Summary
Spatial and temporal expression of specific basic Helix-Loop-Helix (bHLH) transcription factors defines many types of differentiation. We find that the much broader expression of the heterodimer partners of these specific factors is also highly regulated, through a distinct mechanism. A cross-interacting regulatory network links expression of the Drosophila E-protein Daughterless, required to heterodimerize with bHLH proteins, with expression of the Id protein Extramacrochaetae, an antagonist of bHLH proteins. Coupled transcriptional feedback loops maintain the widespread Extramacrochaetae expression that restrains Daughterless expression, opposing bHLH-dependent differentiation while enhancing growth and cell survival. Where extracellular signals repress extramacrochaetae, Daughterless expression is then able to increase. This defines regions of proneural ectoderm, independently from the proneural bHLH genes. Similar regulation is found in multiple Drosophila tissues and in mammalian cells, and may be a conserved general feature of developmental regulation by HLH proteins.
Introduction
Many cell types are defined by expression of Helix-Loop-Helix (HLH) transcription factors. In one of the first examples, the basic HLH (bHLH) protein MyoD was identified as a gene singly able to transform fibroblasts into myoblasts in cultured cells (Lassar et al., 1986; Davis et al., 1987; Weintraub et al., 1989). In parallel, achaete, scute, and lethal of scute, three genes in the Achaete-Scute Complex (AS-C) from Drosophila, were characterized as bHLH proteins both necessary for specification of peripheral neurons in normal development and able when mis-expressed to confer neuronal fate on other ectodermal cells (Garcia-Bellido, 1979; Villares and Cabrera, 1987; Murre et al., 1989a,b; Gomez-Skarmeta et al., 2003; Garcia-Bellido and de Celis, 2009). These and other bHLH transcription factors, such as the MASH gene family, MATH-family, MyoD-family, Myogenin, NeuroD, Neurogenins, eHAND and dHAND, BETA2, Tal1, NSCL-1/2, and Hen1, are now known to regulate conserved programs in myogenesis, neurogenesis, hematopoiesis, heart development, and pancreas development (Massari and Murre, 2000). These have been categorized as ‘Class II’ bHLH factors based on their properties and expression in specific areas during development to confer new developmental fates or potentials (Massari and Murre, 2000).
These powerful transcription factors do not act alone. Each Class II bHLH protein generally functions as a heterodimer with one of the much more broadly expressed ‘Class I’ bHLH proteins. The established paradigm is that highly regulated transcription of the Class II protein confers spatial and temporal specificity, while the broadly-expressed Class I proteins contribute to DNA binding and transcriptional activation (Henthorn et al., 1990; Aronheim et al., 1993). The vertebrate Class I proteins are E12, E47, E2-2 and HEB, collectively required for the Class II-dependent aspects of myogenesis, neurogenesis, hematopoiesis, heart development, and pancreas development, as well as for myelopoiesis, lymphopoiesis, and cell cycle control (Massari and Murre, 2000; Rothschild et al., 2006; Kee, 2009). Drosophila has a single Class I protein, Daughterless (Da), required for the neuronal differentiation, sex determination, and mesoderm development mediated by specific bHLH partners including the Achaete-Scute gene family, Atonal, Amos, SisB, MyoD and others (Murre et al., 1989b; Murre et al., 1994; Goulding et al., 2000; Huang et al., 2000; Massari and Murre, 2000).
Another class of broadly expressed HLH proteins are negative regulators. These Class V HLH proteins include Extramacrochaetae (Emc) in Drosophila, and four Inhibitor of DNA binding (Id) proteins in mammals. Class V HLH proteins lack any basic domain. As a consequence, Class V HLH protein heterodimers with Class I and Class II proteins are unable to bind DNA and cannot function (Benezra et al., 1990; Ellis et al., 1990; Garrell and Modolell, 1990; Campuzano, 2001). Id proteins antagonize Class I and Class II proteins in the processes listed above (Massari and Murre, 2000; Ross et al., 2003; Kee, 2009; Schotte et al., 2010; Lee et al., 2011). In Drosophila, widespread expression of Emc is thought to set a threshold for neurogenesis that only a certain level of AS-C/Da heterodimers can exceed (Cubas and Modolell, 1992; Van Doren et al., 1992).
The highly-regulated transcription of the Class II genes has been studied intensely. The broad expression patterns of Class I and Class V genes have not suggested comparable regulation. Most Drosophila epithelia express both Da and Emc, and many mammalian cells express one or more of each class of protein. It has been suggested that expression levels of Class I and Class V proteins might define precise thresholds for differentiation in response to Class II proteins, but this has not been tested directly (Vaessin et al., 1994; Brown et al., 1995; Ik Tsen Heng and Tan, 2003).
Deletion of the Drosophila Class I gene da precludes function by Class II proteins such as Achaete and Scute, so that da is required for most neurogenesis. Drosophila has a single Class V protein encoded by emc, but studies of emc null mutations have been limited because even clones of cells homozygous for emc null mutations do not survive in imaginal discs, suggesting a role in cell growth or survival (Garcia Alonso, 1988). The conclusion that Emc antagonizes Class II proneural genes is based on studies of partial loss of emc function (Botas et al., 1982; Ellis et al., 1990; Garrell and Modolell, 1990).
Recently, we found that large clones of imaginal disc cells completely null for emc function were recovered when the surrounding cells were heterozygous for a mutation in RpS17 (Bhattacharya and Baker, 2009). This shows that emc is not absolutely required for cell division or survival, although it contributes to the competitive success of cells in vivo. The phenotypes of the emc null mutant clones obtained are stronger than observed with hypomorphic alleles (Bhattacharya and Baker, 2009).
The present study addressed Drosophila eye development and other tissues where ‘proneural regions’ where neural progenitor cells can arise are defined by localized expression of proneural bHLH genes (Gomez-Skarmeta et al., 2003). The Class II bHLH gene for retinal neurogenesis is atonal (ato) whereas many other parts of the Drosophila nervous system are specified by multiple AS-C proneural genes. Transcription of ato, and eye differentiation, begin at the posterior margin of the eye imaginal disc, the epithelial primordium for the adult head. The extracellular signals Hh and Dpp drive a wave of ato expression that spreads anteriorly until the whole retina is differentiating. Notch signaling and other lateral inhibitors restrict ato expression to a spaced array of R8 photoreceptor precursor neurons within the ‘morphogenetic furrow’, an indentation in the epithelium that moves anteriorly as differentiation progresses. Once specified, each R8 neuron recruits multiple other retinal cell types (Wolff, 1993; Roignant and Treisman, 2009). In addition to the relative genetic simplicity of Drosophila eye development, its progressive nature conveniently reveals developmental dynamics, since each eye imaginal disc contains an posterior-to-anterior sequence of cells that initiated the eye differentiation program at progressively later times.
We report a previously unrecognized cross-interacting regulatory network that locks together Class I and Class V gene expression. The circuit is broken by extracellular signaling pathways so that levels of Da and Emc change to accompany differentiation. This novel network provides regulation of HLH gene expression that is essential to proper patterning but parallel to the Class II proneural bHLH genes, eg the proneural region of the eye istill alters expression of Da and Emc even when the proneural gene ato is deleted. Such a regulatory network has potentially wide implications, given the diverse roles of bHLH genes. We find a similar network acting in many Drosophila tissues and in mammalian cells, and we hypothesize that Class I/Class V networks are a general feature of bHLH-dependent differentiation.
