Abstract
Lipid droplets (LDs) are organelles found in most types of cells in the tissues of vertebrates, invertebrates, and plants, as well as in bacteria and yeast. They differ from other organelles in binding a unique complement of proteins and lacking an aqueous core but share aspects of protein trafficking with secretory membrane compartments. In this minireview, we focus on recent evidence supporting an endoplasmic reticulum origin for LD formation and discuss recent findings regarding LD maturation and fusion.
Keywords: Adipocyte, Endoplasmic Reticulum (ER), Lipid Droplet, Lipid Metabolism, Triacylglycerol, Perilipin
Introduction
Eukaryotic cells produce and traffic membranes that maintain intracellular compartments that facilitate regulation of metabolism by controlling the subcellular localization of metabolites, the concentrations of enzymes and substrates to optimize enzyme reactions, and the isolation of toxic metabolites. Fatty acids supply a major source of energy for organisms but also can be toxic. Cells escape cytotoxicity by esterifying fatty acids into neutral lipids and packaging them into lipid droplets (LDs)3 coated with phospholipids and proteins. When cells are exposed to excess fatty acids, protein-coated LDs replete with neutral lipids bud from membranes of the endoplasmic reticulum (ER). When levels of exogenous fatty acids wane, the surface proteins of nascent LDs are exchanged for other proteins that serve various functions on mature LDs. Recent studies have established LDs as a distinct organelle.
The protein components of LDs have been increasingly studied over the past decade. In chordates and flies, members of the perilipin family of proteins are among the most abundant proteins coating LDs (1, 2). Perilipins interact with both lipid and cytosolic proteins, forming an amphipathic interface between stored lipids and the cytosol. LDs in the cells of various vertebrate tissues are coated with two to four types of perilipin; each unique combination of perilipins imparts tissue-specific management of lipid metabolism. The expression of perilipin 1 is limited to adipocytes of white and brown adipose tissue and steroidogenic cells of the adrenal cortex, testes, and ovaries; moreover, subcellular localization of perilipin 1 is essentially restricted to LDs. Perilipin 2 (formerly adipophilin/ADRP (adipose differentiation-related protein)) and perilipin 3 (formerly TIP47 (tail-interacting protein of 47 kDa)) are ubiquitously expressed and hence contribute to the protein coat of LDs in the majority of cells. Perilipin 2 localizes primarily to LDs. Perilipin 3 is stable in the cytoplasm of cells but rapidly associates with nascent LDs when cells are incubated with fatty acids to promote triacylglycerol (TAG) synthesis and storage. Perilipin 4 (formerly S3-12) is expressed primarily in adipocytes of white adipose tissue and localizes to buds of nascent LDs along the ER. Perilipin 5 (formerly MLDP (myocardial lipid droplet protein)/OXPAT (oxidative protein of the PAT family)/LSDP5 (lipid storage droplet protein 5)) is expressed in oxidative tissues, including cardiac and skeletal muscle, and in brown adipocytes. In addition to perilipins, the list of bona fide LD proteins has been growing rapidly and now includes members of the CIDE (cell death-inducing DFF45-like effector) protein family (FSP27 (fat-specific protein of 27 kDa) or CIDEC, CIDEA, and CIDEB), putative methyltransferases METTL7A and METTL7B, and a variety of enzymes required for lipid metabolism (3). We will discuss recent findings on some of these proteins in the context of an emerging model for LD formation. Additionally, we will review key findings that identify protein components of the ER as essential for LD formation and expansion.
Enzymes for Neutral Lipid Synthesis Reside in ER
The majority of cellular membranes comprise the secretory pathway; membrane flow to maintain these compartments originates from the ER. Several distinct organelles are created and maintained by diverting membranes from this stream. Increasing evidence shows that LD constituents, both protein and lipid, are channeled out of ER membrane flow directly onto LDs. Moreover, ultrastructural imaging often shows the ER tightly wrapped around LDs (4, 5), indicative of an intimate relationship between these compartments. This relationship makes sense because the enzymes catalyzing the final steps of neutral lipid synthesis reside on the ER.
