Highlights
► Bacterial expressed human recombinant DI has a structure consistent with that of DI in the published β2GPI crystal structure. ► Mutating residues D8/D9 and R39 do not alter the overall DI protein fold but cause local changes in surface contour. ► Monoclonal aPL-derived antibodies and DI of β2GPI interactions are influenced by specific arginine residues in aPL and particular epitopes in DI.
Abbreviations: aPL, antiphospholipid antibodies; APS, antiphospholipid syndrome; β2GPI, beta-2-glycoprotein I; CL, cardiolipin; DI, domain I of β2GPI; E. coli, Escherichia coli; His6-tag, hexahistidine tag; HSQC, 15N,1H-heteronuclear single quantum correlation; NMR, nuclear magnetic resonance; PL, phospholipids; VH, variable heavy chain of Ig; VL, variable light chain of Ig
Keywords: Antiphospholipid antibodies, Beta-2-glycoprotein I, Domain I, Nuclear magnetic resonance spectroscopy
Abstract
Pathogenic antiphospholipid antibodies (aPL) cause the antiphospholipid syndrome (APS) by interacting with domain I (DI) of beta-2-glycoprotein I (β2GPI). The aPL/β2GPI complex then exerts pathogenic effects on target cells. We previously described periplasmic bacterial expression of native and mutated variants of DI, and reported the presence of immunodominant epitopes at positions 8–9 (D8/D9) and position 39 (R39). Mutations at these positions strongly influenced the ability of recombinant DI to bind patient-derived IgG aPL and to inhibit pathogenic effects of these aPL in a mouse model of APS.
We now describe an improved cytoplasmic bacterial expression system allowing higher yield of DI. We demonstrate that the nuclear magnetic resonance (NMR) spectra of a 15N,13C-isotope-labelled sample of the recombinant DI protein exhibit properties consistent with the structure of DI in crystal structure of intact β2GPI. Mutations at D8/D9 and R39 had limited impact on the NMR spectrum of DI indicating maintenance of the overall fold of the DI domain.
We investigated interactions between five variants of DI and ten monoclonal human IgG antibodies, all derived from the IgG aPL antibody IS4 by sequence manipulation and in vitro expression. Arginine residues at positions 100 and 100g in IS4VH CDR3 play a particularly important role in binding to DI, but this is unlikely to be due to electrostatic interactions with negatively charged amino acids on DI. Both the strength of binding to DI and the ability to discriminate different DI variants varies between the different IgG antibodies tested. There was no simple relationship between these binding properties and antibody pathogenicity.
1. Introduction
Identification of circulating pathogenic antiphospholipid antibodies (aPL) is key to the diagnosis of the antiphospholipid syndrome (APS) (Miyakis et al., 2006). These aPL are recognised to have direct effects on cells causing vascular thrombosis and/or pregnancy morbidity (Hughes, 1983; Hughes et al., 1986). Despite current anticoagulant and antiplatelet therapies for APS, none of which target aPL themselves, the incidence of thrombotic and obstetric complications remains high (Shah et al., 1998; Cervera et al., 2009). Therefore it is critical to develop more targeted therapies to block the effects of aPL through a greater understanding of how pathogenic aPL interact with target antigens.
1.5–5% of the healthy population has circulating aPL but do not develop APS. Such aPL bind directly to neutral and anionic phospholipids (PL) (Greaves et al., 2000). In contrast, pathogenic aPL in patients with APS target anionic PL via interactions with epitopes on PL-binding serum proteins. Although there are patients in whom IgM aPL are pathogenic, clinical features of APS are more commonly associated with the presence of IgG aPL (Alarcon-Segovia et al., 1989; Lynch et al., 1994). Both IgG and IgM aPL are measured in standardised laboratory assays for APS diagnosis (Miyakis et al., 2006). The key antigenic target for pathogenic aPL is beta 2 glycoprotein I (β2GPI) (McNally et al., 1995; Tsutsumi et al., 1996; Kandiah et al., 1998; de Laat et al., 2004), a PL-binding glycoprotein comprising five domains (Iverson et al., 1998; Schwarzenbacher et al., 1999). Studies from several groups using different experimental methods have all identified the N-terminal domain (Domain I or DI) as the major site of epitopes bound by pathogenic aPL (Iverson et al., 1998; Reddel et al., 2000; McNeeley et al., 2001; de Laat et al., 2005).
We developed the first bacterial expression system for DI (Ioannou et al., 2006) and used it to create a number of mutagenised DI variants (Ioannou et al., 2006, 2007). We found that polyclonal IgG purified from patients with APS (APS-IgG) showed significantly higher binding to wild type/native DI (n-DI) than IgG purified from healthy and disease controls. This binding was highly dependent on the presence of an arginine residue at position 39 (R39) in DI and was enhanced by mutagenesis of two aspartic acid residues at positions 8 and 9 to serine and glycine respectively (D8S/D9G). Furthermore, we showed in a mouse model of APS that n-DI and DI(D8S/D9G) but not DI(R39S) inhibited the induction of thrombosis by passive transfer of IgG from a patient with APS (Ioannou et al., 2009). Thus, both recombinant n-DI and DI(D8S,D9G) are potential therapeutic agents in APS, which could be used to block the aPL–DI interaction in vivo. In order to develop this potential, it was necessary to improve the bacterial expression system to give higher yield. This study describes how we have done this by switching from expression of DI in the bacterial periplasm to expression in the form of cytoplasmic inclusion bodies from which DI can be re-folded. It was also important to show that the three-dimensional structure of the recombinant DI produced is not significantly different from that of DI in native whole β2GPI. Any major differences could alter the stability or immunogenicity of the recombinant products, ultimately affecting their potential for use as therapeutic agents. In the current study, we have used nuclear magnetic resonance (NMR) spectroscopy to probe the structure of recombinant n-DI, DI(D8S/D9G) and DI(R39S) in solution.
