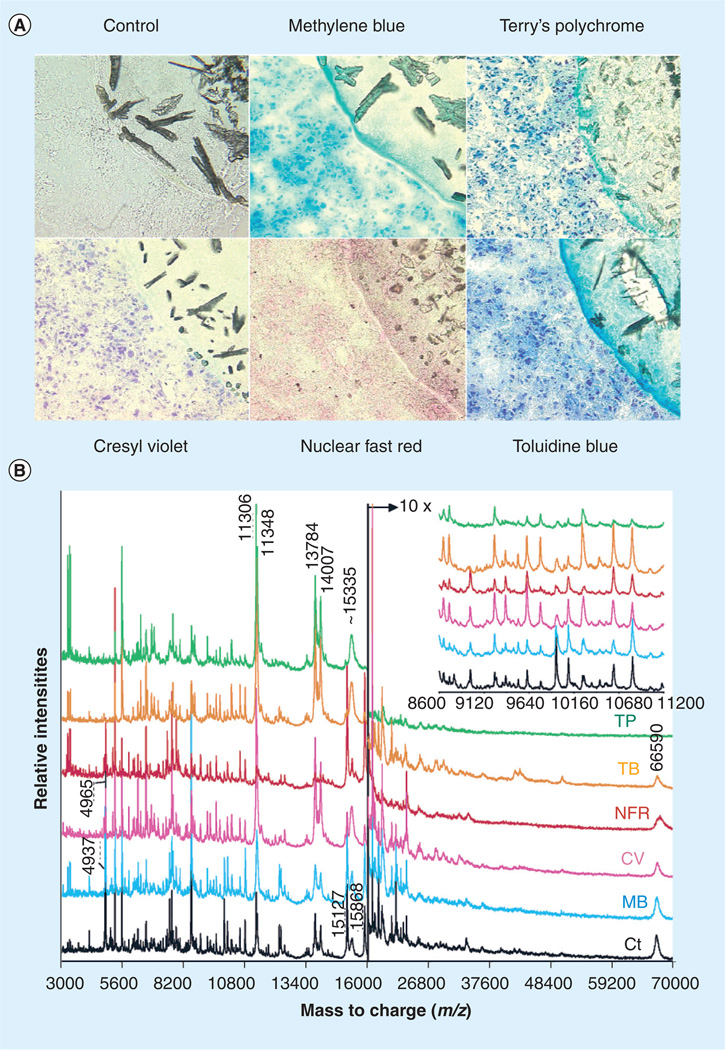
Chaurand et al. have investigated various histological stains and their compatibility with MS when used in the same tissue section [14]. The most popular histopathology stain, H&E, is unfortunately not compatible with MALDI MS analysis [17,18]. On the other hand, histopathological dyes such as cresyl violet and methylene blue are compatible with mass spectrometric analysis. Overall, it has been determined that water-based solvents, such as cresyl violet and Terry’s polychrome, show the strongest deviation from the control, while alcohol-based solvents such as methylene blue demonstrate better agreement with the control [14]. Figure 2 demonstrates various stains that have been tested for compatibility with MALDI MS imaging and the resulting profile spectra from each stained section. Protein distributions that have been obtained by MALDI MS images and cross referenced with histological stained sections that pathologists use to diagnose disease have shown the protein distribution in the MALDI MS image to be predictive of the diagnosis that still has yet to be made [13].
Figure 2. Histological stains and mass spectrometry.
(A) High-magnification (×400) photomicrographs of unstained (control) and stained 10-µm serial human glioma tissue sections. In the upper right corner of each image, the lower left edge of the matrix sample can be seen. (B) MALDI MS protein profiles acquired from unstained (Ct rinsed in 70 and 100% ethanol) and stained grade IV human glioma tissue sections.
Ct: Control section; CV: Cresyl violet; MB: Methylene blue; NFR: Nuclear fast red; TB: Toluidine blue; TP: Terry’s polychrome.
Reprinted with permission from [14]. © 2004 American Chemical Society.
Sample preparation methods for MALDI MS imaging
Sample preparation is an integral step in the MALDI MS imaging process and is often a limiting factor in spatial resolution. Contributing factors to this problem include matrix crystal size and homogeneity and analyte mobility. A strategy for minimizing the diffusion of analytes during matrix deposition involves fixation (chemical treatment) of the tissue before [19,20] or during [21] matrix deposition. It has also recently been shown by Marko-Varga et al. that no pretreatment of the tissue is also advantageous for the detection of the drug compounds erlotinib and gefitinib in adenocarcinoma cell tumors [22].
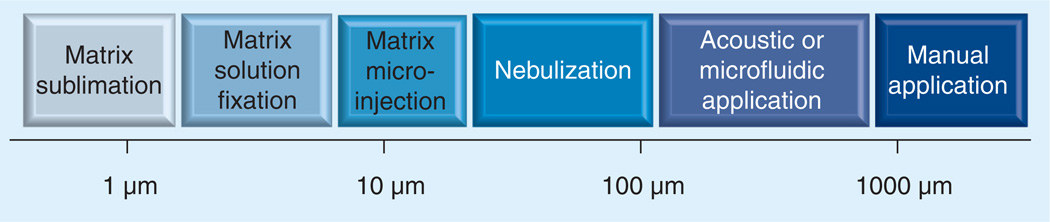
Various matrix deposition methods have been developed that focus on obtaining homogeneous matrix crystals and reducing analyte mobility, thus aiding in obtaining single-cell spatial resolution. Matrix deposition methods include nebulized spray coating [23], matrix sublimation [24], electrospray deposition [25], automated acoustic deposition [26], piezoelectric-based matrix inkjet printers [17,27], spray-droplet deposition [28], and vapor deposition (sublimation) coupled with matrix recrystallization [29]. The resolution limits of these methods are summarized in Figure 3. Reviews have also been published that summarize the advantages and disadvantages of these methods [23,30]. All of these fixation and matrix deposition methods are crucial in the quest to obtain single-cell resolution and each warrant a more in-depth description.
Figure 3. Spatial resolution of matrix deposition methods.
Reprinted from [31] with permission from Springer Science and Business Media.
Chemical treatment/fixation of tissue sections
The chemical treatment of tissue sections for MALDI MS imaging analysis can greatly affect the type of molecules that are detected and the sensitivity of analysis, and is therefore discussed here. Chemical treatment of tissue sections is a vital step in the sample preparation process for MALDI MS imaging of proteins, as it can remove lipids and biological salt adducts that may affect the observed protein signal. Conversely, chemical treatment can also be used to enhance lipid signal, depending upon the solvent used. Some solvents also serve as a way to preserve the tissue by dehydration and fixation, thereby minimizing lateral protein diffusion, one of the main hurdles to overcome in the MALDI MS imaging sample preparation process. Excised tissues are initially snap-frozen in either liquid nitrogen or a dry ice/ethanol bath. Tissue samples are then sectioned on a cryotome and mounted onto a surface such as an indium tin oxide-coated glass slide that can be analyzed by MALDI MS imaging. Fixation of the tissue can then be performed prior to matrix application [19,20] or during the matrix deposition process [21]. This process takes approximately 30 min to complete, depending upon the tissue drying time and solvent fixation time [20].
In addition to the widely accepted histological fixation/dehydration with a graded ethanol series (70% ethanol followed by 95% ethanol fixation), solvents such as acetone, isopropanol, methanol, chloroform, toluene, hexane, xylene, water and tertbutyl methyl ether (t-BME) have been investigated as potential chemical treatments prior to matrix application for MALDI MS imaging [19,20]. Lemaire et al. found an increase in signal detection for m/z >5000 for all solvents tested [19]. They found that all components investigated (signal intensity, signal-to-noise [S/N]) ratio, and number of peaks) were highest when tissue was treated with either xylene (44% increase in signal) or chloroform (40% increase in signal) prior to matrix application, as compared with untreated tissue. Furthermore, they confirmed that the solvents investigated did not perturb cellular structure by staining with known antibodies in serial tissue sections. The organic solvents investigated function as lipid extraction solvents; thus, they will not remove salt adducts from the tissue as a graded ethanol series has been shown to do [23].