Results
da and emc regulate each others expression
Because recent observations in the Drosophila eye indicate that emc has effects where ato is not yet expressed, we examined the effects of emc null mutations on da. Da is the heterodimer partner of Ato (Jarman et al., 1993).
Da is expressed ubiquitously. Its expression level is largely uniform in the imaginal discs, but is elevated in proneural regions where Class II bHLH genes such as ato or members of the AS-C confer neural potential (Figure 1A-C). Compared to normal levels, Da was dramatically elevated in emc null clones(Fig. 1D). In addition, when emc was absent, Da levels were identical within and outside proneural regions. We observed similar results from multiple tissues including eye, antennal, leg and wing imaginal discs and their peripodial epithelia (Figure 1D and S1). These findings established Emc as a general negative regulator of Da expression, and showed that the spatial differences in Da levels seen in normal development depend on emc.
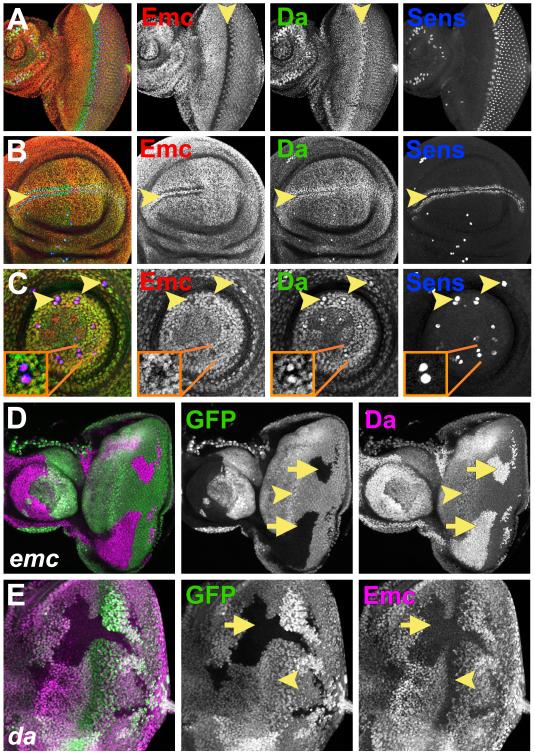
Figure 1. Emc and Da expression depend on each other’s activities.
(A) In eye imaginal discs Emc (red) and Da (green) were expressed in all cells anterior and posterior to the furrow. At the morphogenetic furrow (arrowhead), Emc expression goes down and Da goes up.. Blue labels the differentiating R8 cells with the neural precursor marker Senseless (Sens) (Nolo et al., 2000)).
(B) Emc (red) goes down and Da (green) up in proneural regions of the wing imaginal disc (arrowhead). (C) Emc (red) goes down and Da (green) up in SOP cells in prothoracic leg discs (arrowheads). Insets show enlargement.
In (D-E) homozygous mutant clones lack GFP expression (green).
(D) Clones of homozygous emc null cells had Da (magenta) higher than in the morphogenetic furrow (arrowhead).
(E) In da cells, Emc (magenta) was almost completely lost (arrow). Emc is normally low at the morphogenetic furrow (arrowhead).
Genotypes: (A-C) w1118; (D) ywhsF; emcAP6 FRT80/ [UbiGFP] M(3)67C FRT80; (E) ywhsF; da10 FRT40/ [UbiGFP] FRT40. See also Figure S1.
Emc and Da levels normally vary reciprocally. Emc is lower in proneural regions where Da is higher (Figure 1A-C). We therefore tested whether Da reciprocally inhibits Emc expression. On the contrary, very little Emc expression was detected in da null mutant clones anywhere in the imaginal discs or their peripodial epithelia (Fig. 1E and S1). Thus, da was required for Emc expression. These findings established that Emc and Da proteins were the major regulators of each other’s expression, but that reciprocal changes in Da and Emc levels were not caused by a ‘toggle-switch’ of mutual antagonism. Instead, there was an unsuspected negative feedback loop, in which Da was responsible for expression of its competitive inhibitor, Emc.
A Da-dependent enhancer regulates da transcription
Emc cannot bind DNA and regulates gene expression by blocking DNA binding by bHLH proteins (Massari and Murre, 2000). The fact that Emc repressed Da so widely suggested a role of Da in its own expression. Consistent with this notion, suggested previously on genetic grounds (Smith and Cronmiller, 2001), Da homodimers can bind to DNA (Cabrera and Alonso, 1991; Van Doren et al., 1991), and the homologous mammalian protein E47 forms a homodimeric transcription factor in B-cells (Murre et al., 1989b; Benezra, 1994; Shen and Kadesch, 1995). Emc and its homologs can heterodimerize with Da and its homologs, and prevent their binding to DNA (Benezra et al., 1990; Ellis et al., 1990).
To test a role of Da in da transcription, Da was expressed using a UAS-da transgene that lacks the non-coding first exon (Hinz et al., 1994), which was therefore present only in transcripts from the endogenous gene. When Da was over-expressed in the differentiating portion of the eye-antennal discs using GMR-Gal4, RT-PCR analysis detected 3x more endogenous da mRNA than in GMR-Gal4 controls lacking the UAS-da transgene (Figure 2A-C). This must underestimate the actual stimulation of da transcription, as not all eye disc cells express GMR-Gal4. The emc transcript was elevated 5x in the same experiment, consistent with Da-dependent emc transcription as well (Figure 2C). Note that, since Emc inhibits Da function, Da-induced Emc over-expression potentially limits how much endogenous da and emc genes can respond to Da over-expression. These findings show that Da regulates both da and emc at the transcriptional level.
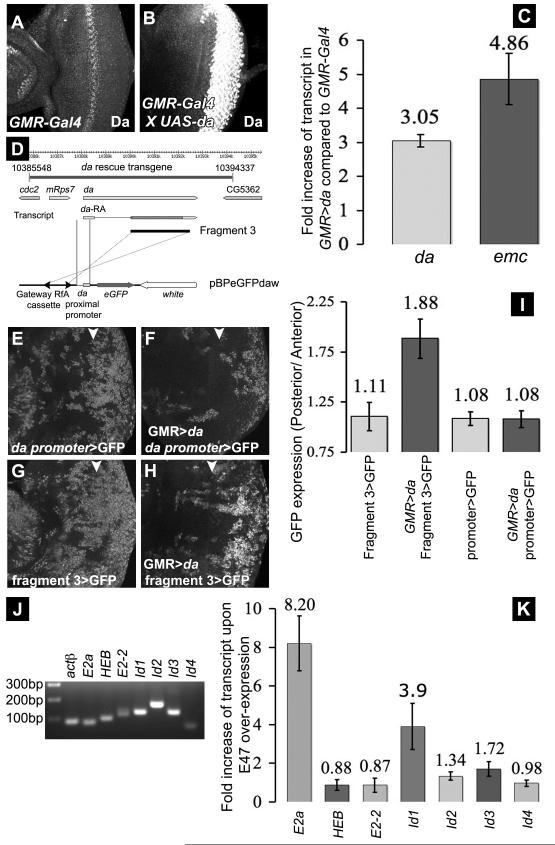
Figure 2. da stimulates its own transcription and that of emc.