LDs emerge from the ER when fatty acids are esterified to glycerol or cholesterol. Given the toxicity of free fatty acids and the need for an esterified fatty acid energy reserve, survival can hinge on esterification capacity; efficiency increases an organism's viability. The enzymes required for neutral lipid synthesis reside in the ER. First, the ER has robust acyl-CoA synthetase (ACS) activity (6) to activate fatty acids for further metabolism. Next, ER enzymes can make TAG from fatty acyl-CoA and glycerol 3-phosphate via glycerol-3-phosphate acyltransferases (GPATs), 1-acylglycerol-3-phosphate acyltransferases (AGPATs), phosphatidic acid phosphohydrolase (PAP; also lipin), and two isoforms of diacylglycerol acyltransferase (DGAT) (Fig. 1) (7–9). Additionally, esterification of fatty acids to cholesterol is catalyzed by the ER transmembrane proteins ACAT1 (acyl-CoA:cholesterol acyltransferase 1) and ACAT2 (10). Neutral lipids are thought to be formed within specific domains of the ER called mitochondria-associated membranes (11, 12). These data point to the ER as the origin for nascent LDs.
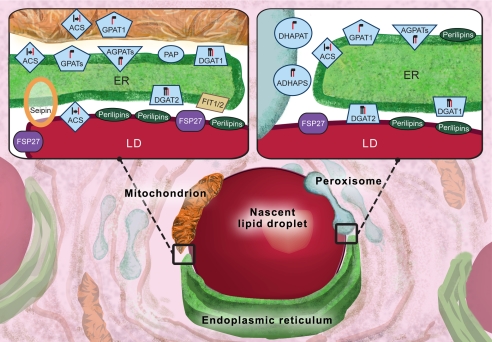
FIGURE 1.
LDs are surrounded by ER, mitochondria, and peroxisomes, which contribute to synthesis of lipids to be assembled into LDs. The insets depict areas where LDs come into close contact with the ER and mitochondria (left) and peroxisomes as well as the ER (right). TAGs are synthesized by enzymes associated with LDs (dark red), the ER (green), and mitochondria (brown). ACSs localize to the ER, LDs, and mitochondria and activate fatty acids for esterification or catabolism. Several isoforms of GPAT localize to the ER and mitochondria and initiate the reactions to build complex lipids. Proteins in the AGPAT family add a second acyl chain (red) to 1-acylglycerol 3-phosphate at the ER. PAP removes the phosphate group so that DGAT can add the final acyl chain to complete the synthesis of TAG. DGAT2 is found on the ER and in close proximity to LDs. TAG is then packaged into the core of LDs. Peroxisomes (cyan) are the sole location for ether lipid synthesis, which requires the enzymes dihydroxyacetone-phosphate acyltransferase (DHAPAT) and alkyldihydroxyacetone-phosphate synthase (ADHAPS). LDs have both ether-linked neutral lipids and ether-linked phospholipids. Members of the perilipin family localize primarily to LDs; perilipins 3 and 4 also localize to the ER, particularly at regions where neutral lipids accumulate. FIT1 and FIT2 localize to the ER and contribute to LD formation and maturation. FSP27 (also called CIDEC) localizes to LDs, where it affects LD size and catabolism of stored TAG. Seipin localizes to contact points between LDs and the ER.