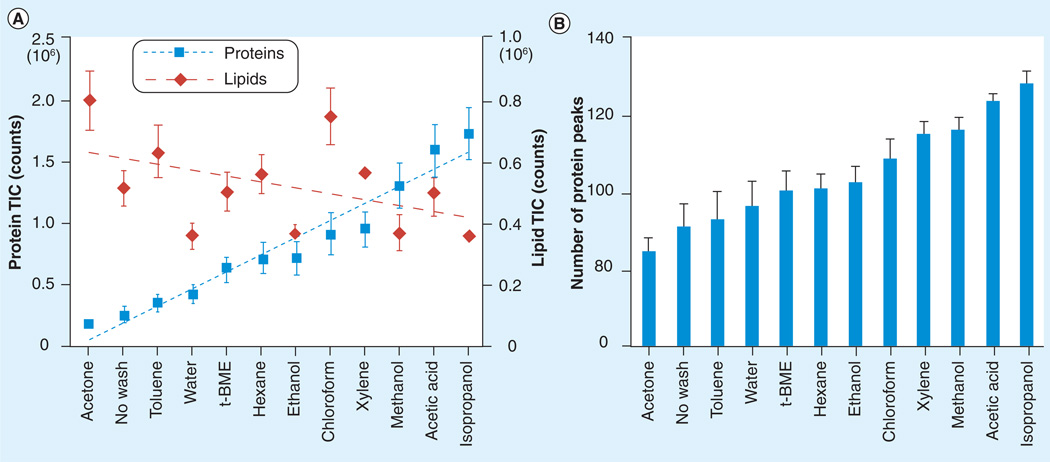
Seeley et al. assessed chemical treatment of tissue sections by comparing the resulting total ion current and number of peaks present in the profile spectra of each section [20]. They determined that alcohol-based washes, such as isopropanol, methanol and ethanol, along with water and acetic acid washes had the most positive effect on protein signal detection. In counterintuitive results, lipid extraction solvents such as chloroform had a substantial effect on enhancing lipid signal detection (Figure 4). They further investigated the effects of ethanol, isopropanol, acetic acid and water washes on tissue morphology and determined that ethanol and isopropanol allowed one to distinguish cell membranes, nuclei and nucleoli after H&E staining at 40× magnification. Acetic acid was found to have negative effects on the tissue, and water was found to completely disrupt the morphology of the tissue section. Their final conclusion was that ethanol and isopropanol were best suited to the coupling of histological staining and MALDI MS imaging.
Figure 4. Chemical treatment of tissue sections.
(A) TIC variations recorded from the MALDI-TOF mass spectrometry protein profiles acquired from serial mouse liver tissue sections either not washed or washed with different solvent systems in the m/z range from 500 to 1100 (lipid component) and m/z range from 2000 to 25,000 (protein component). (B) Number of peak variations as a function of the same washes for the protein component.
t-BME: Tertbutyl methyl ether; TIC: Total ion count.
Reprinted with permission from [20].
Agar et al. have developed a method that fixes the tissue during the matrix deposition process, termed matrix solution fixation [21,31]. Using this method, they observed that their method perturbed subcellular structures such as mitochondria, but preserved the overall tissue integrity. Their sample preparation technique offers decreased sample preparation time (no fixation required previous to matrix application) and is histology compatible.
Methods of matrix deposition
Care must be taken to avoid the lateral diffusion of analytes, which decreases the MALDI MS image resolution, during matrix deposition. Matrix deposition methods with the potential for single-cell analysis are described here.
Nebulized spray coating allows for matrix coverage of the entire tissue section, as opposed to acoustic/induction-based spotting procedures. Nebulized spray coating can also coat the tissue with a homogenous layer of a protease, such as trypsin or pepsin, allowing for on-tissue digestion. In this method, multiple cycles of matrix coating and drying steps are used in order to deposit thin layers of matrix across the entire tissue section while minimizing analyte diffusion [23]. This method can either be carried out manually with a thin-layer chromatography sprayer or with the aid of a manufactured system. If performed manually, the approach is both rapid and cost effective, but results tend to vary from person to person and experiment to experiment. If using a manufactured system such as the ImagePrep™ (Bruker Daltonics Inc., MA, USA) [31,32] or the TM-Sprayer™ (Leap Technologies, Inc., Carrboro, NC, USA), cost may become a limiting factor in the decision-making process for some researchers.
Matrix sublimation is a process by which solid matrix is sublimed upon application of heat in a flat-bottom condenser at reduced pressure. This method was compared with that of electrospray deposition and it was found that sublimation yields a more homogenous crystal layer upon the tissue, leading to the detection of compounds with a higher S/N ratio, and reduces salt adducts [24]. Matrix sublimation is easily one of the most cost-effective techniques other than manual deposition or the use of a thin-layer chromatography sprayer, as there is no need for any type of robotics or instrumentation to perform matrix deposition. The downside of this method is that owing to the lack of solvent, analyte cannot be readily incorporated into the matrix; therefore, this approach is most suited for small molecule/drug studies. In order to adapt this approach to peptide and protein detection, a recrystallization step is suggested when using sublimation as a deposition method [29,33].
Electrospray deposition uses a capillary spray set-up to coat an entire tissue section with matrix. This technique utilizes the potential difference between the charged electrospray emitter and that of the grounded steel MALDI MS target to apply matrix. The tissue section is then coated with multiple layers of matrix prior to drying under vacuum and MALDI MS imaging analysis. This approach has been used to image and identify peptides in invertebrate tissues [25]. Select groups have had trouble obtaining reproducible samples with this approach [23].
Acoustic deposition utilizes picoliter-sized volumes of matrix and matrix seeding in order to achieve high-resolution MALDI MS images in which the resolution is limited by the larger of the spot size and laser diameter, since analyte diffusion is limited to the deposited matrix spot. Picoliter volumes of matrix are spotted on the tissue in a predetermined array using a robotic spotter, yielding matrix spot diameters of approximately 130–150 µm. The resulting matrix crystals are similar to those observed in electrospray deposition [23]. The array of matrix spots is then imaged by selectively analyzing the spotted array using an in-house developed software. Using this method, each matrix spot is uniform in size and can be representative of a single pixel in the final MALDI MS image [26], depending upon the diameter of the matrix spot and lateral resolution in the MALDI MS image. Acoustic deposition also boasts the ability to deposit proteases such as trypsin and pepsin within a defined region on the tissue, allowing for targeted digestion and peptide analysis. Robotics available include the Portrait® (Labcyte Inc., Sunnyvale, CA, USA) and the TM iD™ (Leap Technologies, Inc., Carrboro, NC, USA). These fully automated systems make matrix deposition a facile process, but they are expensive and cost becomes an issue when deciding which method to use. The CHIP-1000™ (Shimadzu Corp., Kyoto, Japan) is an example of a piezoelectric chemical inkjet printer [17,27], a technology that is similar in function to acoustic deposition. The current spot size of approximately 150 µm does not afford single-mammalian cell resolution.
The spray-droplet deposition method introduced by Sugiura et al. combines a sprayed seed coating and acoustic deposition in one method [28]. They found that by combining both methods they were able to achieve, on average, a 9.9-fold increase in S/N ratio compared with conventional acoustic deposition. By first introducing a seed layer, they observed faster crystal formation (60 vs 90 s) compared with tissue that was not treated with a seed layer.
Bouschen et al. have combined matrix sublimation with matrix recrystallization to create a new matrix deposition method termed vapor deposition/recrystallization [29]. In this deposition method, a matrix is first applied to the surface of the tissue by sublimation and then the tissue is placed in a humid, saturated water atmosphere over a 24–72 h period in order to recrystallize the matrix. The researchers found this method to be more reproducible than using either electrospray deposition or pneumatic spray. The downside of this method is the time required to completely coat the tissue section with matrix. In work recently published by Yang and Caprioli, a spatial resolution of 10 µm was achieved using a matrix sublimation/recrystallization for imaging of proteins up to 30 kDa in size. This method requires approximately 10 min to coat and recrystallize matrix on the target of interest [33].
Methods of single-cell analysis
Various methods of single-cell analysis are currently under investigation by groups such as Agar, Agar, Caprioli, Li, Predel and Sweedler. Methods that allow for single-cell analysis include the microinjection of matrix onto the cell of interest in situ [31], micro- and induction-based fluidics (IBF) [34,35], laser capture micro-dissection (LCM) of individual cells [18,36,37], ordered stretching of tissue [38,39], and laser oversampling [40].
Most recently introduced is the in situ labeling of single cells by microinjection of matrix by Agar et al. [31]. This method allows for targeted proteomics of the cell of interest in its native environment. Cells of interest (in this case, motor neurons) are visualized using fluorescence in order to determine the intended location of matrix deposition. Matrix is deposited on the cell of interest with micropipettes through which the matrix is injected with a picoinjector or capillary action. Rhodamine can be added to the matrix solution in order to facilitate the visual detection of the matrix spot within the graphical user interface. The cell of interest can then be analyzed by MALDI-TOF MS. This method restricts analyte diffusion to the matrix spot, but does not require robotics and allows for targeted proteomics and peptidomics of a single cell.