Our previous results using polyclonal IgG from patients with APS (APS-IgG) (Ioannou et al., 2007) led us to hypothesise that pathogenicity of IgG aPL may correlate with strong binding to n-DI and the DI(D8S/D9G) mutant but weak binding to DI in which R39 has been altered to either serine or lysine (DI(R39S) and DI(R39K), respectively). Conversely, non-pathogenic aPL might either fail to bind DI at all or fail to discriminate between the different mutagenised forms. Polyclonal IgG samples from patients contain a mixture of IgG species with different sequences and binding properties, only some of which are pathogenic. Thus studies on polyclonal IgG cannot be used for precise elucidation of the relationship between small sequence changes in antibody or antigen, the ability of antibody to bind DI, and pathogenicity. Such studies require the availability of a panel of monoclonal antibodies whose sequence, binding and pathogenic properties are known. We have exactly such a panel of monoclonal human IgG, generated from our previous extensive mutagenesis and binding studies on IS4, a monoclonal IgG aPL derived from a patient with APS (Zhu et al., 1999) that is thrombogenic in mice (Pierangeli et al., 2000). We previously tested the binding properties of monoclonal antibodies derived from IS4 by mutagenesis of its heavy chain variable region (VH) and/or exchange of the light chain variable region (VL) with VL derived from the same germline gene that differ solely in their pattern of somatic mutations. We found that altering certain arginine residues in IS4VH and/or exchanging the paired VL had dramatic effects on binding to different antigens including various PL, β2GPI and thrombin (Giles et al., 2003, 2005, 2006, 2009). Two such VH/VL combinations were thrombogenic in mice (Giles et al., 2009) and this pathogenicity most closely correlated with binding to thrombin rather than cardiolipin (CL) or β2GPI. In this study we report binding of these pathogenic and non-pathogenic VH/VL combinations to n-DI and mutated DI.
2. Materials and methods
2.1. Human monoclonal IgG antibodies
Generation of the panel of human recombinant monoclonal IgG antibodies derived from IS4 and B3 (a human monoclonal IgG anti-nucleosome antibody) has been described previously (Giles et al., 2003, 2005, 2006). In brief, we studied five mutant forms of IS4VH with particular arginine residues in CDR3 mutated to serine. These mutants were named: IS4VHi&ii containing two arginine to serine mutations at R96 and R97; IS4VHiii and IS4VHiv each containing single mutations of arginine residues R100 and R100g respectively; IS4VHiii&iv with combined R100 and R100g mutations; and IS4VHx containing all four (R96, R97, R100 and R100g) mutations. Native or mutated IS4VH was paired with either IS4VL or B3VL (both derived from the 2a2 germline gene and differing solely in their pattern of somatic mutations) and stable expression of each VH/VL combination was established in CHO cells (Giles et al., 2006). Large-scale production and purification of IgG was outsourced to Chemicon Europe Ltd. (Southampton, UK) and Harlan Ltd. (Leicester, UK). Concentration of IgG was confirmed by ELISA (Giles et al., 2003) and spectrophotometry. The binding and functional properties of the ten monoclonal antibodies used in this study have been previously described (Giles et al., 2003, 2005, 2006). Binding of IgG (at 500 ng/ml) to CL, β2GPI and DI proteins (coated upon nickel plates at 10 μg/ml) was measured by direct ELISA (Giles et al., 2006; Ioannou et al., 2006, 2007).
2.2. Bacterial periplasmic expression of native and mutant recombinant human DI
The system used for native recombinant human DI of β2GPI expression in Escherichia coli and purification using nickel chromatography has been described previously (Ioannou et al., 2006). Briefly, a synthetic gene encoding for recombinant human DI, flanked by NcoI and XhoI restriction sites, was designed (using Juniper http://strubiol.icr.ac.uk/extra/juniper) and synthesised using recursive PCR. DI DNA was cloned into the expression vector pET-26b, encoding an N-terminal pelB leader sequence to transport expressed proteins to the periplasm and a C-terminal hexihistidine tag (His6-tag), and was used to transform BL21(DE3) E. coli cells. Expressed DI protein carried both these appendages and was purified by nickel-immobilised metal ion affinity chromatography (Iverson et al., 2002).
Targeted point mutations at hypothesised immunodominant regions within the human DI sequence were introduced, generating three mutants of DI (DI(D8S/D9G); DI(R39K); DI(G40E)), whilst the addition of the β2GPI DI-DII interlinker region (residues PRVCPF) to the DI sequence generated an extended version of the DI gene (DI(EXT)). All DI variants used for direct binding experiments were expressed and purified in the same way as n-DI (Ioannou et al., 2007). Correct folding was confirmed by Western blotting on a non-denaturing gel, using a murine anti-DI antibody (mAb16; a kind gift from Dr. Mike Iverson and Dr. Matt Linnik, La Jolla Pharmaceuticals, CA, USA) that recognises a conformation-dependent epitope within folded DI (Ioannou et al., 2006).
2.3. Bacterial cytoplasmic expression, purification and in vitro folding of recombinant human DI for NMR
Periplasmic expression is unsuitable for producing amounts of DI sufficient for structural studies. We therefore developed a system for expression of DI in inclusion bodies in the bacterial cytoplasm, followed by in vitro folding by rapid dilution. DI DNA was subcloned into a pET system vector (a kind gift from Gunter Stier, Umea University, Sweden) providing for expression of an N-terminal His6-tag followed by a tobacco etch virus (TEV) protease cleavage site. The vector was transformed into Rosetta (DE3) E. coli cells (Novagen), grown in LB medium with 30 μg/ml kanamycin and 35 μg/ml chloramphenicol (37 °C, 8 h). Following overnight culture in PG medium (Studier, 2005) (basal Eagle medium, vitamins, and antibiotics) cells were harvested and inoculated into 10-times the overnight culture volume in PG medium including ammonium sulphate-15N2 and/or d-glucose-13C6 (Cambridge Isotope Laboratories, Inc.) for isotope labelling. Cells were grown at 37 °C to OD600 ∼ 1.0 and DI expression was induced by 1 mM isopropylthiogalactoside for 4 h. Cells were harvested, lysed by sonication in 50 mM Tris 300 mM NaCl pH8.0, and centrifuged (20 min, 27,000 × g). The cell pellet was resuspended using a mortar and pestle type homogeniser in 0.1 M potassium phosphate, 10 mM Tris–HCl, 6 M guanidine hydrochloride (GdnHCl) pH8.0, sonicated to reduce viscosity and clarified by centrifugation (1 h, 27,000 × g). The supernatant was purified by gravity-flow IMAC (Ni-IDA agarose; Generon). After washing at pH8.0, DI was eluted in 0.1 M potassium phosphate, 10 mM Tris–HCl, 6 M GdnHCl pH4.5. The eluate was concentrated to 4 ml by centrifugal ultrafiltration (Vivaspin20, Vivascience), and the protein was reduced by incubation with 50 mM Tris (2-carboxyethyl) phosphine (TCEP; 2 h, 20 °C). Monomeric DI was isolated by size-exclusion chromatography (SEC; Superdex75, GE Healthcare) in 0.1 M potassium phosphate 10 mM Tris–HCl, 6 M GdnHCl pH4.5 and concentrated to 10 mg/ml. The reduced DI was then diluted batchwise (100 μl every 30 min) ca. 200-fold into 100 mM Tris–HCl, 600 mM arginine, 3 mM cysteine, 0.3 mM cystine pH8.5. After 48 h at 4 °C, folded DI was concentrated to 5 ml and dialysed against PBS. Analysis by analytical SEC and NMR showed that folding was quantitative. The DI(D8S/D9G) and DI(R39S) variants were prepared in an identical fashion to n-DI.