(A) Da expression in wild type eye disc (w1118) (B) Da in GMR-Gal4, UAS-da, recorded in parallel to panel A. (C) Levels of endogenous da and emc transcripts in GMR-Gal4, UAS-da determined using quantitative RT-PCR, compared to their levels in GMR-Gal4 controls. (D) Representation of the da genomic region, showing the da+ genomic transgene. The da proximal promoter region (genomic location 2L: 10387806 - 10388342) was combined with Fragment 3 in GFP-reporter constructs in the pBPeGFPdaw plasmid. Panels (E-I) show analysis of da regulatory DNA in eye discs from transgenic flies. Arrowhead indicate the morphogenetic furrow. All similar transgenes exhibit variegated (patchy) GFP expression, as did a da-Gal4 transgene described previously, perhaps because they lack a probable insulator element at the 3′ end of the endogenous da gene (Figure S2B, C). (E, F) da proximal promoter region transgene. GFP expression was not changed by Da over-expression posterior to the furrow using GMR-Gal4. (G, H) Transgene incorporating Fragment 3 enhancer. GFP levels were elevated posterior to the furrow in GMR-Gal4, UAS-da. (I) Ratios of GFP level (pixel intensity) posterior to anterior of the furrow. (J) Semi-quantitative RT-PCR of actin, E-protein and Id gene transcripts in HEK293T cells. First lane shows size markers. (K) E-protein and id gene transcript levels in HEK293T cells transfected with E47 compared to empty vector controls. . Transfection with very little E47-expression construct was effective. Transfection of 33-fold more DNA is normal and induced E2a transcription by ~200x. See also Figure S2.
To define the mechanism of da transcription in more detail, an 8.8kb genomic transgene was generated that conferred normal Da expression and rescued da null mutant genotypes to normal, fertile adults, showing that it contains all sequences essential for da expression and function (Figure 2D and data not shown). Sections of this genomic DNA were first analyzed for enhancer activity by transient transfection into Drosophila S2 cells (Figure S2A). The ~2.5kb ‘Fragment 3’ segment (Figure 2D) stimulated transcription ~8-fold when a Da-expression construct was also transfected (Figure S2B). Da-dependent transcriptional activation was blocked by co-transfection of an Emc expression construct, suggesting a requirement for DNA-binding by Da protein (data not shown). Fragment 3 is conserved in other Drosophila species, although this is to be expected since it includes much of the da coding region.
The putative Da-dependent enhancer was tested in vivo following Drosophila germline transformation. GFP-reporter constructs made use of the da proximal promoter region including the predicted transcription start site (Figure 2D). This 542 bp promoter conferred broad GFP expression throughout imaginal discs that was unaltered in GMR-Gal4, UAS-da (Figure 2E, F, I). By contrast, incorporating the putative enhancer Fragment 3 led to elevated GFP expression posterior to the furrow in GMR-Gal4, UAS-da (Figure 2G-I). Fragment 3 therefore had properties of an enhancer that responded to Da in manner inhibitable by Emc, consistent with the model that Da regulates its own expression.
A Class I/Class V HLH network in mammalian cells
To investigate whether a similar network acts in mammalian cells, we used the transformed human embryonic kidney cell line HEK293T. All the human paralogs of Da and Emc were expressed in HEK293T cells (Figure 2J). To test the potential of an E protein to regulate expression of these genes, we over-expressed E47 protein in HEK293T cells. Transcription of the endogenous E2a gene (which encodes both E47 and E12 proteins via alternative splicing) was elevated ~8x by transfected E47. The other E-protein genes, HEB and E2-2, were unaffected (Figure 2K). In the same experiment, Id1 transcripts were elevated ~4x by E47 transfection. The level of other id transcripts did not alter significantly (Figure 2K). These data show that E protein-dependent transcription of an E-protein gene and an Id-protein gene occurs in a human cell line, similar to the Da-dependent transcription of da and emc observed in Drosophila.
Extracellular signaling pathways change Emc and Da expression in the morphogenetic furrow
It is puzzling that Emc levels drop where Da levels are highest, eg in the morphogenetic furrow and other proneural regions (Figure 1A-C). We hypothesized that other signals must be active at these locations to alter Da and Emc levels separately. To test the most widely used pathways, we analyzed clones of cells mutated for receptor or signal transduction components of the Dpp, Hh, N, EGFR and Wg pathways for Emc and Da levels in the morphogenetic furrow. Each of these receptor or signal transduction components should be required cell-autonomously if the respective signaling pathway is involved.
A mutation of the signal transduction component Smoothened (smo) was used to assess the contribution of Hh signaling. The smo clones retained some Emc expression in the morphogenetic furrow, and upregulated Da to a lesser degree than wild type cells (Figure 3A). Thus, Hh signaling contributes to changes of Emc and Da levels in the morphogenetic furrow. Mutations of the signal transducer Mad or transcription factor Schnurri (shn) were used to assess the contribution of Dpp signaling. Like smo, both Mad and shn clones retained some Emc expression in the morphogenetic furrow, and also upregulated Da less (Figures 3B, C and Supplemental Figure S3A,B). Thus, Dpp also contributes to changes in the furrow.
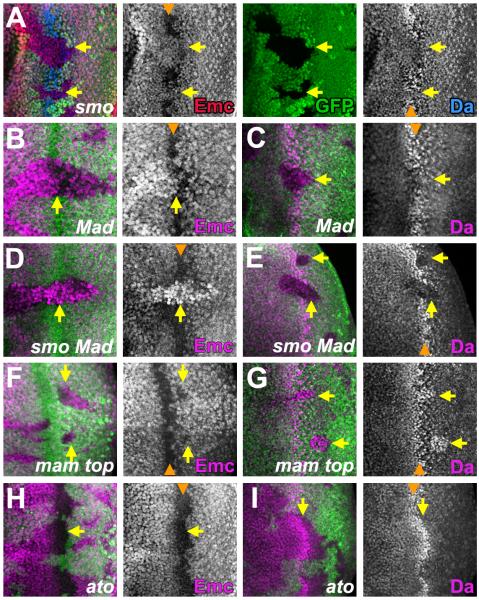
Figure 3. Extracellular signals regulate Emc and Da independent of ato.
Mutant clones lack either GFP or β-gal (green). In (B-I) Emc, Da or Ato proteins are shown in magenta. Orange arrowheads indicate position of the morphogenetic furrow.
(A) In smo cells that cannot respond to Hh, some Emc expression (red) persisted in the furrow and Da (blue) increased less (arrows).
(B,C) In Mad clones that cannot respond to Dpp, Emc expression was maintained for longer in the furrow and Da increased less (arrows).
(D, E) In smo Mad double mutant cells that respond to neither Hh nor Dpp, Emc was higher than that in adjacent cells and Da lower. Neither changed in the furrow (arrows).
(F, G) In mam top clones that lack N and EGFR signaling, Emc and Da levels changed at the furrow but the return of Emc and reduction of Da behind the furrow was delayed(arrows).