LDs, Peroxisomes, and Mitochondria Contribute to Neutral Lipid Synthesis
Several of the enzymes required for neutral lipid synthesis have been localized to LDs. Studies of LD proteomics have identified one or more ACSs on LDs (3). DGAT2 apparently localizes to LDs when fatty acids are added to cells to drive TAG synthesis and storage (12, 13); however, it is currently unclear whether increased TAG synthesis triggers movement of DGAT2 from the ER bilayer to the phospholipid monolayer of LDs or whether DGAT2 remains in the ER, and association of the ER with LDs increases. A major question is how a membrane-spanning protein with one or more luminal domains can localize to LDs, which lack an aqueous compartment. Both DGAT2 and ACAT2 have a single ER luminal domain that is rich in hydrophobic residues (10, 14, 15); it has been proposed that both the putative membrane-spanning and luminal sequences of DGAT2 may be embedded into lipid-filled cores of LDs. If this model is correct, then it would not be surprising to find ACAT2 on LDs; however, neither ACAT2 nor DGAT2 has been found in the proteomes of LDs isolated from a number of different cells, suggesting that these enzymes are embedded into membranes that are in close contact with LDs.
Mitochondria and peroxisomes form both acyl-CoA and complex lipids (6, 16, 17) and may thus contribute to LD assembly. Moreover, both organelles closely abut LDs (Fig. 1) (4, 18). Peroxisomes are semiautonomous from the secretory pathway, yet membranes traffic between these compartments (19). Hence, peroxisomal acylation capacity may contribute to fat storage. Consistent with this notion, the biosynthesis of ether-linked phospholipids occurs only in peroxisomes (20), yet ether linkages are found both in the neutral lipid that fills LDs and in the phospholipid monolayer (21). How ether lipids are transported between peroxisomes and LDs is unstudied, but close physical proximity between the two organelles may play a role in lipid delivery.
Mitochondria are semiautonomous from the secretory pathway and have robust ACS activity (6, 16). The relative contribution of mitochondrial ACS to the synthesis of lipid components of LDs is unknown. Mitochondrial GPAT contributes to the incorporation of fatty acids into TAG (22, 23). Multiple proteins demonstrate AGPAT activity, but all of the AGPATs that contribute to TAG synthesis are thought to reside in the ER (8). Thus, acylglycerol transfer from mitochondria to the ER is required to complete the final three steps of TAG synthesis. This substrate transfer may be assisted by membrane contact between mitochondria and the ER. A recent study has demonstrated that phosphatidylethanolamine synthesized by mitochondrial decarboxylation of phosphatidylserine is a substrate for methyltransferase reactions to form a significant portion of phosphatidylcholine coating LDs in adipocytes (24). Thus, some of the phospholipids on LDs are derived from mitochondrial synthesis.
Interestingly, when cells overexpressing DGAT2 are incubated with fatty acids, mitochondria dramatically reorganize to form a tight network surrounding LDs (12). Furthermore, DGAT2 has been demonstrated to have targeting sequences for mitochondria (12) and, when ER-anchoring transmembrane sequences are deleted, localizes to mitochondria (14). Like the wild-type protein, this deletion mutant promotes TAG storage in LDs, suggesting that mitochondrial association of DGAT2 has a functional consequence. Additionally, deletion of caveolin-1 reduces TAG deposition in the LDs of adipose tissue in mice (25) while reducing the localization of mitochondria to the proximity of LDs (26). It is unclear whether the decreased TAG storage is a consequence of decreased substrate import and esterification or is due to rearrangements in membrane contact sites between cellular organelles and LDs that usually facilitate TAG transfer and packaging into LDs. Finally, a recent report shows that C-terminal sequences of perilipin 5 form contact sites between mitochondria and LDs (27); thus, perilipin 5 may facilitate the exchange of lipids between the two compartments.