Cytoplasmic and periplasmic-expressed n-DI were compared for apparent hydrodynamic size and conformation as previously described (Ioannou et al., 2006, 2007). n-DI expressed in insect cells (a kind gift from Dr. Mike Iverson and Dr. Matt Linnik, La Jolla Pharmaceuticals, CA, USA) was used as a positive control.
2.4. Direct binding assay of aPL to cytoplasmic-expressed DI
Binding of native IS4, and negative and positive control antibodies to cytoplasmic-expressed n-DI, coated on a nickel ELISA plate at 10 μg/ml, was assessed as previously described (Ioannou et al., 2006). The negative control was a non-aPL monoclonal human IgG (expressed in the same way as native IS4, does not bind CL or β2GPI). The positive control was a polyclonal IgG sample from a female patient with APS (with a history of pregnancy morbidity, persistently positive aCL, anti-β2GPI antibodies and lupus anticoagulant). All IgG antibodies were tested at 25, 50 and 100 μg/ml.
2.5. NMR studies of cytoplasmic-expressed in vitro-folded DI proteins
The majority of NMR spectra were acquired at 25 °C on Bruker AVANCE III spectrometers (operating at a nominal 1H frequency of 600 MHz or 700 MHz). Sequence-specific resonance assignments were obtained using standard triple resonance NMR protocols (Cavanagh et al., 2006). NMR spectra were processed using NMRpipe/NMRDraw (Delaglio et al., 1995) and analyzed using CCPN Analysis version 2 (Vranken et al., 2005). 1H, 13C, and 15N chemical shifts were referenced indirectly to sodium 2,2-dimethyl-2-silanepentane-5-sulfonate (DSS), using absolute frequency ratios for the 1H signals (Wishart et al., 1995).
3. Results
3.1. Cytoplasmic expression and immunological characterisation of in vitro folded DI
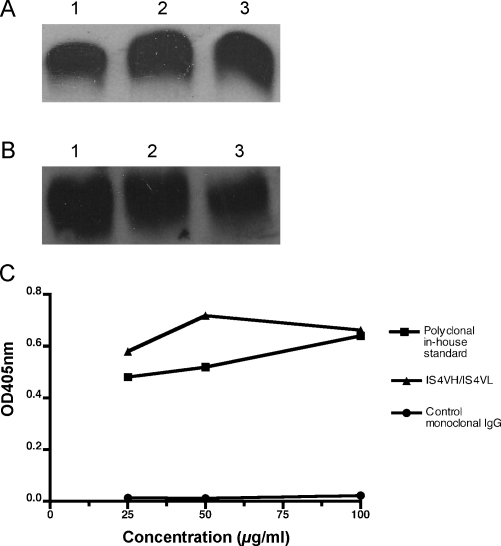
After purification, in vitro folding and dialysis, the yield of his6-tagged DI protein obtained from cytoplasmic expression was approximately 4 mg/L. In comparison the typical yield from the periplasmic expression system was approximately 100 μg/L. Western blotting with anti-His6-tag antibody (Fig. 1A) showed that periplasmic and cytoplasmic n-DI migrate at the same rate indicating a consistent molecular mass (ca. 7 kDa). Furthermore, cytoplasmic-expressed DI binds a murine monoclonal antibody (mAb16) that only recognises folded DI (Fig. 1B). Lastly, IS4VH/IS4VL and polyclonal APS-IgG were shown to bind strongly to cytoplasmic-expressed n-DI, whilst control monoclonal IgG did not (Fig. 1C).
Fig. 1.
Cytoplasmic-expressed DI behaves as periplasmic-expressed DI. (A) All E. coli-expressed DI proteins are of the same size and (B) correct conformation, shown by Western blot. Antibodies against (A) the DI-attached his6tag and (B) correctly folded human DI were used to blot for bacterial cytoplasmic-expressed (lane 1), periplasmic expressed (lane 2) and (insect cell) baculovirus-expressed native DI (n-DI) (lane 3) as a positive control. All DI proteins were recognised by both antibodies, hence were of the same size and conformation. (C) Native DI expressed in the cytoplasm is bound by aPL but not control IgG. Cytoplasmic-expressed n-DI (10 μg/ml) was tested in a direct binding assay against native IS4 (IS4VH/IS4VL), control monoclonal IgG (expressed and purified in the same way as native IS4) and a polyclonal IgG in-house standard (derived from the sera of a female patient with the APS). All IgG samples were tested at varying doses, from 25 to 100 μg/ml, to demonstrate a dose-dependent effect. Concentrations of IgG are shown on the x-axis, with OD at 405 nm on the y-axis. For all IgG tested at 25–100 μg/ml in triplicate, the standard deviation ranged from 0.004 to 0.014, hence error bars are too small to be seen.
3.2. NMR studies of cytoplasmic-expressed native DI, DI(D8S/D9G) and DI(R39S)
Samples of n-DI and the variants DI(D8S/D9G) and DI(R39S) were prepared with either 15N-(variants) or double 15N-/13C-(native) isotope labelling for examination by multidimensional heteronuclear NMR investigation. Two-dimensional (2D) 15N,1H-heteronuclear single quantum correlation (HSQC) spectroscopy is commonly used to assess the folding status of small proteins. A given protein yields a particular pattern of backbone 15N–1H correlation peaks (cross peaks) in the spectrum, the dispersion of which provides a characteristic ‘fingerprint’ corresponding to the local primary, secondary and tertiary structure of the protein. In general, globular folded proteins display significantly higher degree of cross peak chemical shift dispersion than polypeptide chains that are disordered, reflecting the engagement of backbone amide NH groups in stable H-bonding and other non-covalent interactions with the immediate neighbouring atoms in the protein.