(H, I) In ato clones, Emc and Da levels changed at the furrow (arrows) but took longer to return to normal behind the furrow.
Genotypes: (A) ywhsF; smo FRT40/ M(2)24F [arm-lacZ] FRT40; (B and C) ywhsF; Mad FRT40/ M(2)24F [arm-lacZ] FRT40; (D and E)) ywhsF; smo Mad FRT40/ M(2)24F [arm-lacZ] FRT40; (F and G) ywhsF; FRT42 mam top/ FRT42 [arm-lacZ] M(2)56F; (H and I) ywhsF; FRT82 ato/ FRT82 [UbiGFP] M(3)96C. See also Figure S3.
We examined the consequences of blocking Hh and Dpp pathways simultaneously in smo Mad clones. Such cells expressed more Emc than wild type cells ahead of the furrow and maintained this same level in the morphogenetic furrow (Figures 3D). Ahead of the furrow, smo Mad cells expressed Da at levels slightly lower than wild type cells; this Da level remained unchanged in the furrow (Figures 3E). These findings confirm that activity of both Hh and Dpp pathways is required to elevate Da and eliminate Emc from the morphogenetic furrow, and indicate that no changes in Da or Emc levels occur when both these pathways are blocked. Hh and Dpp signaling are already known to peak in the furrow and coordinate several other developmental processes there (Roignant and Treisman, 2009).
None of the N, EGFR or Wg pathways appeared to affect Da or Emc levels in the morphogenetic furrow, because Emc and Da levels did not change there in clones mutant for the Wg co-receptor arrow (arr), the EGF receptor torpedo (top), or the DNA-binding and co-activator proteins for N-regulated transcription, Suppressor-of Hairless (Su(H)) and mastermind (mam) (Figure S3C, D and data not shown). A previous study found that ectopic Wg signaling repressed da and inhibited furrow progression (Cadigan et al., 2002). We found that clones doubly-mutant for both arr and shn had normal Emc and Da patterns, unlike clones mutant for shn alone (Figures S3E, F). This indicated that Wg signaling does affect Emc and Da expression if Dpp signaling is inactivated. Therefore, part of the normal role of Dpp signaling was to prevent Wg from maintaining pre-furrow expression levels. By contrast, clones mutant for smo and arr resembled smo mutant clones (data not shown) and clones mutant for smo, arr and shn resembled smo Mad double-mutant clones (Figure S3G, H), indicating that Hh affected Emc and Da levels independently of Wg.
Although neither N nor EGFR signaling affected Emc and Da levels in the morphogenetic furrow, their combined activity was required for Emc and Da levels to return to normal more posteriorly. In clones mutant for both mam and top, defective for N and EGFR signaling, Da and Emc levels did not return to pre-furrow levels until column 6 or 7 posterior to the furrow (Figure 3F, G). Both N and EGFR signaling are activate posterior to the furrow and known to play many roles in this differentiating region (Roignant and Treisman, 2009). It is possible that EGFR and N signaling act directly on the da or emc genes, or restore Emc and Da to their previous levels by terminating signaling responses to Hh and Dpp (Baker et al 2009).
Taken together, these findings established that the levels of Emc and Da in the morphogenetic furrow differ because of the local activity of particular extracellular signaling pathways.
Emc and Da levels change independently of proneural bHLH activity
Hh and Dpp are already known to promote expression of proneural bHLH gene ato, which peaks in the morphogenetic furrow (Greenwood and Struhl, 1999; Curtiss and Mlodzik, 2000). Ato is required to specify the R8 neurons, and thereby required indirectly for most retinal differentiation (Jarman et al., 1994). Hh and Dpp do not regulate ato through da, because da is not required for initial ato expression (Brown et al., 1996). To test whether Hh and Dpp regulated da and emc through ato, clones of cells lacking Ato function were examined. The ato mutant clones still lost Emc and up-regulated Da in the morphogenetic furrow (Figures 3H, I). Taken together, these findings showed that Hh and Dpp regulated Emc and Da independently of Ato.
Emc is the primary target of extracellular signals
Da and Emc changed reciprocally in all mutant genotypes described above, suggesting their responses might not be independent. Da could rise as a consequence of reduced Emc, given that Emc repressed da expression. If this was correct, maintaining Emc would prevent Da rising. To maintain Emc expression in the morphogenetic furrow, a Flip-on method was used to express Emc in clones from a Gal4-regulated Expressed-P (EP) insertion. The level of Emc obtained in the furrow was comparable to the Emc level normally seen ahead of the furrow (Figure 4A). Maintaining Emc expression cell-autonomously prevented any increase in Da within the furrow (Figure 4B). Outside of the furrow, the small increment in Emc levels had no discernible effect on the Da level. These results indicated that low Emc elevated Da levels in the morphogenetic furrow, and that extracellular signals primarily targeted emc to change the expression of Emc and Da.
Figure 4. Maintaining Emc in the furrow is sufficient to prevent elevation of Da.
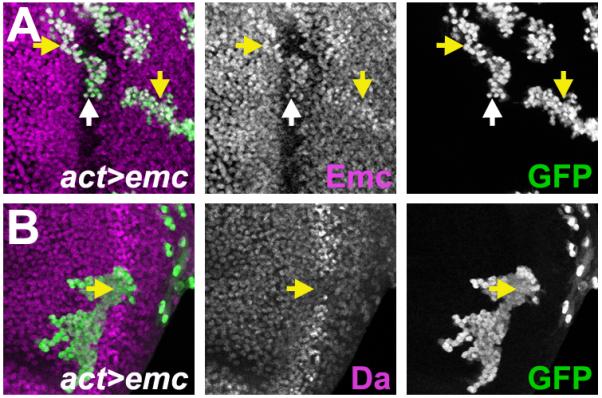
Flip-on clones expressing emc using act-Gal4 and emcEP3620 line are marked by GFP (green).
(A) Emc expression (magenta) from emcEP3620 elevated Emc level slightly (yellow arrow). Within the furrow (white arrow), Emc level was comparable to the normal level ahead of the furrow.
(B) Clonal expression of Emc prevented Da expression from rising in the furrow (yellow arrow). Da expression level was unaffected elsewhere.
Genotype: (A and B) ywhsF; emcEP3620/ act>CD2>Gal4, UAS-GFP.