ER Proteins Are Required for LD Formation and Expansion
The most widely accepted model of LD formation posits the deposition of a lens of neutral lipid between the leaflets of the phospholipid bilayer of the ER, followed by association of LD-specific proteins and eventual scission of the growing LD from the ER. Several ER resident proteins are required for LD formation or expansion, including seipin and the fat storage-inducing transmembrane proteins FIT1 and FIT2 (28, 29). Overexpression of FIT1 or FIT2 in cells or mouse liver increases TAG storage without increasing DGAT activity (28). Conversely, knockdown of FIT2 in cultured adipocytes or zebrafish significantly decreases TAG storage in LDs. Recent studies show that overexpression of FIT2 in mouse skeletal muscle significantly increases storage of TAG in intramyocellular LDs (30). Interestingly, these mice had increased energy expenditure despite decreased β-oxidation of fatty acids in muscle; instead, oxidation of branched-chain amino acids was increased. These observations suggest that FIT proteins play an important role in LD expansion and maintenance, perhaps through increasing transfer of lipids or enzymes from the ER to LDs.
Mutations in the gene for seipin in humans are associated with Berardinelli-Seip congenital lipodystrophy 2, characterized by nearly complete absence of adipose tissue and accumulation of ectopic TAG in liver and muscle (31, 32). Ablation of the gene for seipin in mice produces a comparable phenotype (33). In micrographs of yeast, seipin is detected in puncta at ER-LD junctions (29). Ablation of seipin in yeast results in irregular small and occasional giant LDs, depending upon growth conditions (29, 34). Seipin homologs from Drosophila, mice, and humans complement yeast seipin, suggesting that seipin from multiple organisms has a similar function. Consistent with these observations, Drosophila lacking the seipin homolog store less TAG in the fat body but deposit ectopic TAG in salivary glands, a site that normally lacks stored fat (35). Furthermore, human fibroblasts lacking seipin show a disorganized ER, unusually small LDs, and diffuse staining for lipid and LD proteins throughout the cytoplasm (29, 36). Interestingly, seipin forms a doughnut-shaped homo-oligomer that is functionally important because a nonfunctional mutant protein responsible for Berardinelli-Seip congenital lipodystrophy 2 forms oligomers poorly (37). The function of seipin has not yet been elucidated, but proposed functions include modulation of the metabolism of phosphatidic acid (35), a precursor for both TAG and phospholipid synthesis, and formation of a collar between the ER and LDs that directs organized assembly of LDs (37).
Strong evidence for an ER origin of LDs has been obtained through study of the assembly of oleosins onto LDs in the seeds of plants. Oleosins are inserted into the ER during translation and anchor into plant LDs via a hydrophobic spike of ∼75 amino acids with a central proline knot motif (38, 39). It is thought that the central hydrophobic spike requires insertion into areas where the membrane bilayers of the ER expand to accommodate TAG because the sequence is longer than the ER membrane bilayer is thick, and hydrophobic amino acids in this motif are incompatible with the aqueous lumen of the ER. Thus, the hydrophobic spike assures targeting of oleosins to patches of ER where nascent LDs are forming and retention of oleosins on LDs as they sever from the ER and become autonomous organelles. These examples support the concept that the ER has discrete protein-mediated and regulated exit sites for LDs.
Diacylglycerol Is Required for Formation of LDs and Recruitment of Exchangeable Perilipins to Nascent Droplets
Perilipins are abundant structural proteins associated with LDs of chordates, where they function in the control of lipolysis (1). The perilipins can be segregated into two functional groups based on subcellular localization. Exchangeable perilipins, including perilipins 3–5, are stable both in the cytoplasm and at the surfaces of LDs; constitutive perilipins, including perilipins 1 and 2, are unstable in the cytoplasm and associate primarily with LDs (2). In adipocytes, the earliest deposits of neutral lipid are coated with perilipins 3 and 4 (Fig. 2), which are recruited from the cytosol (40, 41). Little is known about how perilipins recognize nascent LDs forming along the ER, but locally increased diacylglycerol (DAG) content is a likely determinant (42). Incubation of cells with a cell-permeable form of DAG, such as 1,2-dioctanoylglycerol or 1-oleoyl-2-acetylglycerol, increases recruitment of perilipin 3 to ER membranes (42); this effect is augmented by the addition of a DAG lipase inhibitor to prevent catabolism of DAG. In contrast, overexpression of DGAT1 reduces perilipin 3 recruitment to the ER, unless triacsin C, an inhibitor of ACS, is added to inhibit activation of fatty acids for esterification. Similarly, perilipins 4 and 5 are also recruited to the ER when the local DAG concentration is elevated (42). Furthermore, the importance of DAG in LD formation has been shown in yeast, which lacks perilipins; DAG produced by PAP is critical to LD formation, even when the yeast enzymes required for the synthesis of TAG are deleted and LDs contain primarily sterol esters (43). In the absence of PAP, neutral lipids accumulate in the ER, presumably within the membrane bilayer, in a disorganized arrangement. This requirement for DAG is significant because DAG is the metabolic intermediate for either TAG or phospholipid synthesis or hydrolysis to release a source of fatty acids for β-oxidation. Thus, DAG is a branch point for determining the use of fatty acids for storage, growth, or energy production.