Micro- and IBF allow for nanoliter (nl)-volume matrix spots. Both methods are based upon the same concept of minimizing analyte diffusion to the spot itself rather than across the tissue section. The main distinction between the two is that IBF utilizes a charged capillary tip to charge the matrix solution and ‘launch’ the droplet onto the tissue specimen, which is located on a grounded steel MALDI target. The resulting signal intensity using IBF was found to be approximately tenfold greater than that observed by manually depositing 0.5 µl of matrix solution by pipette [34].
Laser capture microdissection of individual cells is a method that allows the cell of interest to be excised from the tissue and analyzed separately, and is often used in proteomic analysis [41]. This is accomplished by using an infrared laser to accumulate the cells of interest. Xu et al. have used this technique and determined that a single excision with the LCM system (30 µm diameter tissue spot acquisition) yielded ten mouse crypt cel1ls. The excised cells were placed onto a MALDI target using conductive double-sided tape and a limited amount of matrix was then applied (100 pl–10 nl). Cells were then subjected to MALDI-TOF analysis. These ten mouse colon crypt cells were sufficient to generate a MALDI mass spectrum [18].
In the stretched sample approach (ordered stretching of tissue), glass beads are embedded on a Parafilm membrane that the tissue is then mounted upon. The Parafilm is then stretched, allowing for separation of the tissue section based upon the size of the glass bead. This method reduces analyte migration by physically tearing the tissue into cell-sized pieces for imaging analysis. The image can then be reconstructed through computational means [38,39]. This method was used to analyze the abdominal ganglion of Aplysia californica and the resulting spectra were found to be quite similar to those obtained from isolated single cells [39].
Laser oversampling involves moving the laser by spatial increments that are smaller than the diameter of the laser beam. If the matrix is ablated at a given location, the effect is that a 100 µm laser, when moved in 25 µm increments, yields 25 µm spatial resolution. In laser oversampling, resolution is not limited by the dimensions of the laser beam. Spatial resolution of approximately 25 µm was achieved using a laser with dimensions of approximately 100 µm × 200 µm. Sweedler and coworkers have used this method to investigate the distribution of acidic peptide and egg-laying hormone in isolated peptidergic neurons from the Aplysia californica CNS [40].
Micromanipulation techniques, such as the dissection of individual neurons, have been performed on various systems. Sweedler and coworkers have extensively researched the neuropeptidome of Aplysia californica (sea slug) through the use of microdissection and on-plate microextraction [40,42–49]. Other organisms studied using this method include Drosophila melanogaster (fruit fly) [50,51], Periplaneta americana (cockroach) [52,53], Helicoverpa zea and Manduca sexta (moth) [54,55], Lymnaea stagnalis (freshwater snail) [56–60], Xenopus laevis (frog) [61], Ixodes ricinus and Boophilus microplus (ticks) [62], Homarus americanus (crustacean) [63], neurons from the supraoptic nucleus of rats [64,65], individual cells from the rat pituitary [66] and mouse bone marrow-derived mast cells [67]. Most recently, research has also been carried out on single neurons of the parasitic nematode, Ascaris suum [68].
Analysis directed by cell-specific fluorescence
The coupling of MALDI MS imaging with fluorescence is especially pertinent for studies of neurodegenerative disorders such as Parkinson’s disease, Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), motor neuron disease and multiple sclerosis. In these disorders the death of a relatively small population of neurons begins the disease process. MALDI MS imaging studies have been conducted on a mouse model of Parkinson’s disease [12,69,70] and Alzheimer’s disease [71,72]. Transgenic mice exhibiting expression of fluorescent proteins, such as cyan fluorescent protein, yellow fluorescent protein (YFP), green fluorescent protein and so on, have been crossed with neurodegenerative disease-associated mice [73] in order to further visualize the cells that are affected by the disease. Obtaining MALDI MS images of these transgenic animals that exhibit fluorescence in a specific cell type would make a more targeted study possible, which could contribute to further elucidating neurodegenerative diseases at the cellular level.
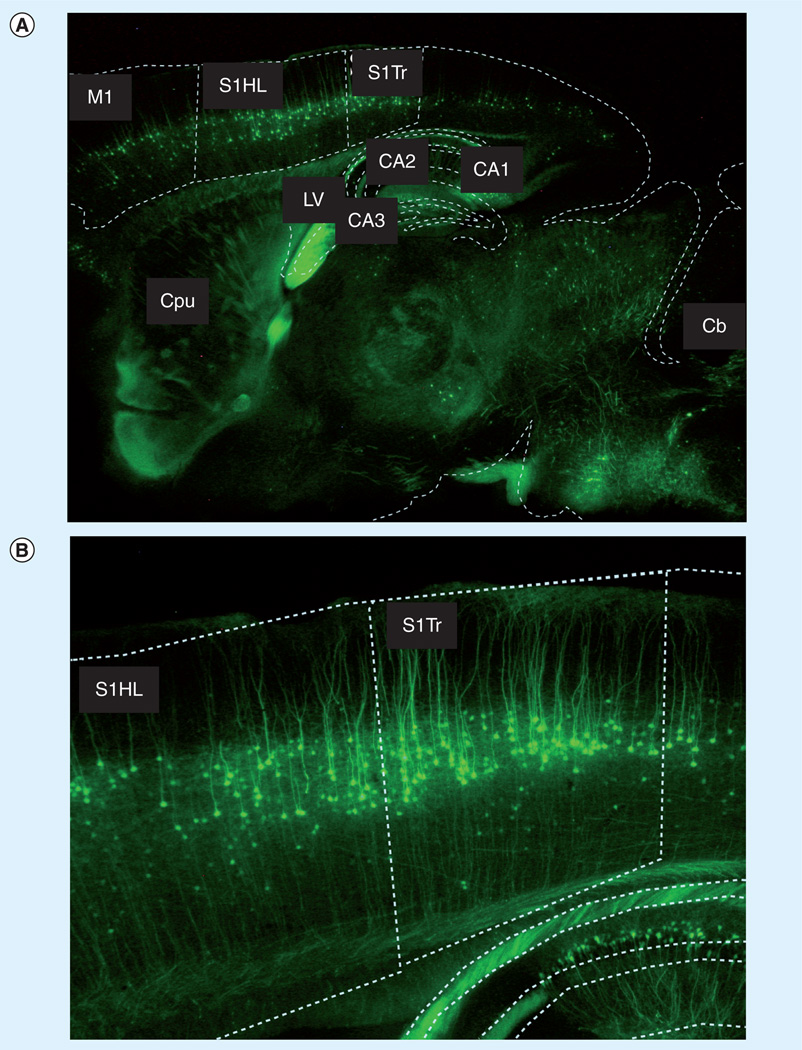
Amyotrophic lateral sclerosis is a disease that is characterized by the death of both upper (motor cortex) and lower (spinal cord) neurons. Through the combination of microinjection of matrix [31] with a transgenic mouse model that exhibits cell-specific fluorescence, it is possible to perform targeted proteomics studies on specific cells of interest, achieving cellular resolution of approxmiately 20 µm. In unpublished results, we have bred mice overexpressing YFP in motor neurons (B6.Cg-Tg(Thy1-YFP)16Jrs/J [74]) with hSOD1 (B6SJL-Tg(SOD1)2Gur/J [75]) [31], hSOD1G93A (B6.Cg-Tg(SOD1-G93A)1Gur/J [75]) [73], and hSOD1G85R (B6.Cg-Tg(SOD1-G85R)148Dwc/J [76]) ALS transgenic mice, allowing for the investigation of the molecular signature of the diseased motor neurons. All work was carried out in accordance with state and federal regulations and under the direction of the Foster Biomedical Research Laboratory (Brandeis University, MA, USA). It is estimated that the cell-specific fluorescence of the transgenic mice used offers fiduciary markers that can be used to identify a given region (e.g., the motor cortex) to within 1 mm. To identify regions of interest, we compare fluorescence-delineated anatomical boundaries with the anatomical boundaries delineated in the Mouse Brain Atlas (Figure 5) [77]. An example of a single-neuron spectrum from a layer V motor neuron in a YFP transgenic mouse is shown in Figure 6.
Figure 5. Registration of a fluorescence microscopy image with anatomical atlas of a mouse brain.
Fluorescence microscopy images of a sagittal brain section of a B6. Cg-Tg(Thy1-YFP)16Jrs/J mouse. Dashed white lines delineate different regions of the brain and are in the exact proportions of the anatomical boundaries of the 1.66 mm lateral sagittal section of the Mouse Brain Atlas. The degree of overlap between the experimentally determined, fluorescence-based anatomical boundaries and those of the Mouse Brain Atlas gives an estimate of location to within 1 mm. (A) M1: primary motor cortex; S1Tr: primary somatosensory cortex, trunk region; S1HL: primary somatosensory cortex, hind limb region; CPu: caudate putamen (striatum); LV: lateral ventricle; CA1–CA3: fields CA1–CA3 of the hippocampus, respectively. (B) Primary somatosensory cortex trunk region and hind limb region.