Emc affects development through Da
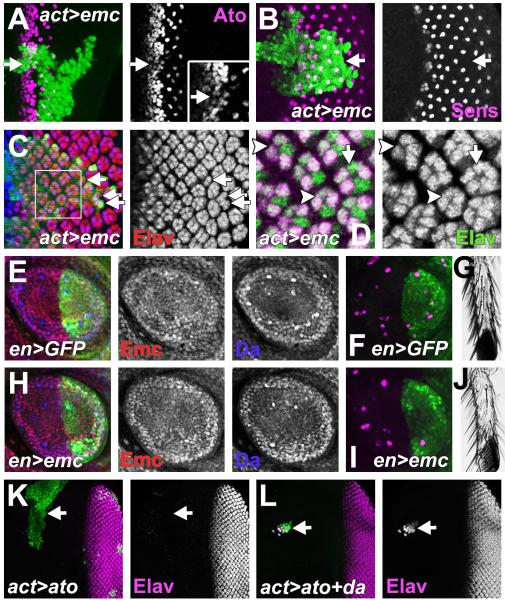
Our findings suggest that the effects of mutating emc might be due to over-expression of Da. Clones of cells over-expressing Da were studied to evaluate this. The effects were indeed similar to emc clones. Homozygous emc null cells only survive in imaginal discs when induced in a background heterozygous for the Minute (M) mutation RpS17 (Bhattacharya and Baker, 2009). The emc clones accelerate the morphogenetic furrow (Bhattacharya and Baker, 2009) and often initiated ectopic morphogenetic furrows from the lateral eye margin (Figure 5A). Clones were also associated with sporadic neuronal differentiation anterior to the furrow and in the peripodial membrane (Figure 5A and data not shown)), although such ectopic neurons did not adopt the R8 photoreceptor cell fate (Figure S4A). Similarly, da over-expressing clones grew poorly (Figure 5E-G) and were rare anterior to the morphogenetic furrow, but like emc clones, recovery was enhanced in a M/+ background (Figures 5B, H). Like emc clones, da over-expressing clones showed some ectopic neuronal differentiation, and could accelerate progression of the furrow (Figure 5B).
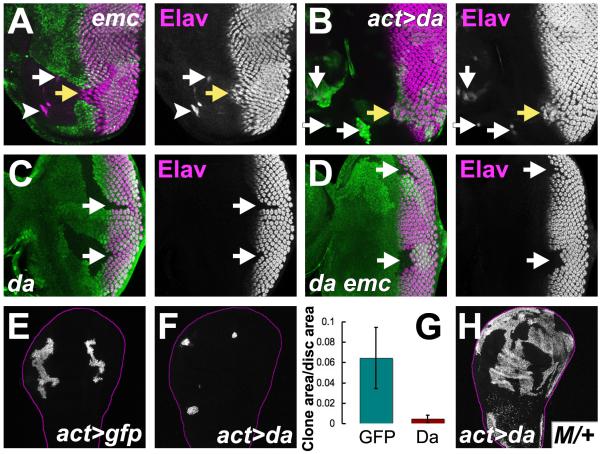
Figure 5. The emc phenotype is caused by Da over-expression.
In (A-D) differentiation is marked by Elav protein (magenta). In (A, C, D) mutant clones lack GFP (green). In (B, E, F, H) mutant clones express GFP (green).
(A) Furrow progression was accelerated (yellow arrow) in emc clones. Ectopic furrows could arise from lateral disc margins (white arrowhead). Sporadic neuronal differentiation also occurred ahead of the morphogenetic furrow (white arrow).
(B) Da over-expression accelerated furrow progression (yellow arrow). Ectopic neuronal differentiation seen ahead of the furrow (white arrows).
(C) Ommatidia do not begin differentiation in da clones (arrows). Note that R1, R6 & R7 cell types can differentiate in mosaic ommatidia at clone borders (Brown et al., 1996).
(D) Like da clones, da emc double mutant clones lacking differentiation were recovered in non-Minute backgrounds (arrows).
(E) Control clones in third instar wing imaginal discs.
(F) Clones over-expressing Da grew less.
(G) Area of Da over-expressing clones compared to controls (normalized against total wing disc area).
(H) Clones over-expressing Da (labeled for GFP) grew extensively in a M/+ background. Genotypes: (A) ywhsF; emcAP6 FRT80/ [UbiGFP] M(3)67C FRT80; (B) ywhsF; UAS-da/+; act>CD2>Gal4, UAS-GFP/+ (C) ywhsF; da10 FRT40/ [UbiGFP] FRT40; (D) ywhsF; da3; P{da+, w+}68A4 [UbiGFP] FRT80/ emcAP6 FRT80; (E) ywhsF; act>CD2>Gal4, UAS-GFP/+; (F) ywhsF; UAS-da/+; act>CD2>Gal4, UAS-GFP/+; (H) ywhsF, UAS-GFP; UAS-da/ act-Gal4; FRT82 Gal80 M(3)96C/ FRT82. See also Figure S4.
Phenotypic effects of Da over-expression were weaker than seen in emc null clones. This could indicate that emc has additional functions besides regulating da. Alternatively, the effects of ectopic Da may be mitigated by elevated expression of the endogenous emc gene,, which cannot occur in emc null cells. To distinguish these possibilities, clones simultaneously mutant for both emc and da were examined. Any differences between da emc mutant cells and da mutant cells would indicate Da-independent roles for Emc. The da mutant clones grow in a non-M/+ background but do not initiate neurogenesis (Figure 5C) (Brown et al., 1996). The da emc double mutant clones also survived in non-M/+ backgrounds and lacked neuronal differentiation (Figure 5D and S4B). Neither ectopic neuronal differentiation ahead of the furrow nor ectopic morphogenetic furrow initiation from the eye margins was observed (Figure 5D and S4B). Therefore, removing da function rescued all the defects associated with emc null mutant clones, suggesting they are all due to Da over-expression.
Precise levels of Emc and Da are important for neurogenesis
Although it is known that Da levels are higher in proneural regions, the importance of the higher levels has not been determined. Similarly, although it is known that mutating emc leads to ectopic differentiation, it is not known whether the lesser reduction in Emc levels seen at proneural regions contributes to wild type neurogenesis. The expression changes normally seen in the morphogenetic furrow were prevented to address these questions. As described above (Figure 4), maintaining Emc levels also prevented any increase in Da, so that both proteins were maintained at their pre-furrow levels. Under such circumstances, the proneural gene ato began expression normally, but did not then rise to the levels expected (Figure 6A) (Jarman et al., 1994). This is consistent with a reduction in the ato autoregulation that depends on Ato/Da heterodimers (Jarman et al., 1995; Brown et al., 1996; Sun et al., 1998; Melicharek et al., 2008). The expression of scabrous (sca), a direct target of the Ato/Da heterodimer (Mlodzik et al., 1990; Singson et al., 1994), was also lower (Figure S5A).
Figure 6. Emc and Da levels are important for neurogenesis.
(A-C) Flip-on clones expressing emc from emcEP3620 line were marked by GFP expression (green).
(A) Where Emc levels were maintained, Ato expression (magenta) began but did not reach normal levels (white arrow). Inset shows enlargement.
(B) 46% of clones maintaining Emc expression had missing R8 cells (n=13) eg white arrow. R8 cells labeled for Sens protein (magenta).
(C) Photoreceptor cells R1-R8 express ELAV (red), cells R2-R5 also express Ro (blue) (Kimmel et al., 1990). Maintaining emc expression led to ommatidia with fewer photoreceptor cells (arrows). Ommatidial rotation was also affected.
(D) Enlargement from panel (C). White arrow indicates a cell in the R4 position lacking Ro. Arrowheads indicate ommatidia with missing cells.
(E) Central portion of 3rd instar prothoracic leg disc. Emc was lower and Da higher in developing SOP cells.
(F) Sens (magenta) labels SOP’s in both compartments.
(G) Posterior view of the tarsus (T1) of the adult prothoracic leg. 20.9±1.4 posterior sensory bristles (n = 16).