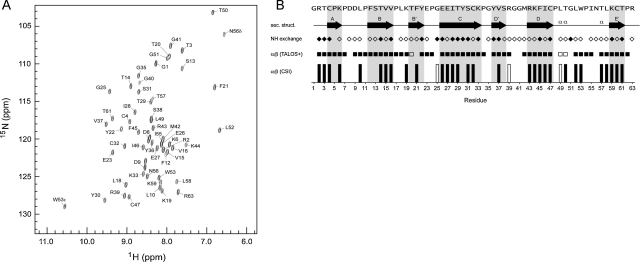
2D 15N,1H HSQC NMR yielded a well resolved spectrum for n-DI, with excellent chemical shift dispersion unambiguously characteristic of a folded globular domain (Fig. 2A). Full sequence-specific assignments of the n-DI amide NH cross peaks were obtained using standard triple resonance 3D NMR spectroscopy (Cavanagh et al., 2006). The assignments allow for prediction of the local secondary structure (α vs. β conformation) along the n-DI chain using both the Chemical Shift Index (Wishart and Sykes, 1994) (CSI) and TALOS+ (Shen et al., 2009) computer programs. These programs exploit databases of secondary chemical shifts for proteins with known 3D structure (a secondary chemical shift represents the difference between the observed chemical shift for a given nucleus and the corresponding chemical shift for a similar nucleus in a peptide with random coil conformation). In Fig. 2B we plot the output of the CSI and TALOS+ analysis (where those programs provide an high confidence prediction), along with the pattern of observed backbone NH/solvent exchange characteristics (lack of observed exchange indicates that the amide NH is likely engaged in regular H-bonded secondary structure), and align these parameters with the secondary structure of the n-DI domain in the X-ray crystal structure of intact β2GPI (PDB codes 1QUB (Bouma et al., 1999) and 1C1Z (Schwarzenbacher et al., 1999)). The pattern of the NMR-derived parameters is entirely consistent with the β-rich secondary structure of the protein in the crystal structure. In addition, the n-DI structure is constrained by two disulphide cross-links (confirmed by characteristic Cβ chemical shifts in the NMR spectra of the oxidized Cys residues (Sharma and Rajarathnam, 2000; Wang et al., 2006)). Preliminary calculations of the 3D structure of n-DI based upon inter-proton NOE contacts yield a 3D conformer bundle that superposes well with the crystal structure.
Fig. 2.
NMR analysis of β2GPI native DI. (A) The 2D 15N,1H-HSQC spectrum of 15N,13C-labelled β2GPI native DI. The sequence-specific backbone amide NH cross peak assignments obtained by 3D triple resonance methods are indicated. The dispersion of cross peaks is typical of a well-ordered globular protein domain. (B) Analysis of the β2GPI native DI NMR data with reference to the conformation in the X-ray structure of intact β2GPI. We plot two indicators on a per-residue basis of the local backbone torsion angles based upon the backbone atom chemical shift assignments for β2GPI DI: the chemical shift index (CSI; bottom) and the secondary structure prediction of the program TALOS+. In each case the prediction is either for extended strand (filled symbol) or α-helical (empty symbol) conformation. Also indicated is the qualitative assessment of the rate of backbone amide NH/solvent exchange due to H-bonding based upon the presence or absence of an exchange NH/solvent water cross peak in a 3D 15N-separated 1H-NOESY spectrum of β2GPI DI. The overall pattern of these parameters is consistent with the β-strand-rich secondary structure of β2GPI DI (top) as seen in the crystal structure (PDB code 1QUB32).
In summary, these NMR spectroscopy data indicate that in solution the n-DI protein, obtained by refolding of material expressed in inclusion bodies, adopts the protein fold anticipated from the X-ray structure of the intact protein purified from citrated human plasma. Conversely, we can be confident that the atomic coordinates of the crystal structure provide an excellent model for the conformation of the n-DI protein in solution, and a reliable framework for computer modelling of the effects of surface residue substitutions, without the necessity to produce a highly refined solution structure by relatively laborious NMR study. However the ‘fingerprint spectrum’ provided by 15N,1H-HSQC NMR provides an efficient and robust basis for the assessment of the effects of the substitution of surface residue side chains in DI.
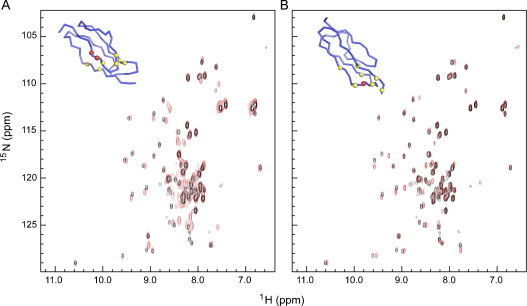
The 2D 15N,1H HSQC spectra of the DI(D8S/D9G) and DI(R39S) variants superpose well with the spectrum of n-DI (Fig. 3A and B respectively). Whilst the majority of resolved cross peaks are unperturbed by the mutations, there are small chemical shift differences for a small subset of cross-peaks. Using the chemical shift assignments of n-DI it is clear that in each case these are limited to residues at or around the mutation site in the overall 3D structure of DI. This is illustrated in the diagrams of the polypeptide backbone of DI(D8S/D9G) and DI(R39S) in Fig. 3A and B. In each case, the residues showing chemical shifts (shown in yellow) are clustered around the sites of mutation (shown in red). Therefore, relative to n-DI, the overall polypeptide folding topology of the DI(D8S/D9G) and DI(R39S) variants is maintained and the effects of the introduced mutations upon interaction with antibodies must be determined by local effects on the surface of DI rather than by any gross change in the overall protein fold.
Fig. 3.
NMR analysis and comparison of native and mutant DI spectra. Superposition of the 2D 15N,1H HSQC spectra of (A) D8S/D9G and (B) R39S (red contours) with that of native DI (black contours) of β2GPI. Inserts show a backbone Cα tracing of the β2GPI DI structure highlighting the site of mutation (red spheres) and residues whose cross peaks are shifted in the spectrum of the mutant protein by greater than the cross peak linewidth (yellow spheres). Note that the majority of cross peaks are not shifted by the mutations, and that the shifts are for residues local to the mutation site, indicating maintenance of the overall 3D fold of the variant proteins. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.3. Substitution of VH CDR3 arginine residues in the panel of ten monoclonal VH/VL combinations alters binding to native DI and CL in a similar pattern
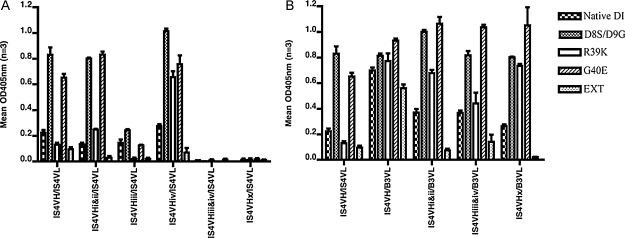
We studied ten monoclonal VH/VL combinations with arginine to serine mutations at four different sites (R96S, R97S, R100S and R100gS) in IS4VHCDR3, paired with either IS4VL or B3VL (Table 1). Our previous work focused on the binding of these variants to a range of antigens (Giles et al., 2003, 2006, 2009), effects on cultured endothelial cells in vitro, and the ability to promote thrombosis in a mouse model in vivo (Giles et al., 2009). We do not reproduce those published results in this paper, but the known sequence and key binding properties of the variants are summarised here in Table 1. Previously, we showed that introducing any of these four mutations into IS4VH/IS4VL reduced binding to both CL and β2GPI (Giles et al., 2006). We found a similar effect of these mutations on binding to n-DI, with a pattern that most closely mirrors binding to CL (binding to CL, β2GPI and DI are summarised in Table 1, binding to DI variants is graphically shown in Fig. 4A and B). In the presence of IS4VL (Fig. 4A) any mutation in IS4VH reduced binding to n-DI, apart from R100gS (IS4VHiv/IS4VL) which displayed reduced binding to CL and β2GPI but not to n-DI. In particular, simultaneous mutation of R100S and R100gS (IS4VHiii&iv/IS4VL and IS4VHx/IS4VL) had the most profound effects with abrogation of binding to n-DI (Fig. 4A). Therefore, the combination of R100 and R100g residues in native IS4 is important for the recognition of CL, β2GPI and n-DI.