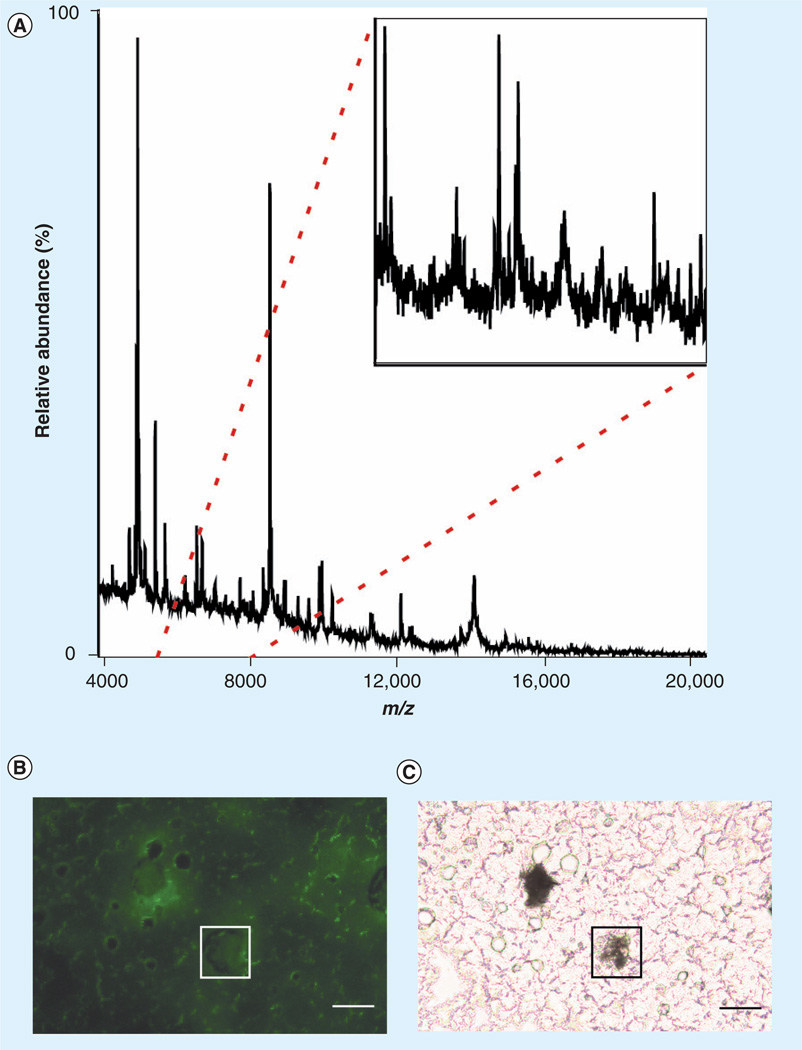
Figure 6. MALDI mass spectrometry of a single neuron.
(A) Raw data from a positive-ion mode mass spectrum of a single motor neuron from layer V of a B6. Cg-Tg(Thy1-YFP)16Jrs/J transgenic mouse motor cortex. Inset: 5730–8030 m/z. Inset depicts complexity of sample. (B) Using a modified microinjection setup, a single neuron was labeled with MALDI matrix (sinapinic acid). Fluorescence is used to visualize the location of the matrix in conjunction with the location of the neuron. In this image, fluorescence is not observed due to the matrix that is deposited on the neuron, as visualized in (C). Scale bar is equal to 100 µm. (C) Same microscopic field as in (B), visualized with bright field microscopy. Scale bar is equal to 100 µm.
We performed a microinjection of matrix on 97 neurons from layer V of the motor cortex of YFP transgenic mice for this study, with matrix spot size diameters in the range of 25–500 µm. Profile spectra were acquired on an UltrafleXtreme™ (Bruker Daltonics, Inc.) MALDI-TOF/TOF system equipped with a 1kHz Smartbeam™ Laser. All spectra were processed with FlexAnalysis and ClinProTools™ (Bruker Daltonics, Inc.).
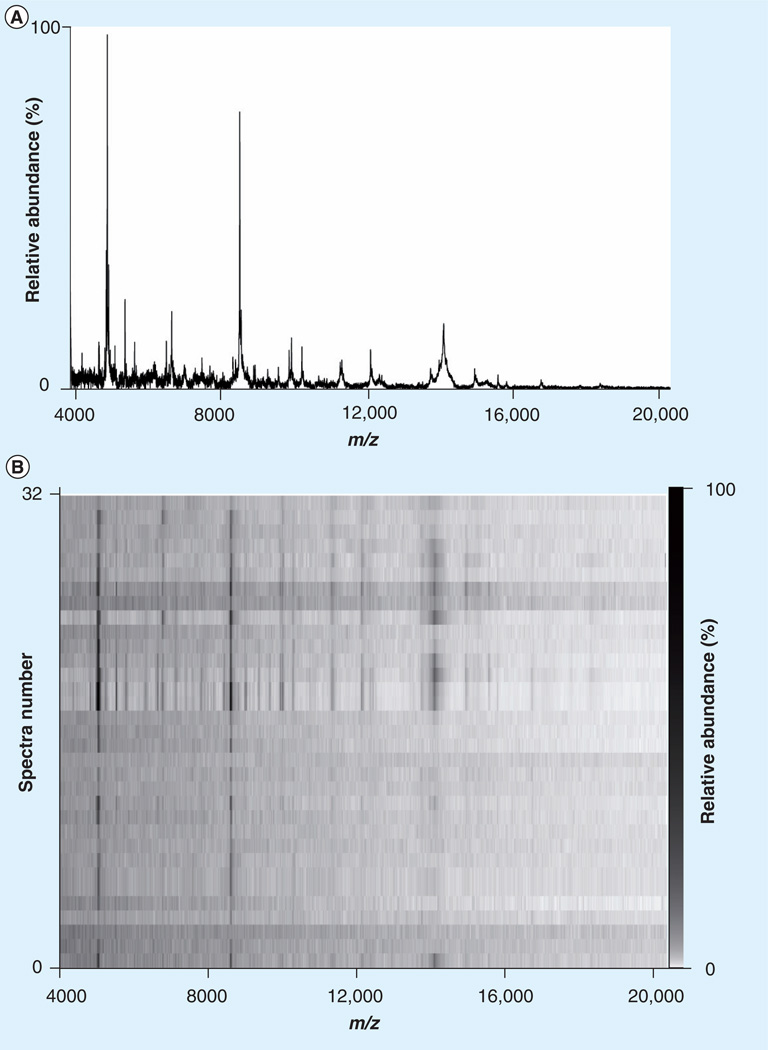
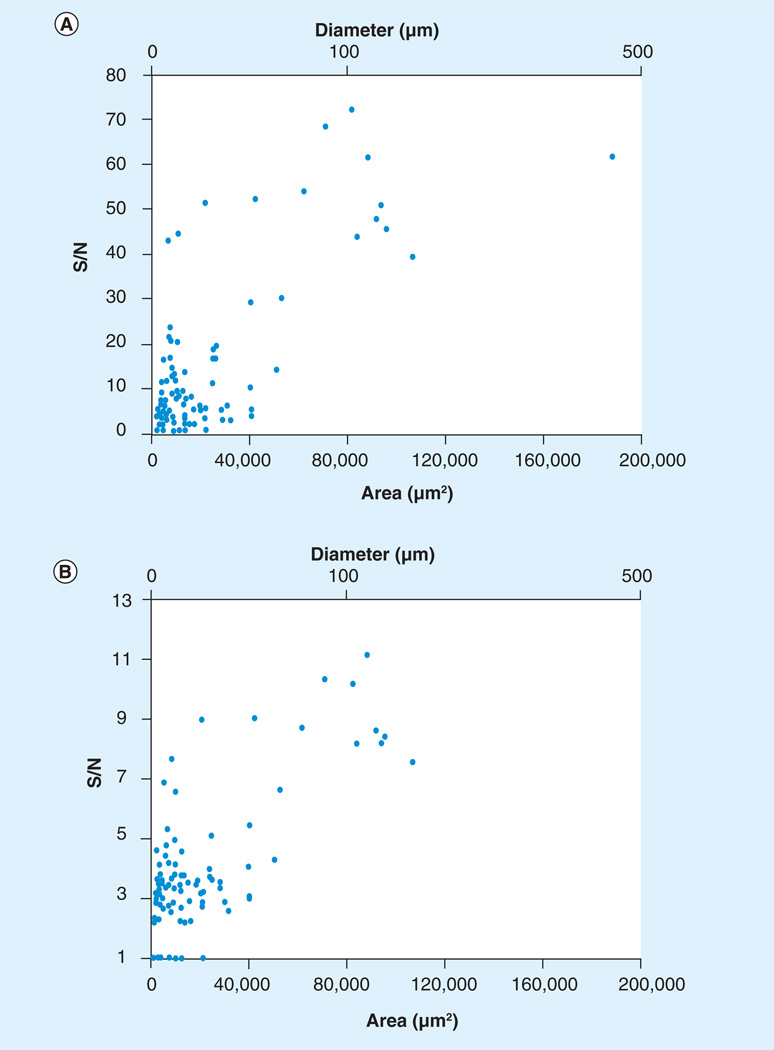
An average spectrum of 32 single-neuron spectra and the related pseudo-gel view depicting all 32 spectra are shown in Figure 7. In total, 91% of microinjected matrix spots yielded spectra having at least one peak with a S/N ratio greater than 2, and 36% yielded spectra had at least one peak with a S/N ratio greater than 10 (Figure 8). We found that neurons that were analyzed within 2 h of matrix deposition yielded spectra with a higher S/N than from preparations stored at −80°C overnight (data not shown). We find that although this method is valuable for performing targeted proteomics/peptidomics, the approach is limited by the distribution of surrounding cells and the reproducibility of the method, as demonstrated in Figure 8. The approach is viable for studying cells such as pyramidal cells that are approxmately 50 µm in diameter, but would not be suited to smaller organelles or subcellular imaging without further method development.