(H) Central portion of 3rd instar prothoracic leg disc from en-Gal4 UAS-GFP emcEP3620. SOP’s with altered Emc and Da levels were not evident in the posterior compartment.
(I) Fewer SOP’s in the posterior compartment (Sens in magenta).
(J) The adult tarsus (T1) had only 5.5 ± 1.3 bristles (n = 24; compare panel (G))
(K) GFP (green) marks clone expressing Ato ectopically. No neuronal differentiation resulted (arrow).
(L) GFP (green) marks clone ectopically expressing Ato and Da. Almost all cells undergo neuronal differentiation (arrow).
Genotypes: (A-D) ywhsF; emcEP3620/ act>CD2>Gal4, UAS-GFP; (E-G) en-Gal4, UAS-GFP/+; (H-J) en-Gal4, UAS-GFP/+; emcEP3620/+; (K) ywhsF; UAS-ato/+; act>CD2>Gal4, UAS-GFP/+; (F) ywhsF; UAS-da/ UAS-ato; act>CD2>Gal4, UAS-GFP/+. See also Figures S5.
Ato specifies the R8 photoreceptor precursors that initiate neural differentiation in the retina (Jarman et al., 1994). After R8 specification, the Ato level still affects the recruitment of other retinal cell types by R8 (White and Jarman, 2000); some of these cells also require Da independently of Ato (Brown et al., 1996). When pre-furrow levels of Emc and Da were maintained, 46% of the clones were associated with missing R8 cells (Figure 6B). Interestingly, R8 cells were missing more often close to the posterior margin of the clone. Even when the R8 cells were present, 53% (n=101) of the ommatidia had one or more cells from the R2, R3, R4, and R5 classes either missing, or failing to express the transcription factor Rough (Ro) (Figure 6C, D). On average ommatidia differentiated only 6.8 photoreceptor cells instead of the usual 8. In addition, development of two classes of still-later differentiating retinal cells, the R7 photoreceptor cells and the non-neuronal cone cells, was delayed by 2 columns (about 3 h) when changes in constant Emc/Da levels were maintained (Figure S5B). These findings show that modulating the levels of Da and Emc within the morphogenetic furrow is important for multiple aspects of retina differentiation.
To test whether precise levels of Emc and Da were important in other organs, Emc expression was maintained in the posterior compartments of leg imaginal discs using en-Gal4 and the emc EP line. At 23°C this did not affect Emc levels other than to maintain Emc at the sites of presumptive sensory organ precursors (SOPs)(Figure 1C, H and 6E). This prevented Da upregulation in the SOPs (Figure 6E and H). Maintaining Emc and Da levels reduced SOP selection, and then sensory bristle numbers in the posterior compartment of adult legs, by 70% (27.8±3.3 bristles in the posterior compartments of prothoracic femur, tibia and tarsus (T1)(n = 21), compared to 92.9 ± 1.4 in the en-Gal4 control (n = 15) (Figure 6F-G and I-J, Figure S5C-H). These findings establish that the modulation of Da and Emc levels is even more important for neurogenesis in the developing leg than in the eye.
Regulation of Emc and Da synergizes with proneural gene regulation
Parallel definition of proneural regions by both proneural proteins and by Emc and Da could make development more reliable if both are required for differentiation. To explore this, proneural genes were expressed ectopically either alone or together with Da. Ectopic Ato anterior to the morphogenetic furrow, prior to its normal expression, led to no premature differentiation (Figure 6K). Da over-expression ahead of the furrow led to neuronal differentiation by 14% of the cells (n=146) (Figure S5I). By contrast, 81% of cells overexpressing both Ato and Da ahead of the furrow differentiated as neurons (n=52) (Figure 6L). Thus, Ato expression was insufficient for premature neuronal differentiation unless Da levels were elevated also. Similar results were obtained when the proneural gene sc was overexpressed in the developing thorax. Neither Sc nor Da induced ectopic neuronal differentiation in the notum region of third instar wing discs, but co-expression of Sc and Da together induced neuronal fate in almost all cells (Figure S5J-L). These results support the idea that parallel regulation of Emc/Da and of proneural bHLH genes is required for effective neuronal differentiation.
DISCUSSION
A Class I/Class V HLH network that helps define proneural regions
We describe a simple and unexpected regulatory network that is important to the function of bHLH genes (Figure 7A). This network contributes to both the definition and function of proneural regions, those areas of the ectoderm where proneural bHLH genes are expressed and neural precursor cells are specified. Studies of Drosophila eye and leg development, and of human cells suggest the conclusions apply to many tissues.
Figure 7. Models.
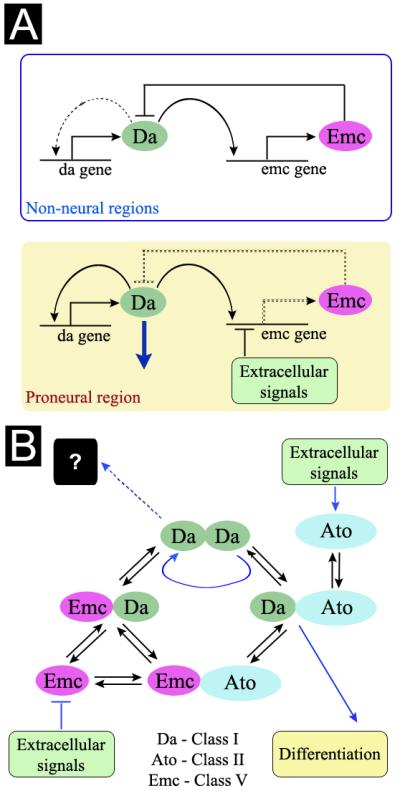
A. Class I/Class V networks differ in proneural ectoderm and non-neural ectoderm. In non-neural ectoderm Da protein regulates emc transcription, then Emc protein feeds back to inhibit self-stimulation of da transcription. Basal da transcription is maintained by promoter-proximal regulatory elements that are Da-independent. In proneural ectoderm, extracellular signaling pathways block emc expression. Hh and Dpp are responsible in the morphogenetic furrow of the eye imaginal disc. Loss of feedback inhibition then permits Da levels to rise by self-stimulation. The non-neural network can be restored by resumption of emc expression, which in the Drosophila eye is mediated by Notch and Ras signaling. This distinction between neural and non-neural ectoderm is independent of proneural genes.
B. Interplay between the Class I/Class V network and HLH protein interactions. Extracellular signals both activate transcription of classic proneural bHLH genes (eg Ato in the developing eye), and block expression of Emc. Differentiation is thereby promoted by driving Da and Ato towards Class I/Class II heterodimer formation and away from heterodimers with Emc. It is possible that Da may have functions independent of Class II proteins, for example as a Da homodimer, or in a complex with other proteins (not shown), making high Da levels detrimental for growth.
The well-known, specific transcription of proneural Class II bHLH genes (eg achaete, sc, ato) is only one feature of neurogenic ectoderm. To this is now added the regulation of the levels of the proneural genes’ heterodimer partners, defined independently by the interaction of positional signals with a regulatory network that links emc and da (Figure 7). The key feature is that da and emc are both transcriptional targets of the Da protein, making Emc a negative feedback regulator that prevents runaway self-stimulation of da gene expression (Figure 7A). The altered levels of Da and Emc in proneural regions are important for proper neural precursor specification. Two other conclusions of our study are that most or all changes in da expression are actually consequences of altered Emc levels, and that most or all phenotypic effects of mutating emc are actually due to runaway expression of da. Outside of proneural regions, the Class I/Class V network maintains low Da levels. This is necessary because high Da levels are detrimental during growth.