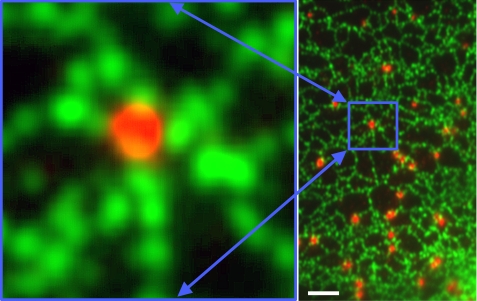
FIGURE 2.
Earliest deposits of neutral lipid are found along ER. OP9 preadipocytes ectopically expressing HA epitope-tagged AGPAT2 were incubated with fatty acids to induce LD biogenesis. Cells were fixed and stained with anti-HA antibody to reveal the ER (green) and with anti-perilipin 3 antibody to detect LDs (red). Scale bar = 2 μm.
ER-LD Continuity or Distinct Organelles?
Recent imaging experiments in yeast suggest that LDs remain intimately associated with the ER (29, 44, 45); it has not been fully resolved whether the limiting phospholipid monolayer of the yeast LD is contiguous with the membrane bilayer of the ER or, alternatively, the two membranes are distinct but share contact points. Experiments with LD-associated proteins, such as Erg6, a sterol methyltransferase in the biosynthetic pathway for ergosterol, suggest that LDs in yeast are contiguous with the ER (44). Erg6 localizes uniformly to the ER when genes encoding neutral lipid synthesis enzymes are deleted in yeast but rapidly transfers to LDs when a gene encoding an enzyme that synthesizes TAG is re-induced. Moreover, when GFP-Erg6 on and near LDs is photobleached, GFP-Erg6 fluorescence on LDs recovers with a t½ of 2 min in the absence of translation of new proteins. These data suggest that Erg6 transfers between the ER and LDs via diffusion through contiguous membranes. Similar data have been obtained for yeast DGAT, suggesting that it also traffics readily between the ER and LDs (44). Because these proteins display uniform distribution over the entire LD, it is thought that ER membranes are continuous with the phospholipid monolayer of LDs in yeast; alternatively, yeast LDs are fully enwrapped in the ER. This raises the possibility that TAG transfers via diffusion through the ER to LDs in yeast.
In animal cells, experimental evidence suggests that once nascent LDs accumulate sufficient neutral lipid, they dissociate from the ER in a budding process; the protein composition of LDs is distinctly different from that of ER membranes. The mechanism for budding has been minimally elucidated but presumably requires a complex of LD coat proteins, including perilipins. Recent data suggest that heterotrimeric G-proteins and GTP hydrolysis are required for LD formation (42); the incubation of cells with aluminum fluoride together with fatty acids increases recruitment of perilipin 3 to the ER while preventing the budding of nascent LDs. Hence, the budding of both LDs and aqueous vesicles shares common mechanistic features. Furthermore, functionally segregating the ER from the rest of the secretory pathway through the incubation of cells with brefeldin A does not inhibit TAG deposition or LD formation (46). Thus, secretory compartments distal to the ER are not required for TAG storage, suggesting that neutral lipid can traffic directly from the ER to LDs.