Figure 7. MALDI mass spectrometry of single neurons.
(A) Average spectrum of 32 cortical neurons from a B6.Cg-Tg(Thy1-YFP)16Jrs/J transgenic mouse. (B) Pseudo-gel view representation of the spectra of the 32 cortical neurons that were averaged for the spectra above. All spectra were baseline subtracted using a top-hat algorithm and were not smoothed.
Figure 8. Sensitivity of microdeposition.
(A) Maximum S/N versus microdeposition spot area. (B) Average S/N versus microdeposition spot area. Data points include samples that were transferred on both dry ice and at room temperature.
S/N: Signal-to-noise ratio.
Current advances in single-cell imaging
As stated previously, obtaining single-cell resolution in MALDI MS imaging is influenced by various factors such as matrix crystal size/homogeneity, analyte diffusion and current instrumentation. McDonnell et al. extensively review sensitivity in relation to MALDI MS imaging in their recent review. Of note is their determination of the amount of protein present in an image as a function of pixel size, tissue thickness and protein concentration [78]. Increasing spatial resolution implies sampling from smaller areas. In fact, the area and amount of analyte decreases as the square of the spatial resolution, requiring extremely sensitive instrumentation. In our own studies of single cells, instrument sensitivity is now the limiting factor.
High-resolution mass spectrometers, such as the Fourier transform ion cyclotron resonance mass spectrometer, are often coupled to a MALDI MS source and can produce images with a lateral resolution of 0.6 × 0.5 µm (when coupled to a scanning microprobe MALDI source) [79]. Various in-house mass spectrometers have also been fabricated that are able to attain single-cell resolution. One example of such an instrument is that presented by Spengler and Hubert. They introduce scanning microbe MALDI MS instrumentation that has a lateral resolution of 0.6–1.5 µm [80]. Luxembourg et al. have used ion microscopy coupled to a MALDI mass spectrometer to attain lateral resolution of 4 µm [81]. Chaurand et al. have modified a commercially available MALDI-TOF mass spectrometer with a two-stage ion source and obtained images at a scanning resolution less than 10 µm [82].
Much work has recently been carried out by Bruker Daltonics Inc. in the introduction of their new MALDI-TOF/TOF system, the UltrafleXtreme™ and the Smartbeam™ laser. The Smartbeam is a solid state laser with an adjustable repetition rate of up to 1 kHz. Using the UltrafleXtreme, Lagarrigue et al. have achieved 20 µm lateral resolution [83]. They are able to visualize various stages of germ cell development in testicular seminiferous tubules. To our knowledge, this is the highest resolution achieved on commercially available (as compared with in-house built as previously described) instrumentation.
Clinical impact of single-cell MALDI imaging
Since its inception, MALDI MS imaging has been used for biological, pathological and drug discovery applications. Examples of biological applications include one of the first MALDI MS imaging experiments performed by Caprioli et al. Researchers mapped the location of insulin in an islet of the rat pancreas, hormone peptides contained within the rat pituitary, and identified multiple proteins and peptides from human buccal mucosa cells [84]. Chaurand et al. have obtained MALDI MS images of a rat brain, along with profile spectra of various sections of mouse prostate tissue and human brain tissue [85]. Research has also been carried out on mapping proteins and peptides within the mouse brain and human glioblastomas [86]. Caldwell et al. have also identified an isoform of histone H4 in a MALDI MS image of a mouse brain with a tumor [87]. Molecular signatures produced by MS imaging of tissues have been shown to discriminate tumor grade [13,88], and could potentially be used for diagnostic and prognostic applications. A potential advantage of using MS imaging platforms for tissue evaluation is the ability to identify molecular changes associated with the tissue that are not yet detectable from histopathological evaluation. Numerous studies have been performed on cancerous tissues including lung [13,89,90], breast/mammary [91–94], oral [95], pancreatic [96], gastric [97], renal [98], colon [99], prostate [16,100,101], ovarian [102–104], Hodgkin’s lymphoma [105] and brain [88,106,107]. Resolving molecular information from cancerous tissue to the single-cell level will allow building of reference systems with more specific signatures of disease and healthy states. Single-cell imaging should consequently improve sensitivity and specificity in the classification of clinical specimens through the refinement of reference systems.
Along with diagnostic research, drug metabolism and small-molecule studies have also been conducted using MALDI MS imaging. This is arguably the fastest-growing MS application used by the pharmaceutical industry. The developing applications are based on the ability of MALDI MS imaging to differentiate drugs from their metabolites, and to determine to what extent the drug has penetrated its intended target. Whole-body images of rodents have been acquired, allowing the researcher to follow the drug and its metabolites throughout the entire animal without the need to label drugs [108–111]. Numerous studies have also been published regarding single-cell metabolomics [112–115]. In addition, to provide simultaneous molecular and spatial resolution for pharmacokinetics studies, the MS imaging experiment has the potential to render information regarding the drug effect at the molecular level. While the drug target might not always be readily detectable, it is very likely that the analysis could detect an overall change in cellular signatures indicative of drug effect. Recent advances in single-cell metabolomics are covered by Svatoš [116], emphasizing the importance of the ability to analyze at the single-cell level. This information would be useful in understanding how drug metabolites are processed at the cellular level.
Another utility of single-cell imaging exists in the analysis of tumor stem cells (TSCs). TSCs have been isolated and characterized for a number of neoplasms [117] and one of their defining characteristics is their tumor-initiating potential. The TSCs initiate and maintain tumor growth by producing a more differentiated state of cancer cells and sustaining their own niche, giving rise to a heterogeneous population of TSCs and cancer cells. With a growing arsenal of therapies targeting such mechanisms, the task rests on the analytical community to increase classification resolution. MS imaging has proven itself to be capable of discriminating healthy from diseased in many systems, and single-cell resolution could potentially enable the realization of personalized treatment by providing detailed information on the molecular mechanisms driving a tumor at the cellular level.
Expert commentary & five-year view
A better MS microscope
The current limiting factor in ultrahigh-resolution MS is instrument sensitivity, and our ‘crystal ball’ is clouded by the secrecy that surrounds new instrument development. If new, ultrasensitive mass spectrometers are developed, laser focal diameters approaching the diffraction limit (~170 nm for N2 lasers) should become possible, enabling resolution of organelles. Oversampling should improve resolution by approximately fivefold [40], which would then require piezoelectric X–Y positioning, and would result in approximately 30 nm resolution, approaching that of large molecules and membranes. As more groups perform top-down MS studies, the molecular identities of what are currently only m/z values in the mass spectrum will become known, adding biological relevance to analysis.
Widespread adoption by the drug-development industry
In addition, the growth in high m/z resolution mass spectrometer-based (Fourier transform MS) MALDI MS imaging is enabling the detection of a drug and approximately ten metabolites. Decoupling of a drug from its metabolites is not easily achieved by autoradiography, the current industry standard for drug imaging, and we therefore expect the widespread adoption of high m/z resolution MALDI MS imaging for industry absorption, distribution, metabolism, excretion and toxicity studies.
Key issues.
Single-cell MALDI mass spectrometry (MS) imaging resolution is currently limited by the size of laser spot diameter, matrix crystal size/homogeneity, displacement of analytes during matrix deposition, and by instrument sensitivity at high spatial resolution.