Properties of the Class I/Class V network
The network topology observed for HLH genes may merit discussion. Why are da and emc expression regulated by coupled feedback loops, when a bistable network would also change Da and Emc levels reciprocally? Why are da and emc regulated in parallel to the proneural bHLH genes, when da or emc could be their targets?
Coupled positive and negative feedback loops can be homeostatic,, oscillate, or show ‘excitable’ behavior, in which an increase in the level of one gene accelerates until brought under control by the other (Suel et al., 2006; Alon, 2007). Emc and Da exhibit both constant and dynamic expression, like other such networks (Ozaki et al., 2005; Suel et al., 2006; Alon, 2007). In the Class I/Class V network, however, extracellular signals act as circuit breakers, rewiring the network by removing the negative feedback loop to prevent adaptation and homeostasis in proneural regions. In eye development, Da expression seems to be restored after the morphogenetic furrow by Notch and EGFR signaling and not solely by network dynamics, although the emc gene is also required.
HLH gene networks for circadian rhythms or somite development oscillate because of time delays between activatory and inhibitory responses (Alon, 2007; Shoval and Alon, 2010). Without suitable time delays, an extrinsic signal seems the only way to change network output so that Da expression can behave differently inside and outside proneural regions.
The regulation of da and emc independently of proneural genes may guard against inappropriate differentiation by requiring parallel differentiation signals for two pathways. Even though Class II bHLH proteins are iconic examples of master regulatory genes whose expression is sufficient to confer a differentiated fate, at best a small fraction of cells that express proneural genes ectopically differentiated neural fates in our experiments, as in previous studies (Hinz et al., 1994; Giagtzoglou et al., 2003; Villa-Cuesta et al., 2003; Pi et al., 2004; Wildonger & Mann, 2005). By contrast, expression of Da with Ato or Sc led to neuronal differentiation by virtually all co-expressing cells.
We propose that the general outline shown in Figure 7A applies to all proneural regions that use Class II bHLH proneural genes, except that the particular extracellular signals acting on emc are likely to be different in each case, just as the particular prepatterns initiating proneural gene expression are also distinct for each proneural region. Hh and Dpp repress emc in the developing eye; other signals are likely to be important in other body regions.
Conservation of the Class I/Class V network
Homologous Class I/Class V regulatory networks may regulate other bHLH-regulated processes, such as myogenesis, and may be conserved beyond Drosophila. A transcriptional Class I/Class V network was found in HEK293T cells, in which the human E47 protein was an activator both of its own expression through the E2a gene, and of its antagonist Id1. Since Id1 heterodimerizes with E47 and blocks its function (Benezra et al., 1990), Id1 appears to be a feedback inhibitor of E47 in these human cells, just as Emc is a feedback inhibitor of Da in Drosophila. Published microarray datasets contain evidence for similar networks in other mammalian cells. Thus E2a, Id1 and Id2 appear to be transcriptional targets of E47 in the T-cell lineage, in MDCK cells, and in neural cells (Schwartz et al., 2006; Rothschild et al., 2006; Jorda et al., 2007). The absence of Id2 impairs NK-cell specification in an E2a-dependent manner (Boos et al., 2007), so that the phenotype of this Class V gene mutant is due to over-expression of a Class I gene, just as the phenotype of emc mutants is due to over-expression of da.
The multiple mammalian Class I and Class V family members have partially redundant functions and overlapping and distinct expression domains, and were not all E47 targets in HEK293T cells. It may be that it is E2a, Id1, and in some cases Id2, that retain the Class I/Class V network regulation, or it may be that different family members are involved in distinct cell types, perhaps with other regulatory inputs superimposed. It will be interesting to determine the contribution of Class I/Class V networks to the regulation of differentiation and other processes by mammalian E proteins and Id proteins.
EXPERIMENTAL PROCEDURES
Genetic methods
Mosaic clones were obtained using the FLP/FRT-mediated mitotic recombination technique (Golic, 1991; Xu and Rubin, 1993). For details of the specific procedures and genotypes, see Extended Experimental Procedures online. A set of da+ transgenics contained an 8.8kb genomic region extending from the EcoRI site in the cdc2 gene to the KpnI site in the CG5362 gene (Figure 2D), cloned into the pattB vector and integrated into the attP2 site at 68A4 on chromosome arm 3L (Groth et al., 2004). emc da double mutant clones were obtained using this transgene to rescue a strain homozygous for da3. Since this P{da+, w+}68A4 transgene and the emc gene were both on 3L, they segregated together during mitotic recombination at FRT80. Genotypes are described in Figure legends. Details of all the alleles and transgenes used are available online in the Extended Experimental Procedures.
Immunochemistry
Antibody labeling were performed as described previously (Firth et al., 2006; Bhattacharya and Baker, 2009). Images were recorded using either BioRad Radiance 2000 or Leica SP2 Confocal microscopes and processed using NIH Image J. Channels for different antibodies were recorded separately in time and usually adjusted independently using Levels and Curves in Adobe Photoshop 10.0.1. A list of antibodies is available online in the Extended Experimental Procedures.