Abundant evidence shows that LDs are closely associated with multiple membrane compartments and readily receive both protein and lipid components from these membranes. Common mechanisms mediate protein trafficking between the ER and LDs or vesicles of the secretory pathway. In genome-wide screens conducted in Drosophila (47, 48) and yeast (29) cells, knockdown of components of the Arf1-COPI vesicular transport machinery yields dispersed LDs of larger than normal size. This protein complex is required for trafficking of vesicles from the Golgi apparatus to the ER. Studies revealed that Arf1-COPI complex proteins are required for delivery of adipose triglyceride lipase (ATGL) to LDs from the ER (49) and efficient lipolysis (47, 49). Similarly, delivery of perilipin 2 to LDs requires Arf1-COPI complex proteins (49), suggesting that this trafficking mechanism is common to several proteins that define LDs.
Maturation of LDs and LD Fusion
Like other intracellular organelles, LDs are coated with proteins that link them to cytosolic machinery, including motor proteins and cytoskeletal elements to position and anchor the organelle, trafficking factors to transport lipid cargo, and enzymes to process lipid substrates. As LDs mature, they gain neutral lipid but also alter the composition of associated proteins. Although nascent LDs of adipocytes are coated with perilipins 3 and 4, as LDs gain TAG and enlarge, these perilipins are replaced with perilipin 2 and, ultimately, perilipin 1 (40, 41). This is significant because perilipin 1 plays a major role in regulation of lipolysis of adipose TAG stores by controlling the access of lipases and lipase cofactors to stored lipids (50). In adipocytes of fed animals, perilipin 1 sequesters CGI-58/ABHD5 (51, 52), a coactivator of ATGL, preventing the activation of TAG hydrolysis. When ATP is needed during fasting or exercise, catecholamines bind to β-adrenergic receptors on the cell surfaces of adipocytes, triggering a G-protein-mediated signaling pathway that activates lipolysis through increasing cAMP levels and activating cAMP-dependent protein kinase. Subsequent phosphorylation of perilipin 1 activates lipolysis through at least two mechanisms: 1) CGI-58/ABHD5 is released from the perilipin scaffold (51, 53–55), facilitating a protein-protein interaction between CGI-58/ABHD5 and ATGL (54, 56), which activates TAG hydrolase activity (57–59); and 2) phosphorylated perilipin 1 recruits and docks phosphorylated hormone-sensitive lipase on LDs via a protein-protein interaction (53, 60, 61). The latter interaction is required for the lipase to gain access to lipid substrates, primarily DAG. Thus, proteins acquired during the maturation of LDs in adipocytes include both perilipins and components of the lipolytic pathway.
When LDs expand to accommodate newly synthesized neutral lipids, the phospholipid content of LDs must also increase. Phosphatidylcholine is the major phospholipid in the limiting monolayer of LDs, followed by phosphatidylethanolamine, phosphatidylinositol, and ether-linked species with choline and ethanolamine headgroups (21, 62). It is thought that the phospholipid monolayer of nascent LDs is derived from leaflets of the ER during early deposition of neutral lipids; however, it is unclear how phospholipids are added to maturing LDs once they have budded off from the ER. A recent study shows that expanding LDs recruit CTP:phosphocholine cytidylyltransferase (CCT), the rate-limiting enzyme for phosphatidylcholine synthesis; binding of CCT to LDs activates the enzyme (63). Prior studies have shown that CCT binds to membranes depleted of phosphatidylcholine and enriched in anionic phospholipids (64). Thus, as the LD expands through acquisition of TAG, relative depletion of phosphatidylcholine creates an ideal binding environment for CCT. The resulting activation of CCT drives increased phosphatidylcholine synthesis, which provides lipids to replenish the phospholipid monolayer of LDs. However, the final step in phosphatidylcholine synthesis is catalyzed by CDP-choline:1,2-diacylglycerol cholinephosphotransferase, an integral membrane protein in the ER, so newly synthesized phosphatidylcholine must transfer between compartments. The mechanisms of transfer are unknown but may include diffusion of lipids through ER contact sites with LDs or transport through the cytosol on phospholipid transfer proteins. Regardless of the mechanism, this new study raises the intriguing possibility that the lipid composition of LDs exerts control over cellular lipid metabolism.