Matrix deposition and cell isolation methods available for single-cell MALDI MS imaging include nebulized spray coating, matrix sublimation, electrospray deposition, automated acoustic deposition, piezoelectric-based matrix inkjet printers, spray-droplet deposition, vapor deposition coupled with matrix recrystallization, microdeposition of matrix onto the cell of interest in situ, micro- and induction-based fluidics, laser capture microdissection of individual cells, ordered stretching of tissue and laser oversampling.
Single-cell MALDI MS images can be coupled with other imaging modalities, such as immunohistochemistry and fluorescence microscopy, to further elucidate molecular changes.
Acknowledgements
The authors would like to thank members of the Agar laboratory for critical reading and comments on this manuscript. The authors would also like to thank Roman Pavlyuk for initial technical work on the YFP mouse colony. We acknowledge the Brandeis University Animal Care Facility for care of transgenic mice used in this study.
This work was supported by the Amyotrophic Lateral Sclerosis Association (grant number 1856) (to Jeffrey N Agar), and the Brain Science Foundation and the Daniel E Ponton Fund for the Neurosciences (to Nathalie YR Agar). Matrix solution fixation is the subject of a patent pending with Jeffrey N Agar and Nathalie YR Agar as inventors.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.Sinha TK, Khatib-Shahidi S, Yankeelov TE, et al. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat. Methods. 2008;5(1):57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCombie G, Staab D, Stoeckli M, Knochenmuss R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal. Chem. 2005;77(19):6118–6124. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- 3.Deininger SO, Ebert MP, Futterer A, Gerhard M, Rocken C. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J. Proteome Res. 2008;7(12):5230–5236. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- 4.Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. Vitamin E imaging and localization in the neuronal membrane. J. Am. Chem. Soc. 2005;127(35):12152–12153. doi: 10.1021/ja051223y. [DOI] [PubMed] [Google Scholar]
- 5.Lechene C, Hillion F, McMahon G, et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J. Biol. 2006;5(6):20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 7.Talaty N, Takats Z, Cooks RG. Rapid in situ detection of alkaloids in plant tissue under ambient conditions using desorption electrospray ionization. Analyst. 2005;130(12):1624–1633. doi: 10.1039/b511161g. [DOI] [PubMed] [Google Scholar]
- 8.Shrestha B, Vertes A. In situ metabolic profiling of single cells by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 2009;81(20):8265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 9.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal. Chem. 2007;79(21):8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 10.Mayer P. Mitteilungen aus der Zoologischen station zu neapel (Vol. 12) Berlin, Germany: 1896. [Google Scholar]
- 11.Lillie RD. Histopathologic Technic and Practical Histochemistry. NY, USA: McGraw-Hill Book Co.; 1965. [Google Scholar]
- 12.Pierson J, Norris JL, Aerni HR, Svenningsson P, Caprioli RM, Andren PE. Molecular profiling of experimental Parkinson’s disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J. Proteome Res. 2004;3(2):289–295. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- 13.Yanagisawa K, Shyr Y, Xu BJ, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362(9382):433–439. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 14.Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal. Chem. 2004;76(4):1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 15.Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16(7):1093–1099. doi: 10.1016/j.jasms.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Schwamborn K, Krieg RC, Reska M, Jakse G, Knuechel R, Wellmann A. Identifying prostate carcinoma by MALDI-Imaging. Int. J. Mol. Med. 2007;20(2):155–159. [PubMed] [Google Scholar]
- 17.Nakanishi T, Ohtsu I, Furuta M, Ando E, Nishimura O. Direct MS/MS analysis of proteins blotted on membranes by a matrix-assisted laser desorption/ionization-quadrupole ion trap-time-of-flight tandem mass spectrometer. J. Proteome Res. 2005;4(3):743–747. doi: 10.1021/pr0497834. [DOI] [PubMed] [Google Scholar]
- 18.Xu BGJ, Caprioli RM. Direct analysis of laser capture microdissected cells by MALDI mass spectrometry. J. Am. Soc. Mass Spectr. 2002;13(11):1292–1297. doi: 10.1016/S1044-0305(02)00644-X. [DOI] [PubMed] [Google Scholar]
- 19.Lemaire R, Wisztorski M, Desmons A, et al. MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal. Chem. 2006;78(20):7145–7153. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- 20. Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J. Am. Soc. Mass Spectrom. 2008;19(8):1069–1077. doi: 10.1016/j.jasms.2008.03.016.. • Assessment of the effect of chemical washes prior to MALDI imaging analysis as a means to increase signal-to-noise of proteins and/or lipids
- 21.Agar NY, Yang HW, Carroll RS, Black PM, Agar JN. Matrix solution fixation: histology-compatible tissue preparation for MALDI mass spectrometry imaging. Anal. Chem. 2007;79(19):7416–7423. doi: 10.1021/ac071460e. [DOI] [PubMed] [Google Scholar]
- 22.Marko-Varga G, Fehniger TE, Rezeli M, Dome B, Laurell T, Vegvari A. Drug localization in different lung cancer phenotypes by MALDI mass spectrometry imaging. J. Proteomics. 2011;74(7):982–992. doi: 10.1016/j.jprot.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 2003;38(7):699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 24.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007;18(9):1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse R, Sweedler JV. Spatial profiling invertebrate ganglia using MALDI MS. J. Am. Soc. Mass Spectrom. 2003;14(7):752–759. doi: 10.1016/S1044-0305(03)00288-5. [DOI] [PubMed] [Google Scholar]
- 26.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 2006;78(3):827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 27.Sloane AJ, Duff JL, Wilson NL, et al. High throughput peptide mass fingerprinting and protein macroarray analysis using chemical printing strategies. Mol. Cell. Proteomics. 2002;1(7):490–499. doi: 10.1074/mcp.m200020-mcp200. [DOI] [PubMed] [Google Scholar]
- 28.Sugiura Y, Shimma S, Setou M. Two-step matrix application technique to improve ionization efficiency for matrix-assisted laser desorption/ionization in imaging mass spectrometry. Anal. Chem. 2006;78(24):8227–8235. doi: 10.1021/ac060974v. [DOI] [PubMed] [Google Scholar]
- 29.Bouschen W, Schulz O, Eikel D, Spengler B. Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun. Mass Spectrom. 2010;24(3):355–364. doi: 10.1002/rcm.4401. [DOI] [PubMed] [Google Scholar]
- 30. Kaletas BK, van der Wiel IM, Stauber J, et al. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9(10):2622–2633. doi: 10.1002/pmic.200800364.. • Broad review of MALDI imaging methods, including a comparison of various robotic spotting and spraying techniques
- 31.Agar NY, Kowalski JM, Kowalski PJ, Wong JH, Agar JN. Tissue preparation for the in situ MALDI MS imaging of proteins, lipids, and small molecules at cellular resolution. Methods Mol. Biol. 2010;656:415–431. doi: 10.1007/978-1-60761-746-4_24. [DOI] [PubMed] [Google Scholar]
- 32.Schuerenberg M, Luebbert C, Deininger S, Ketterlinus R, Suckau D. MALDI tissue imaging: mass spectrometric localization of biomarkers in tissue slices. Nat. Methods. 2007;4:iii–iv. [Google Scholar]
- 33.Yang J, Caprioli RM. Matrix sublimation/ recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 2011;83(14):5728–5734. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu T, Sauter AD, Jr, Sauter AD 3rd, Gross ML. Improving the signal intensity and sensitivity of MALDI mass spectrometry by using nanoliter spots deposited by induction-based fluidics. J. Am. Soc. Mass Spectrom. 2008;19(8):1086–1090. doi: 10.1016/j.jasms.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Tu T, Gross ML. Miniaturizing sample spots for matrix-assisted laser desorption/ ionization mass spectrometry. Trends Analyt. Chem. 2009;28(7):833–841. doi: 10.1016/j.trac.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer-Toy DE, Sarracino DA, Sgroi D, LeVangie R, Leopold PE. Direct acquisition of matrix-assisted laser desorption/ ionization time-of-flight mass spectra from laser capture microdissected tissues. Clin. Chem. 2000;46(9):1513–1516. [PubMed] [Google Scholar]