Supplementary Material
Acknowledgements
This work was enabled by the gift of polyclonal anti-Emc antibody by Yuh-Nung Jan. We also thank Claire Cronmiller for monoclonal anti-Da antibody, Konrad Basler and Christos Delidakis for strains and other reagents, Matthew Scharff and Richard Chahwan for help with HEK293T cell culture, Model System Genomics for embryo injections, John Fullard and David Lubensky for advice, and Hannes Buelow, Claude Desplan, Scott Emmons, Andreas Jenny, Richard Mann, Ertugrul Ozbudak, Julie Secombe, Jessica Treisman, and Lan-Hsin Wang for comments on the manuscript. Supported by grant GM47892 from the NIH and by an Unrestricted Grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences. Confocal microscopy was performed at the AIF, AECOM. Data in this paper are from a thesis to be submitted in partial fulfillment of the requirement for the degree of Doctor of Philosophy in the Graduate Division of Biomedical Sciences, Albert Einstein College of Medicine, Yeshiva University, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Aronheim A, Shiran R, Rosen A, Walker MD. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc Natl Acad Sci U S A. 1993;90:8063–8067. doi: 10.1073/pnas.90.17.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Bhattacharya A, Firth LC. Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Dev Biol. 2009;335:356–366. doi: 10.1016/j.ydbio.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE. The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev Biol. 2009;327:288–300. doi: 10.1016/j.ydbio.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botas J, Moscoso del Prado J, Garcia-Bellido A. Gene-dose titration analysis in the search of trans-regulatory genes in Drosophila. EMBO J. 1982;1:307–310. doi: 10.1002/j.1460-2075.1982.tb01165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Paddock SW, Sattler CA, Cronmiller C, Thomas BJ, Carroll SB. daughterless is required for Drosophila photoreceptor cell determination, eye morphogenesis, and cell cycle progression. Dev Biol. 1996;179:65–78. doi: 10.1006/dbio.1996.0241. [DOI] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Paddock SW, Carroll SB. Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell. 1995;80:879–887. doi: 10.1016/0092-8674(95)90291-0. [DOI] [PubMed] [Google Scholar]
- Cabrera CV, Alonso MC. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Jou AD, Nusse R. Wingless blocks bristle formation and morphogenetic furrow progression in the eye through repression of Daughterless. Development. 2002;129:3393–3402. doi: 10.1242/dev.129.14.3393. [DOI] [PubMed] [Google Scholar]
- Campuzano S. Emc, a negative HLH regulator with multiple functions in Drosophila development. Oncogene. 2001;20:8299–8307. doi: 10.1038/sj.onc.1205162. [DOI] [PubMed] [Google Scholar]
- Cochrane SW, Zhao Y, Welner RS, Sun XH. Balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions. Blood. 2009;113:1016–1026. doi: 10.1182/blood-2008-06-164996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Modolell J. The extramacrochaetae gene provides information for sensory organ patterning. Embo J. 1992;11:3385–3393. doi: 10.1002/j.1460-2075.1992.tb05417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Domingos PM, Brown S, Barrio R, Ratnakumar K, Frankfort BJ, Mardon G, Steller H, Mollereau B. Regulation of R7 and R8 differentiation by the spalt genes. Dev Biol. 2004;273:121–133. doi: 10.1016/j.ydbio.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Spann DR, Posakony JW. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nat Rev Immunol. 2001;1:193–199. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- Firth LC, Li W, Zhang H, Baker NE. Analyses of RAS regulation of eye development in Drosophila melanogaster. Methods Enzymol. 2006;407:711–721. doi: 10.1016/S0076-6879(05)07056-4. [DOI] [PubMed] [Google Scholar]
- Garcia Alonso LA, Garcia-Bellido A. Extramacrochaetae, a trans-acting gene of the achaete-scute complex of Drosophila involved in cell communication. Roux Arch dev Biol. 1988;197:328–338. doi: 10.1007/BF00375952. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A. Genetic Analysis of the Achaete-Scute System of DROSOPHILA MELANOGASTER. Genetics. 1979;91:491–520. doi: 10.1093/genetics/91.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A, de Celis JF. The complex tale of the achaete-scute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics. 2009;182:631–639. doi: 10.1534/genetics.109.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrell J, Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N, Alifragis P, Koumbanakis KA, Delidakis C. Two modes of recruitment of E(spl) repressors onto target genes. Development. 2003;130:259–270. doi: 10.1242/dev.00206. [DOI] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Campuzano S, Modolell J. Half a century of neural prepatterning: the story of a few bristles and many genes. Nat Rev Neurosci. 2003;4:587–598. doi: 10.1038/nrn1142. [DOI] [PubMed] [Google Scholar]
- Goulding SE, zur Lage P, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Greenwood S, Struhl G. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development. 1999;126:5795–5808. doi: 10.1242/dev.126.24.5795. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- Hinz U, Giebel B, Campos-Ortega JA. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Huang ML, Hsu CH, Chien CT. The proneural gene amos promotes multiple dendritic neuron formation in the Drosophila peripheral nervous system. Neuron. 2000;25:57–67. doi: 10.1016/s0896-6273(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Ik Tsen Heng J, Tan SS. The role of class I HLH genes in neural development--have they been overlooked? Bioessays. 2003;25:709–716. doi: 10.1002/bies.10299. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- Jorda M, Vinyals A, Marazuela A, Cubillo E, Olmeda D, Valero E, Cano A, Fabra A. Id-1 is induced in MDCK epithelial cells by activated Erk/MAPK pathway in response to expression of the Snail and E47 transcription factors. Exp Cell Res. 2007;313:2389–2403. doi: 10.1016/j.yexcr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- Kimmel BE, Heberlein U, Rubin GM. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- Lee EC, Hu X, Yu SY, Baker NE. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16:1179–1188. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Hao E, Kiselyuk A, Shapiro J, Shields DJ, Lowy A, Levine F, Itkin-Ansari P. The id3/e47 axis mediates cell-cycle control in human pancreatic ducts and adenocarcinoma. Mol Cancer Res. 2011;9:782–790. doi: 10.1158/1541-7786.MCR-10-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melicharek D, Shah A, DiStefano G, Gangemi AJ, Orapallo A, Vrailas-Mortimer AD, Marenda DR. Identification of novel regulators of atonal expression in the developing Drosophila retina. Genetics. 2008;180:2095–2110. doi: 10.1534/genetics.108.093302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989a;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989b;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Ozaki Y, Sasagawa S, Kuroda S. Dynamic characteristics of transient responses. J Biochem. 2005;137:659–663. doi: 10.1093/jb/mvi084. [DOI] [PubMed] [Google Scholar]
- Pi H, Huang SK, Tang CY, Sun YH, Chien CT. phyllopod is a target gene of proneural proteins in Drosophila external sensory organ development. Proc Natl Acad Sci U S A. 2004;101:8378–8383. doi: 10.1073/pnas.0306010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Rothschild G, Zhao X, Iavarone A, Lasorella A. E Proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol Cell Biol. 2006;26:4351–4361. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte R, Dontje W, Nagasawa M, Yasuda Y, Bakker AQ, Spits H, Blom B. Synergy between IL-15 and Id2 promotes the expansion of human NK progenitor cells, which can be counteracted by the E protein HEB required to drive T cell development. J Immunol. 2010;184:6670–6679. doi: 10.4049/jimmunol.0901508. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CP, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval O, Alon U. SnapShot: network motifs. Cell. 2010;143:326–e321. doi: 10.1016/j.cell.2010.09.050. [DOI] [PubMed] [Google Scholar]
- Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- Smith JE, 3rd, Cronmiller C. The Drosophila daughterless gene autoregulates and is controlled by both positive and negative cis regulation. Development. 2001;128:4705–4714. doi: 10.1242/dev.128.23.4705. [DOI] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Brand M, Jan LY, Jan YN. daughterless is essential for neuronal precursor differentiation but not for initiation of neuronal precursor formation in Drosophila embryo. Development. 1994;120:935–945. doi: 10.1242/dev.120.4.935. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Powell PA, Pasternak D, Singson A, Posakony JW. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- Villa-Cuesta E, de Navascues J, Ruiz-Gomez M, Diez del Corral R, Dominguez M, de Celis JF, Modolell J. Tufted is a gain-of-function allele that promotes ectopic expression of the proneural gene amos in Drosophila. Genetics. 2003;163:1403–1412. doi: 10.1093/genetics/163.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villares R, Cabrera CV. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NM, Jarman AP. Drosophila atonal controls photoreceptor R8-specific properties and modulates both receptor tyrosine kinase and Hedgehog signalling. Development. 2000;127:1681–1689. doi: 10.1242/dev.127.8.1681. [DOI] [PubMed] [Google Scholar]
- Wildonger J, Mann RS. Evidence that nervy, the Drosophila homolog of ETO/MTG8, promotes mechanosensory organ development by enhancing Notch signaling. Dev Biol. 2005;286:507–520. doi: 10.1016/j.ydbio.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.