Another recent study has shown that two isoforms of lysophosphatidylcholine acyltransferase (LPCAT) localize to LDs (65). LPCAT forms phosphatidylcholine from lysophosphatidylcholine and acyl-CoA. This reaction is an important component of the Lands cycle, in which lysophosphatidylcholine is derived from phospholipase A2 activity on phosphatidylcholine to remove the sn-2-position fatty acid; LPCAT activity then regenerates phosphatidylcholine with an altered sn-2-position acyl chain. Because both long-chain fatty acyl-CoAs and lysophosphatidylcholine are somewhat water-soluble, delivery of these two substrates to LD-bound LPCAT likely contributes to phosphatidylcholine synthesis during LD expansion.
In adipocytes and hepatocytes, a third pathway contributes to synthesis of phosphatidylcholine. Phosphatidylethanolamine N-methyltransferase (PEMT) assembles methyl groups onto the ethanolamine headgroup to produce phosphatidylcholine. A recent study has shown that PEMT localizes to either LDs or ER or mitochondria-associated membranes closely associated with LDs in cultured adipocytes (24). Radiolabeled metabolites were used to demonstrate that PEMT contributes significantly to the phosphatidylcholine content of adipocyte LDs. Further studies are required to determine whether PEMT contributes to LD assembly in hepatocytes.
How does the size of a LD affect the metabolism of stored neutral lipids? Knockdown of the expression of genes encoding enzymes of the biosynthetic pathway for phosphatidylcholine, but not phosphatidylethanolamine, leads to abnormally enlarged LDs in cultured cells (48, 63). Moreover, when CCT was deleted in Drosophila, leading to decreased phosphatidylcholine synthesis, unusually large LDs were found in the fat body (63), the tissue that serves as adipose tissue in flies. These large LDs were more slowly metabolized, hence less susceptible to lipases, and conferred a survival advantage during starvation. In contrast, small LDs provide higher surface area relative to volume, which favors lipase binding and TAG hydrolysis; the size of LDs decreases as lipolysis progresses (66). However, size is only part of the story because proteins associated with LDs also control lipase access to substrates.
In animals, the LDs of adipocytes in white adipose tissue are unilocular and can be 100 μm in diameter or larger. In contrast, adipocytes in brown adipose tissue have smaller multilocular LDs and maintain high levels of stored TAG. Most other cells of the body have relatively tiny LDs and low TAG levels. What determines LD size? Because phosphatidylcholine is the most abundant phospholipid on LDs in most or all types of cells, phospholipid composition cannot be the only determinant of LD size. Time-lapse photography of cultured adipocytes has revealed that the large LDs of these cells are formed, at least in part, by fusion of smaller LDs (67). Because most non-adipose cells have only tiny dispersed LDs, fusion of LDs probably does not occur in all types of cells. The mechanism of LD fusion is poorly understood. In one study, mechanisms promoting fusion of membrane vesicles inspired the investigation of whether similar mechanisms contribute to the fusion of LDs (68). This study provided evidence that SNAP23 (soluble N-ethylmaleimide-sensitive factor attachment protein of 23 kDa) fractionates with purified LDs and suggested a common mechanism requiring SNAP23 for fusion of membrane vesicles or LDs. A recent study has challenged these findings, showing that knockdown of SNAP23 in cultured cells fails to alter the average size of LDs (63). A good candidate for a LD-associated fusion-mediating protein is FSP27/CIDEC. In wild-type mice, expression of FSP27 is limited to adipocytes in white and brown adipose tissue (69, 70), where the largest LDs are found. Furthermore, the Fsp27 gene is also turned on in steatotic livers of leptin-deficient (ob/ob) obese mice (70, 71), where LDs are correspondingly enlarged. Expression of FSP27 in cultured cells that normally lack the protein increases the size of LDs, whereas knockdown of FSP27 in adipocytes decreases LD size (71–73). Moreover, deletion of the Fsp27 gene in mice leads to smaller multilocular LDs in white adipose tissue and is accompanied by decreases in TAG storage and increased catabolism, altering energy homeostasis (69, 70). Thus, the correlation of Fsp27 expression with the formation of large LDs in cells makes FSP27 a good candidate for a LD fusion factor. The mechanism by which LDs fuse is unknown but, given the surfactant role of the phospholipid monolayer and LD-associated proteins, may require depletion of phosphatidylcholine and thinning of the protein coat at the site of fusion.