Table 1.
The ability of monoclonal aPL to bind native antigens.
| Heavy chain | Position 96 | Position 97 | Position 100 | Position 100g | Light chain | CL | β2GPI | Native DI |
|---|---|---|---|---|---|---|---|---|
| IS4VH | R | R | R | R | IS4VL | ++ | + | + |
| IS4VHi&ii | S | S | R | R | IS4VL | + | − | + |
| IS4VHiii | R | R | S | R | IS4VL | + | + | + |
| IS4VHiv | R | R | R | S | IS4VL | + | + | + |
| IS4VHiii&iv | R | R | S | S | IS4VL | − | − | − |
| IS4VHx | S | S | S | S | IS4VL | − | − | − |
| IS4VH | R | R | R | R | B3VL | ++++ | ++ | ++ |
| IS4VHi&ii | S | S | R | R | B3VL | +++ | + | + |
| IS4VHiii&iv | R | R | S | S | B3VL | +++ | + | + |
| IS4VHx | S | S | S | S | B3VL | + | − | + |
A panel of 10 monoclonal aPL, all comprised of native or mutated IS4VH paired with either IS4VL or B3VL, were tested against CL, β2GPI, and native DI. Variants of IS4VH are named using Roman numerals to represent positions at which arginine (R) residues in IS4VHCDR3 have been replaced by serine (S) as indicated. Each VH/VL combination was tested at 500 ng/ml in triplicate and the degree of binding was defined from the mean absorbance as follows: −, OD < 0.1; +, OD 0.1–<0.4; ++, OD 0.4–<0.8; +++, OD 0.8–<1.2; and ++++, OD ≥ 1.2. CL and β2GPI binding has previously been described in full (Giles et al., 2006). Abbreviations: β2GPI, beta 2 glycoprotein I; CDR, complementarity determining region; CL, cardiolipin; DI, Domain I of β2GPI; VH, variable heavy chain sequence; VL, variable light chain sequence.
Fig. 4.
Altering the pattern of arginine residues in aPL affects their ability to bind native and mutated DI of β2GPI. (A) Mutating arginine residues in IS4VH affects the ability of native IS4 (IS4VH/IS4VL) to recognise DI variants. Native IS4VH and five IS4VH mutants, each with one or more arginine residues mutated, were paired with IS4VL and tested against five DI variants. (B) B3VL is dominant over IS4VL in allowing monoclonal aPL to bind DI variants. Native IS4VH and three variants were paired with B3VL instead of IS4VL. All antibodies were tested at 500 ng/ml, in triplicate. Mean OD at 405 nm is shown in the y-axis, with standard deviation bars. The ability of each antibody to bind native DI (as per Table 1) is also shown here. A summary of how each antibody bound each antigen is listed in Table 2.
As observed with CL and β2GPI binding, the presence of B3VL (Fig. 4B) enhanced the ability of native and mutated IS4VH chains to bind n-DI compared with equivalent IS4VH/IS4VL pairings (Fig. 4A and B). In the presence of IS4VL, R100S and R100gS mutations clearly had a much larger effect than R96S and R97S on binding to n-DI, however this pattern was not observed in combinations containing B3VL. In these B3VL combinations (IS4VHiii&iv/B3VL; IS4VHx/B3VL; and IS4VHi&ii/B3VL), all mutations introduced into IS4VH reduced binding to n-DI by a similar degree.
3.4. IS4-derived monoclonal antibodies differ in their ability to discriminate variants of DI
Previously, we showed by both direct and fluid-phase inhibition ELISA that polyclonal APS-IgG bound strongly to n-DI and DI(D8S/D9G), but displayed reduced binding to the DI variants DI(R39K) and DI(R39S) (Ioannou et al., 2007). Binding to the DI(G40E) and DI(EXT) variants was difficult to interpret since direct ELISA results suggested an increase in binding to APS-IgG, but inhibition ELISAs failed to confirm this finding. In this study, we found that the native combination IS4VH/IS4VL was very similar to APS-IgG in its pattern of direct binding to most of the DI variants. Compared to its ability to bind n-DI, IS4VH/IS4VL bound 3.7-fold more strongly to DI(D8S/D9G), 2.9-fold more strongly to DI(G40E) and almost 2-fold less strongly to DI(R39K) (Table 2 and Fig. 4A). Binding to the DI(EXT) variant was 2.5-fold less strong as to n-DI, consistent with the results seen in fluid-phase binding to APS-IgG (Ioannou et al., 2007).
Table 2.
Tabular presentation of the results graphically illustrated in Fig. 4.
| Native DI | D8S/D9G | R39K | G40E | EXT | |
|---|---|---|---|---|---|
| IS4VH/IS4VL | 100% (1) | 3.7 | 0.6 | 2.9 | 0.4 |
| IS4VHi&ii/IS4VL | 0.6 | 3.6 | 1.1 | 3.7 | 0.1 |
| IS4VHiii/IS4VL | 0.6 | 1.1 | 0.1 | 0.6 | 0.1 |
| IS4VHiv/IS4VL | 1.2 | 4.5 | 2.9 | 3.4 | 0.3 |
| IS4VHiii&iv/IS4VL | 0 | 0 | 0 | 0 | 0.1 |
| IS4VHx/IS4VL | 0 | 0.1 | 0.1 | 0.1 | 0 |
| IS4VH/B3VL | 3.1 | 3.6 | 3.5 | 4.2 | 2.5 |
| IS4VHi&ii/B3VL | 1.7 | 4.5 | 3.0 | 4.8 | 0.3 |
| IS4VHiii&iv/B3VL | 1.7 | 3.7 | 2.0 | 4.6 | 0.6 |
| IS4VHx/B3VL | 1.2 | 5.2 | 3.3 | 4.7 | 0.1 |
Binding of native IS4 (IS4VH/IS4VL) to native DI was determined as 100% activity (equivalent to 1-fold), and was used to calculate a fold increase or decrease in the ability of each aPL to bind each of the five DI variants used in this study.