- 37.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J. Mass Spectrom. 2001;36(4):355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 38.Zimmerman TA, Monroe EB, Sweedler JV. Adapting the stretched sample method from tissue profiling to imaging. Proteomics. 2008;8(18):3809–3815. doi: 10.1002/pmic.200800331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monroe EB, Jurchen JC, Koszczuk BA, Losh JL, Rubakhin SS, Sweedler JV. Massively parallel sample preparation for the MALDI MS analyses of tissues. Anal. Chem. 2006;78(19):6826–6832. doi: 10.1021/ac060652r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. J. Am. Soc. Mass Spectrom. 2005;16(10):1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Mustafa D, Kros JM, Luider T. Combining laser capture microdissection and proteomics techniques. Methods Mol. Biol. 2008;428:159–178. doi: 10.1007/978-1-59745-117-8_9. [DOI] [PubMed] [Google Scholar]
- 42.Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. Excess salt removal with matrix rinsing: direct peptide profiling of neurons from marine invertebrates using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 1996;31(10):1126–1130. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Garden RW, Romanova EV, Sweedler JV. In situ sequencing of peptides from biological tissues and single cells using MALDI-PSD/CID analysis. Anal. Chem. 1999;71(24):5451–5458. doi: 10.1021/ac9907181. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Romanova EV, Rubakhin SS, et al. Peptide profiling of cells with multiple gene products: combining immunochemistry and MALDI mass spectrometry with on-plate microextraction. Anal. Chem. 2000;72(16):3867–3874. doi: 10.1021/ac000260z. [DOI] [PubMed] [Google Scholar]
- 45.Rubakhin SS, Garden RW, Fuller RR, Sweedler JV. Measuring the peptides in individual organelles with mass spectrometry. Nat. Biotechnol. 2000;18(2):172–175. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- 46.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal. Chem. 2003;75(20):5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 47.Rubakhin SS, Sweedler JV. Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal. Chem. 2008;80(18):7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubakhin SS, Li L, Moroz TP, Sweedler JV. Characterization of the Aplysia californica cerebral ganglion F cluster. J. Neurophysiol. 1999;81(3):1251–1260. doi: 10.1152/jn.1999.81.3.1251. [DOI] [PubMed] [Google Scholar]
- 49.Garden RW, Shippy SA, Li L, Moroz TP, Sweedler JV. Proteolytic processing of the Aplysia egg-laying hormone prohormone. Proc. Natl Acad. Sci. USA. 1998;95(7):3972–3977. doi: 10.1073/pnas.95.7.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neupert S, Johard HA, Nassel DR, Predel R. Single-cell peptidomics of drosophila melanogaster neurons identified by Gal4-driven fluorescence. Anal. Chem. 2007;79(10):3690–3694. doi: 10.1021/ac062411p. [DOI] [PubMed] [Google Scholar]
- 51.Neupert S, Predel R. Peptidomic analysis of single identified neurons. Methods Mol. Biol. 2010;615:137–144. doi: 10.1007/978-1-60761-535-4_11. [DOI] [PubMed] [Google Scholar]
- 52.Neupert S, Predel R. Mass spectrometric analysis of single identified neurons of an insect. Biochem. Biophys. Res. Commun. 2005;327(3):640–645. doi: 10.1016/j.bbrc.2004.12.086. [DOI] [PubMed] [Google Scholar]
- 53.Neupert S, Schattschneider S, Predel R. Allatotropin-related peptide in cockroaches: identification via mass spectrometric analysis of single identified neurons. Peptides. 2009;30(3):489–494. doi: 10.1016/j.peptides.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Ma PW, Garden RW, Niermann JT, O’ Connor M, Sweedler JV, Roelofs WL. Characterizing the Hez-PBAN gene products in neuronal clusters with immunocytochemistry and MALDI MS. J. Insect Physiol. 2000;46(3):221–230. doi: 10.1016/s0022-1910(99)00174-2. [DOI] [PubMed] [Google Scholar]
- 55.Neupert S, Huetteroth W, Schachtner J, Predel R. Conservation of the function counts: homologous neurons express sequence-related neuropeptides that originate from different genes. J. Neurochem. 2009;111(3):757–765. doi: 10.1111/j.1471-4159.2009.06361.x. [DOI] [PubMed] [Google Scholar]
- 56.Jimenez CR, Li KW, Dreisewerd K, et al. Direct mass spectrometric peptide profiling and sequencing of single neurons reveals differential peptide patterns in a small neuronal network. Biochemistry. 1998;37(7):2070–2076. doi: 10.1021/bi971848b. [DOI] [PubMed] [Google Scholar]
- 57.El Filali Z, Hornshaw M, Smit AB, Li KW. Retrograde labeling of single neurons in conjunction with MALDI high-energy collision-induced dissociation MS/MS analysis for peptide profiling and structural characterization. Anal. Chem. 2003;75(13):2996–3000. doi: 10.1021/ac034057q. [DOI] [PubMed] [Google Scholar]
- 58.Jimenez CR, van Veelen PA, Li KW, et al. Neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption ionization mass spectrometry of single neurons. J. Neurochem. 1994;62(1):404–407. doi: 10.1046/j.1471-4159.1994.62010404.x. [DOI] [PubMed] [Google Scholar]
- 59.de With ND, Li KW, Jimenez CR, et al. Intracellular degradation of C-peptides in molluscan neurons producing insulin-related hormones. Peptides. 1997;18(6):765–770. doi: 10.1016/s0196-9781(97)00020-x. [DOI] [PubMed] [Google Scholar]
- 60.Li KW, Hoek RM, Smith F, et al. Direct peptide profiling by mass spectrometry of single identified neurons reveals complex neuropeptide-processing pattern. J. Biol. Chem. 1994;269(48):30288–30292. [PubMed] [Google Scholar]
- 61.van Strien FJ, Jespersen S, van der Greef J, Jenks BG, Roubos EW. Identification of POMC processing products in single melanotrope cells by matrix-assisted laser desorption/ionization mass spectrometry. FEBS Lett. 1996;379(2):165–170. doi: 10.1016/0014-5793(95)01503-5. [DOI] [PubMed] [Google Scholar]
- 62.Neupert S, Predel R, Russell WK, Davies R, Pietrantonio PV, Nachman RJ. Identification of tick periviscerokinin, the first neurohormone of Ixodidae: single cell analysis by means of MALDI-TOF/TOF mass spectrometry. Biochem. Biophys. Res. Commun. 2005;338(4):1860–1864. doi: 10.1016/j.bbrc.2005.10.165. [DOI] [PubMed] [Google Scholar]
- 63.Li L, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV, Marder E. Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs. J. Comp. Neurol. 2002;444(3):227–244. doi: 10.1002/cne.10139. [DOI] [PubMed] [Google Scholar]
- 64.Millet LJ, Bora A, Sweedler JV, Gillette MU. Direct cellular peptidomics of supraoptic magnocellular and hippocampal neurons in low-density co-cultures. ACS Chem. Neurosci. 2010;1(1):36–48. doi: 10.1021/cn9000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bora A, Annangudi SP, Millet LJ, et al. Neuropeptidomics of the supraoptic rat nucleus. J. Proteome Res. 2008;7(11):4992–5003. doi: 10.1021/pr800394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat. Protoc. 2007;2(8):1987–1997. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- 67.Shimizu M, Ojima N, Ohnishi H, Shingaki T, Hirakawa Y, Masujima T. Development of the single-cell MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) mass-spectroscopic assay. Anal. Sci. 2003;19(1):49–53. doi: 10.2116/analsci.19.49. [DOI] [PubMed] [Google Scholar]
- 68.Jarecki JL, Andersen K, Konop CJ, Knickelbine JJ, Vestling MM, Stretton AO. Mapping neuropeptide expression by mass spectrometry in single dissected identified neurons from the dorsal ganglion of the nematode. Ascaris suum. ACS Chem. Neurosci. 2010;1(7):505–519. doi: 10.1021/cn1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skold K, Svensson M, Nilsson A, et al. Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J. Proteome Res. 2006;5(2):262–269. doi: 10.1021/pr050281f. [DOI] [PubMed] [Google Scholar]
- 70.Stauber J, Lemaire R, Franck J, et al. MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J. Proteome Res. 2008;7(3):969–978. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- 71.Stoeckli M, Staab D, Staufenbiel M, Wiederhold KH, Signor L. Molecular imaging of amyloid β peptides in mouse brain sections using mass spectrometry. Anal. Biochem. 2002;311(1):33–39. doi: 10.1016/s0003-2697(02)00386-x. [DOI] [PubMed] [Google Scholar]
- 72.Rohner TC, Staab D, Stoeckli M. MALDI mass spectrometric imaging of biological tissue sections. Mech. Ageing Dev. 2005;126(1):177–185. doi: 10.1016/j.mad.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 73.Ozdinler PH, Benn S, Yamamoto TH, Guzel M, Brown RH, Jr, Macklis JD. Corticospinal motor neurons and related subcerebral projection neurons undergo early and specific neurodegeneration in hSOD1G(3)A transgenic ALS mice. J. Neurosci. 2011;31(11):4166–4177. doi: 10.1523/JNEUROSCI.4184-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng G, Mellor RH, Bernstein M, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28(1):41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 75.Gurney ME, Pu H, Chiu AY, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264(5166):1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 76.Bruijn LI, Becher MW, Lee MK, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18(2):327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 77.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 78.McDonnell LA, Corthals GL, Willems SM, van Remoortere A, van Zeijl RJ, Deelder AM. Peptide and protein imaging mass spectrometry in cancer research. J. Proteomics. 2010;73(10):1921–1944. doi: 10.1016/j.jprot.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 79.Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B. A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer. Rapid. Commun. Mass Spectrom. 2008;22(20):3275–3285. doi: 10.1002/rcm.3733. [DOI] [PubMed] [Google Scholar]