In contrast, none of the IS4VH/IS4VL variants with mutations introduced in IS4VHCDR3 showed the same degree of similarity to APS-IgG as native IS4VH/IS4VL (Table 2 and Fig. 4A). Introducing the R100S mutation reduced (IS4VHiii/IS4VL) or abrogated (IS4VHiii&iv/IS4VL and IS4VHx/IS4VL) binding to all DI variants. Double R96S and R97S or single R100gS mutations (IS4VHi&ii/IS4VL and IS4VHiv/IS4VL) had a more subtle effect. These variants were still able to bind strongly to DI(D8S/D9G) and DI(G40E) variants, but unlike IS4VH/IS4VL they bound to DI(R39K) better than n-DI, indicating a reduced ability to discriminate the residue at position 39 on DI. This phenomenon was particularly marked for IS4VHiv/IS4VL, which bound very strongly to DI(R39K). A similar lack of selectivity of binding was seen more clearly in B3VL combinations (Table 2 and Fig. 4B). All such combinations displayed enhanced binding to variant forms of DI – except DI(EXT) – compared to n-DI. IS4VH/B3VL bound n-DI, and all its variants (including DI(R39K)) to a similar degree. Combinations of B3VL with IS4VH containing any arginine to serine mutation showed excellent binding to DI(D8S/D9G), DI(R39K) and DI(G40E). However, introduction of these arginine–serine mutations into IS4VH/B3VL did reduce binding to n-DI and DI(EXT).
4. Discussion
This study has resulted in two advances important to the potential development of recombinant DI as a treatment for APS. First, we established cytoplasmic expression of recombinant human DI in E. coli in high yield. Secondly, we confirmed by NMR analysis that the structures of in vitro folded DI and two variants are consistent with its crystal structure. Therefore, the effects of D8-D9 and R39 substitutions on antigen–antibody interactions are dominated by local effects on the surface properties of DI and do not cause major changes in the three-dimensional structure of the domain. This confirms the predictions of the computer models of the structure of DI published in our previous paper (Ioannou et al., 2007).
We have recently used the murine femoral vein pinch model of APS to show that bacterially expressed human recombinant n-DI and DI(D8S/D9G) but not DI(R39S) significantly reduced thrombus size induced by APS-IgG in vivo (Ioannou et al., 2009). The inhibitory effect of DI was dose dependent and at lower doses DI(D8S/D9G) was a more potent inhibitor than n-DI. Furthermore, DI inhibited APS-IgG induction of VCAM-1 on the aortic endothelial surface and production of tissue factor by murine macrophages (Ioannou et al., 2009). Hence n-DI, as well as DI(D8S/D9G), could be valuable as future therapeutics directly targeting pathogenic aPL, as opposed to current treatments which were not developed specifically for the APS but are generic anticoagulants such as warfarin and heparin. There are numerous expression systems currently used to produce human recombinant proteins for therapeutic purposes, such as bacterial, yeast, insect and mammalian cells. E. coli were the first such expression systems to be successfully exploited for drug production, leading to recombinant human insulin production to treat diabetes (Kroeff et al., 1989). Bacterial expression has important advantages in terms of larger scale and lower expense, but is best used for proteins that lack glycosylation, such as recombinant tissue plasminogen activator, used to treat patients with acute myocardial infarction. DI is not glycosylated in vivo and is thus a good candidate for bacterial expression, as achieved in this study.
It is widely accepted that monoclonal aPL accumulate arginine to serine mutations, and these arginine residues allow recognition of antigenic targets. We previously showed that the R100 and R100g residues in IS4VHCDR3 played a critical role in binding of IS4 to PL and β2GPI, whereas R96 and R97 residues did not (Giles et al., 2003, 2005, 2006). Our current findings are that mutations at R100 and R100g in IS4VH are also more effective at altering binding to DI, compared to mutations at R96 and R97. This result supports the theory that DI contains an important epitope, that apparently drove the accumulation of R100 and R100g in the B cell clone that originally produced IS4. From this study, we now see a distinction between R100 and R100g that was not evident before. Any IS4VH/IS4VL mutant carrying the R100S mutation displayed reduced/no binding to all DI variants tested. In contrast, introducing the single R100gS mutation into IS4VH/IS4VL enhanced binding to DI(D8S/D9G), DI(R39K), and DI(G40E) mutants.
Therefore, we propose that the presence of R100 is critical in enabling IS4-DI binding whilst R100g is not as important but plays a role in the interaction with the R39 epitope. In particular, the R39K mutation in DI dramatically enhances the binding of IS4VHiv/IS4VL, hence the residues at position 100g in IS4VHCDR3 and position 39 in DI may interact closely in this antigen/antibody complex. Since both of these residues are positively charged arginines in the native sequences, any interaction between them is unlikely to be entirely electrostatic in nature. In fact, our findings do not suggest a major role for interaction of positive residues in IS4VHCDR3 with surface-exposed negative residues on the surface of DI. For example, we found that mutation of R100g to serine in IS4VH/IS4VL enhanced binding to n-DI compared to native IS4. Furthermore, most VH/VL combinations regardless of the pattern of arginine to serine mutations displayed enhanced binding to DI(D8S/D9G) which has a reduction in overall surface charge. Therefore, alternative mechanisms such as hydrogen bond or hydrophobic contact formation may be important in the aPL–DI interaction.
We previously showed that IS4VH/B3VL binds strongly to PL and β2GPI but is not pathogenic in mice whereas the closely related IS4VHi&ii/B3VL and IS4VH/IS4VL are pathogenic (Giles et al., 2009). We postulated whether this difference occurs because IS4VHi&ii/B3VL and IS4VH/IS4VL bind thrombin whereas IS4VH/B3VL does not, and/or because of some other difference in binding selectivity. Our current study explores the hypothesis that this difference in pathogenicity may also reflect differences between the ways in which DI is bound by these three monoclonal antibodies. Our results provide some support for this hypothesis. In particular, IS4VH/B3VL displayed enhanced binding to all DI variants with very little discrimination between native and mutant DI – a completely different pattern to that seen with IS4VH/IS4VL. The properties of IS4VHi&ii/B3VL binding to DI are intermediate between those of IS4VH/IS4VL and IS4VH/B3VL (see Fig. 4), which is consistent with our previous studies examining binding to PL and β2GPI (Giles et al., 2006, 2009). We previously showed that of six PL tested, IS4VH/B3VL bound strongly to all six (including neutral PL), IS4VHi&ii/B3VL bound four of six (and not to neutral PL) and IS4VH/IS4VL bound two of six PL tested (not neutral PL). In the current study, IS4VH/B3VL binds strongly to all DI variants, IS4VHi&ii/B3VL does not bind the DI(EXT) variant and has lower binding to DI(R39K), whereas IS4VH/IS4VL binds only n-DI, DI(D8S/D9G) and DI(G40E) (i.e. with a similar profile to polyclonal IgG derived from the blood of patients with APS). The lack of selectivity of binding displayed by IS4VH/B3VL is striking and unique amongst this panel of ten IgG antibodies. However there is no simple relationship between pathogenicity and ability to discriminate different DI variants. Thus IS4VHi&ii/B3VL and IS4VHi&ii/IS4VL show very similar patterns of binding to the five DI variants studied, but only the first was pathogenic in mice (Giles et al., 2009), and only four of these ten VH/VL combinations have been tested for pathogenicity in the mouse model. A full understanding of the structure–activity profile for these antibodies will require further progress in obtaining high resolution structural data for one or more of them in complex with the relevant cognate antigen(s).
In summary we have developed a new bacterial expression system enabling the production of high yields of n-DI and DI(D8S/D9G) in the form of cytoplasmic inclusion bodies from which DI proteins can be isolated and refolded in vitro. NMR studies indicate that these recombinant molecules have very similar structure to that of DI found as part of whole β2GPI in vivo. Both n-DI and DI(D8S/D9G) have been shown to inhibit binding and pathogenicity of IgG aPL derived from patients with APS. Thus the production of these molecules in a high-yield bacterial system has clear implications for the feasibility of developing recombinant n-DI and/or DI(D8S/D9G) as therapeutic agents for the APS.
Disclosure
The authors have no conflicting financial interests.
Acknowledgements
We are indebted to Dr. Pojen Chen for donating the hybridoma line of native IS4. Monoclonal IgG expression vectors were a kind gift from Dr. Katy Kettleborough and Dr. Tarran Jones (Aeres Biomedical, London, UK). We are thankful to Dr. Siobhan ÒBrien and Dr. Alison Levy for their help and advice on the assembly of IgG constructs for expression. The murine monoclonal anti-DI antibody (mAb16) used in Western blot studies was a kind gift from Dr. Mike Iverson and Dr. Matt Linnik (La Jolla Pharmaceuticals, CA, USA). We are grateful for access to the MRC Centre for Biomedical NMR at Mill Hill. This work was supported by Arthritis Research, UK and the Oliver Bird Rheumatism Programme, Nuffield Foundation. The work in the Driscoll group is supported by the MRC (file reference U117574559). Dr. Ian Giles acknowledges support from the Rosetrees Foundation.
References
- Alarcon-Segovia D., Deleze M., Oria C.V., Sanchez-Guerrero J., Gomez-Pacheco L., Cabiedes J., Fernandez L., Ponce de Leon S. Antiphospholipid antibodies and the antiphospholipid syndrome in systemic lupus erythematosus. A prospective analysis of 500 consecutive patients. Medicine (Baltimore) 1989;68(6):353–365. doi: 10.1097/00005792-198911000-00003. [DOI] [PubMed] [Google Scholar]
- Bouma B., de Groot P.G., van den Elsen J.M., Ravelli R.B., Schouten A., Simmelink M.J., Derksen R.H., Kroon J., Gros P. Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure. EMBO J. 1999;18(19):5166–5174. doi: 10.1093/emboj/18.19.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother W.J., Palmer A.G., III, Skelton N.J., Rance M. second ed. Academic Press; 2006. Protein NMR Spectroscopy: Principles and Practice. [Google Scholar]
- Cervera R., Khamashta M.A., Shoenfeld Y., Camps M.T., Jacobsen S., Kiss E., Zeher M.M., Tincani A., Kontopoulou-Griva I., Galeazzi M., Bellisai F., Meroni P.L., Derksen R.H., de Groot P.G., Gromnica-Ihle E., Baleva M., Mosca M., Bombardieri S., Houssiau F., Gris J.C., Quere I., Hachulla E., Vasconcelos C., Roch B., Fernandez-Nebro A., Piette J.C., Espinosa G., Bucciarelli S., Pisoni C.N., Bertolaccini M.L., Boffa M.C., Hughes G.R. Morbidity and mortality in the antiphospholipid syndrome during a 5-year period: a multicenter prospective study of 1,000 patients. Ann. Rheum. Dis. 2009;68(9):1428–1432. doi: 10.1136/ard.2008.093179. [DOI] [PubMed] [Google Scholar]
- de Laat B., Derksen R.H., Urbanus R.T., de Groot P.G. IgG antibodies that recognize epitope Gly40–Arg43 in domain I of beta 2-glycoprotein I cause LAC, and their presence correlates strongly with thrombosis. Blood. 2005;105(4):1540–1545. doi: 10.1182/blood-2004-09-3387. [DOI] [PubMed] [Google Scholar]
- de Laat H.B., Derksen R.H., Urbanus R.T., Roest M., de Groot P.G. Beta2-glycoprotein I-dependent lupus anticoagulant highly correlates with thrombosis in the antiphospholipid syndrome. Blood. 2004;104(12):3598–3602. doi: 10.1182/blood-2004-03-1107. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Giles I., Haley J., Nagl S., Latchman D., Chen P., Chukwuocha R., Isenberg D., Rahman A. Relative importance of different human aPL derived heavy and light chains in the binding of aPL to cardiolipin. Mol. Immunol. 2003;40:49–60. doi: 10.1016/s0161-5890(03)00100-7. [DOI] [PubMed] [Google Scholar]
- Giles I., Lambrianides N., Latchman D., Chen P., Chuckwuocha R., Isenberg D., Rahman A. The critical role of arginine residues in the binding of human antiphospholipid antibodies to cardiolipin. Arthritis Res. Ther. 2005;7(1):R47–R56. doi: 10.1186/ar1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles I., Lambrianides N., Pattni N., Faulkes D., Latchman D., Chen P., Pierangeli S., Isenberg D., Rahman A. Arginine residues are important in determining the binding of human monoclonal antiphospholipid antibodies to clinically relevant antigens. J. Immunol. 2006;177(3):1729–1736. doi: 10.4049/jimmunol.177.3.1729. [DOI] [PubMed] [Google Scholar]
- Giles I., Pericleous C., Liu X., Ehsanullah J., Clarke L., Brogan P., Newton-West M., Swerlick R., Lambrianides A., Chen P., Latchman D., Isenberg D., Pierangeli S., Rahman A. Thrombin binding predicts the effects of sequence changes in a human monoclonal antiphospholipid antibody on its in vivo biologic actions. J. Immunol. 2009;182(8):4836–4843. doi: 10.4049/jimmunol.0804241. [DOI] [PubMed] [Google Scholar]
- Greaves M., Cohen H., MacHin S., Mackie I. Guidelines on the investigation and management of the antiphospholipid syndrome. Br. J. Haematol. 2000;109(4):704–715. doi: 10.1046/j.1365-2141.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- Hughes G.R. Thrombosis, abortion, cerebral disease, and the lupus anticoagulant. Br. Med. J. 1983;287(6399):1088–1089. doi: 10.1136/bmj.287.6399.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes G.R., Harris N.N., Gharavi A.E. The anticardiolipin syndrome. J. Rheumatol. 1986;13(3):486–489. [PubMed] [Google Scholar]