- 80.Spengler B, Hubert M. Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J. Am. Soc. Mass Spectrom. 2002;13(6):735–748. doi: 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- 81.Luxembourg SL, Mize TH, McDonnell LA, Heeren RM. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal. Chem. 2004;76(18):5339–5344. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 82.Chaurand P, Schriver KE, Caprioli RM. Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J. Mass Spectrom. 2007;42(4):476–489. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- 83.Lagarrigue M, Becker M, Lavigne R, et al. Revisiting rat spermatogenesis with MALDI imaging at 20-microm resolution. Mol. Cell Proteomics. 2011;10(3) doi: 10.1074/mcp.M110.005991. M110 005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 85.Chaurand P, Schwartz SA, Caprioli RM. Imaging mass spectrometry: a new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr. Opin. Chem. Biol. 2002;6(5):676–681. doi: 10.1016/s1367-5931(02)00370-8. [DOI] [PubMed] [Google Scholar]
- 86.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 2001;7(4):493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 87.Caldwell RL, Caprioli RM. Tissue profiling by mass spectrometry: a review of methodology and applications. Mol. Cell Proteomics. 2005;4(4):394–401. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Schwartz SA, Weil RJ, Thompson RC, et al. Proteomic-based prognosis of brain tumor patients using direct-tissue matrix-assisted laser desorption ionization mass spectrometry. Cancer Res. 2005;65(17):7674–7681. doi: 10.1158/0008-5472.CAN-04-3016. [DOI] [PubMed] [Google Scholar]
- 89.Groseclose MR, Massion PP, Chaurand P, Caprioli RM. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics. 2008;8(18):3715–3724. doi: 10.1002/pmic.200800495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yanagisawa K, Xu BJ, Carbone DP, Caprioli RM. Molecular fingerprinting in human lung cancer. Clin. Lung Cancer. 2003;5(2):113–118. doi: 10.3816/CLC.2003.n.025. [DOI] [PubMed] [Google Scholar]
- 91.Bauer JA, Chakravarthy AB, Rosenbluth JM, et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin. Cancer Res. 2010;16(2):681–690. doi: 10.1158/1078-0432.CCR-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie L, Xu BJ, Gorska AE, et al. Genomic and proteomic analysis of mammary tumors arising in transgenic mice. J. Proteome Res. 2005;4(6):2088–2098. doi: 10.1021/pr050214l. [DOI] [PubMed] [Google Scholar]
- 93.Rauser S, Marquardt C, Balluff B, et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J. Proteome Res. 2010;9(4):1854–1863. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- 94.Reyzer ML, Caldwell RL, Dugger TC, et al. Early changes in protein expression detected by mass spectrometry predict tumor response to molecular therapeutics. Cancer Res. 2004;64(24):9093–9100. doi: 10.1158/0008-5472.CAN-04-2231. [DOI] [PubMed] [Google Scholar]
- 95.Patel SA, Barnes A, Loftus N, et al. Imaging mass spectrometry using chemical inkjet printing reveals differential protein expression in human oral squamous cell carcinoma. Analyst. 2009;134(2):301–307. doi: 10.1039/b812533c. [DOI] [PubMed] [Google Scholar]
- 96.Djidja MC, Claude E, Snel MF, et al. MALDI-ion mobility separation-mass spectrometry imaging of glucose-regulated protein 78 kDa (Grp78) in human formalin-fixed, paraffin-embedded pancreatic adenocarcinoma tissue sections. J. Proteome Res. 2009;8(10):4876–4884. doi: 10.1021/pr900522m. [DOI] [PubMed] [Google Scholar]
- 97.Morita Y, Ikegami K, Goto-Inoue N, et al. Imaging mass spectrometry of gastric carcinoma in formalin-fixed paraffin-embedded tissue microarray. Cancer Sci. 2010;101(1):267–273. doi: 10.1111/j.1349-7006.2009.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oppenheimer SR, Mi D, Sanders ME, Caprioli RM. Molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J. Proteome Res. 2010;9(5):2182–2190. doi: 10.1021/pr900936z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chaurand P, DaGue BB, Pearsall RS, Threadgill DW, Caprioli RM. Profiling proteins from azoxymethane-induced colon tumors at the molecular level by matrix-assisted laser desorption/ionization mass spectrometry. Proteomics. 2001;1(10):1320–1326. doi: 10.1002/1615-9861(200110)1:10<1320::AID-PROT1320>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 100.Cazares LH, Troyer D, Mendrinos S, et al. Imaging mass spectrometry of a specific fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin. Cancer Res. 2009;15(17):5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- 101.Masumori N, Thomas TZ, Chaurand P, et al. A probasin-large T antigen transgenic mouse line develops prostate adenocarcinoma and neuroendocrine carcinoma with metastatic potential. Cancer Res. 2001;61(5):2239–2249. [PubMed] [Google Scholar]
- 102.Kang S, Shim HS, Lee JS, et al. Molecular proteomics imaging of tumor interfaces by mass spectrometry. J. Proteome Res. 2010;9(2):1157–1164. doi: 10.1021/pr900666q. [DOI] [PubMed] [Google Scholar]
- 103.Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J. Proteome Res. 2007;6(4):1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 104.El Ayed M, Bonnel D, Longuespee R, et al. MALDI imaging mass spectrometry in ovarian cancer for tracking, identifying, and validating biomarkers. Med. Sci. Monit. 2010;16(8):BR233–BR245. [PubMed] [Google Scholar]
- 105.Schwamborn K, Krieg RC, Jirak P, et al. Application of MALDI imaging for the diagnosis of classical Hodgkin lymphoma. J. Cancer Res. Clin. Oncol. 2010;136(11):1651–1655. doi: 10.1007/s00432-010-0823-x. [DOI] [PubMed] [Google Scholar]
- 106.Agar NY, Malcolm JG, Mohan V, et al. Imaging of meningioma progression by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Anal. Chem. 2010;82(7):2621–2625. doi: 10.1021/ac100113w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schwartz SA, Weil RJ, Johnson MD, Toms SA, Caprioli RM. Protein profiling in brain tumors using mass spectrometry: feasibility of a new technique for the analysis of protein expression. Clin. Cancer Res. 2004;10(3):981–987. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- 108.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem. 2006;78(18):6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 109.Stoeckli M, Staab D, Schweitzer A, Gardiner J, Seebach D. Imaging of a b-peptide distribution in whole-body mice sections by MALDI mass spectrometry. J. Am. Soc. Mass Spectrom. 2007;18(11):1921–1924. doi: 10.1016/j.jasms.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 110.Trim PJ, Henson CM, Avery JL, et al. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal. Chem. 2008;80(22):8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- 111.Chaurand P, Cornett DS, Angel PM, Caprioli RM. Perspective: from whole-body sections down to cellular level, multiscale imaging of phospholipids by MALDI mass spectrometry. Mol. Cell Proteomics. 2010;10(2) doi: 10.1074/mcp.O110.004259. O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R. Single-cell MALDI-MS as an analytical tool for studying intrapopulation metabolic heterogeneity of unicellular organisms. Anal. Chem. 2010;82(17):7394–7400. doi: 10.1021/ac1015326. [DOI] [PubMed] [Google Scholar]
- 113.Amantonico A, Urban PL, Zenobi R. Analytical techniques for single-cell metabolomics: state of the art and trends. Anal. Bioanal. Chem. 2010;398(6):2493–2504. doi: 10.1007/s00216-010-3850-1. [DOI] [PubMed] [Google Scholar]
- 114.Heinemann M, Zenobi R. Single cell metabolomics. Curr. Opin. Biotechnol. 2010;22(1):26–31. doi: 10.1016/j.copbio.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 115.Miura D, Fujimura Y, Yamato M, et al. Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Anal. Chem. 2010;82(23):9789–1996. doi: 10.1021/ac101998z. [DOI] [PubMed] [Google Scholar]
- 116.Svatos A. Single-cell metabolomics comes of age: new developments in mass spectrometry profiling and imaging. Anal. Chem. 2011;83(13):5037–5044. doi: 10.1021/ac2003592. [DOI] [PubMed] [Google Scholar]
- 117.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 118.Schwamborn K, Caprioli RM. MALDI imaging mass spectrometry – painting molecular pictures. Mol. Oncol. 2010;4(6):529–538. doi: 10.1016/j.molonc.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]