- Ioannou Y., Giles I., Lambrianides A., Richardson C., Pearl L.H., Latchman D.S., Isenberg D.A., Rahman A. A novel expression system of domain I of human beta2 glycoprotein I in Escherichia coli. BMC Biotechnol. 2006;6:8. doi: 10.1186/1472-6750-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou Y., Pericleous C., Giles I., Latchman D.S., Isenberg D.A., Rahman A. Binding of antiphospholipid antibodies to discontinuous epitopes on domain I of human beta(2)-glycoprotein I: mutation studies including residues R39 to R43. Arthritis Rheum. 2007;56(1):280–290. doi: 10.1002/art.22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou Y., Romay-Penabad Z., Pericleous C., Giles I., Papalardo E., Vargas G., Shilagard T., Latchman D.S., Isenberg D.A., Rahman A., Pierangeli S. In vivo inhibition of antiphospholipid antibody-induced pathogenicity utilizing the antigenic target peptide domain I of beta2-glycoprotein I: proof of concept. J. Thromb. Haemost. 2009;7(5):833–842. doi: 10.1111/j.1538-7836.2009.03316.x. [DOI] [PubMed] [Google Scholar]
- Iverson G.M., Jones D.S., Marquis D., Linnik M.D., Victoria E.J. A chemically defined, toleragen-based approach for targeting anti-beta2-glycoprotein I antibodies. Lupus. 1998;7(Suppl. 2):S166–S169. doi: 10.1177/096120339800700236. [DOI] [PubMed] [Google Scholar]
- Iverson G.M., Reddel S., Victoria E.J., Cockerill K.A., Wang Y.X., Marti-Renom M.A., Sali A., Marquis D.M., Krilis S.A., Linnik M.D. Use of single point mutations in domain I of beta 2-glycoprotein I to determine fine antigenic specificity of antiphospholipid autoantibodies. J. Immunol. 2002;169(12):7097–7103. doi: 10.4049/jimmunol.169.12.7097. [DOI] [PubMed] [Google Scholar]
- Kandiah D.A., Sali A., Sheng Y., Victoria E.J., Marquis D.M., Coutts S.M., Krilis S.A. Current insights into the antiphospholipid syndrome: clinical, immunological, and molecular aspects. Adv. Immunol. 1998;70:507–563. doi: 10.1016/s0065-2776(08)60393-4. [DOI] [PubMed] [Google Scholar]
- Kroeff E.P., Owens R.A., Campbell E.L., Johnson R.D., Marks H.I. Production scale purification of biosynthetic human insulin by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;461:45–61. doi: 10.1016/s0021-9673(00)94274-2. [DOI] [PubMed] [Google Scholar]
- Lynch A., Marlar R., Murphy J., Davila G., Santos M., Rutledge J., Emlen W. Antiphospholipid antibodies in predicting adverse pregnancy outcome. A prospective study. Ann. Intern. Med. 1994;120(6):470–475. doi: 10.7326/0003-4819-120-6-199403150-00004. [DOI] [PubMed] [Google Scholar]
- McNally T., Mackie I.J., Machin S.J., Isenberg D.A. Increased levels of beta 2 glycoprotein-I antigen and beta 2 glycoprotein-I binding antibodies are associated with a history of thromboembolic complications in patients with SLE and primary antiphospholipid syndrome. Br. J. Rheumatol. 1995;34(11):1031–1036. doi: 10.1093/rheumatology/34.11.1031. [DOI] [PubMed] [Google Scholar]
- McNeeley P.A., Dlott J.S., Furie R.A., Jack R.M., Ortel T.L., Triplett D.A., Victoria E.J., Linnik M.D. Beta2-glycoprotein I-dependent anticardiolipin antibodies preferentially bind the amino terminal domain of beta2-glycoprotein I. Thromb. Haemost. 2001;86(2):590–595. [PubMed] [Google Scholar]
- Miyakis S., Lockshin M.D., Atsumi T., Branch D.W., Brey R.L., Cervera R., Derksen R.H., De Groot P.G., Koike T., Meroni P.L., Reber G., Shoenfeld Y., Tincani A., Vlachoyiannopoulos P.G., Krilis S.A. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS) J. Thromb. Haemost. 2006;4(2):295–306. doi: 10.1111/j.1538-7836.2006.01753.x. [DOI] [PubMed] [Google Scholar]
- Pierangeli S.S., Liu X., Espinola R., Olee T., Zhu M., Harris N.E., Chen P.P. Functional analyses of patient-derived IgG monoclonal anticardiolipin antibodies using in vivo thrombosis and in vivo microcirculation models. Thromb. Haemost. 2000;84(3):388–395. [PubMed] [Google Scholar]
- Reddel S.W., Wang Y.X., Sheng Y.H., Krilis S.A. Epitope studies with anti-beta 2-glycoprotein I antibodies from autoantibody and immunized sources. J. Autoimmun. 2000;15(2):91–96. doi: 10.1006/jaut.2000.0427. [DOI] [PubMed] [Google Scholar]
- Schwarzenbacher R., Zeth K., Diederichs K., Gries A., Kostner G.M., Laggner P., Prassl R. Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. EMBO J. 1999;18(22):6228–6239. doi: 10.1093/emboj/18.22.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N.M., Khamashta M.A., Atsumi T., Hughes G.R. Outcome of patients with anticardiolipin antibodies: a 10 year follow-up of 52 patients. Lupus. 1998;7(1):3–6. doi: 10.1191/096120398678919624. [DOI] [PubMed] [Google Scholar]
- Sharma D., Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR. 2000;18(2):165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Tsutsumi A., Matsuura E., Ichikawa K., Fujisaku A., Mukai M., Kobayashi S., Koike T. Antibodies to beta 2-glycoprotein I and clinical manifestations in patients with systemic lupus erythematosus. Arthritis Rheum. 1996;39(9):1466–1474. doi: 10.1002/art.1780390905. [DOI] [PubMed] [Google Scholar]
- Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang C.C., Chen J.H., Yin S.H., Chuang W.J. Predicting the redox state and secondary structure of cysteine residues in proteins using NMR chemical shifts. Proteins. 2006;63(1):219–226. doi: 10.1002/prot.20875. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Sykes B.D. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4(2):171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6(2):135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Zhu M., Olee T., Le D.T., Roubey R.A., Hahn B.H., Woods V.L., Jr., Chen P.P. Characterization of IgG monoclonal anti-cardiolipin/anti-beta2GP1 antibodies from two patients with antiphospholipid syndrome reveals three species of antibodies. Br. J. Haematol. 1999;105(1):102–109. [PubMed] [Google Scholar]