Concluding Remarks
LDs have become the subject of increasing numbers of studies relevant to lipid metabolism, cell biology, and the pathophysiology of chronic diseases. Over the past 5 years, major progress has been made in defining the ER as the origin of LDs. Genome-wide screens of yeast and Drosophila have identified novel pathways for trafficking of lipids and proteins to LDs and for assembly and maturation of LDs. Recent studies have revealed that some proteins and lipids traffic to LDs via routes that have been defined by the study of other membrane compartments. Other aspects of LD formation, maturation, and dissolution are unique. There are still numerous unanswered questions. As more LD-associated proteins are identified and examined, we will learn more about the origins and functions of this interesting and dynamic organelle.
Acknowledgment
We thank R. Hasney for the artwork in Fig. 1.
This work was supported, in whole or in part, by National Institutes of Health Grants DK054797 (to D. L. B.) and DK088206 (to N. E. W.). This is the first article in the Thematic Minireview Series on the Lipid Droplet, a Dynamic Organelle of Biomedical and Commercial Importance.
- LD
- lipid droplet
- ER
- endoplasmic reticulum
- TAG
- triacylglycerol
- ACS
- acyl-CoA synthetase
- GPAT
- glycerol-3-phosphate acyltransferase
- AGPAT
- 1-acylglycerol-3-phosphate acyltransferase
- PAP
- phosphatidic acid phosphohydrolase
- DGAT
- diacylglycerol acyltransferase
- DAG
- diacylglycerol
- ATGL
- adipose triglyceride lipase
- CCT
- CTP:phosphocholine cytidylyltransferase
- LPCAT
- lysophosphatidylcholine acyltransferase
- PEMT
- phosphatidylethanolamine N-methyltransferase.
REFERENCES
- 1. Brasaemle D. L. (2007) J. Lipid Res. 48, 2547–2559 [DOI] [PubMed] [Google Scholar]
- 2. Wolins N. E., Brasaemle D. L., Bickel P. E. (2006) FEBS Lett. 580, 5484–5491 [DOI] [PubMed] [Google Scholar]
- 3. Hodges B. D., Wu C. C. (2010) J. Lipid Res. 51, 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., Londos C. (1995) J. Lipid Res. 36, 1211–1226 [PubMed] [Google Scholar]
- 5. Cushman S. W. (1970) J. Cell Biol. 46, 326–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krisans S. K., Mortensen R. M., Lazarow P. B. (1980) J. Biol. Chem. 255, 9599–9607 [PubMed] [Google Scholar]
- 7. Carman G. M., Han G. S. (2009) J. Biol. Chem. 284, 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takeuchi K., Reue K. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E1195–E1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yen C. L., Stone S. J., Koliwad S., Harris C., Farese R. V., Jr. (2008) J. Lipid Res. 49, 2283–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang T. Y., Li B. L., Chang C. C., Urano Y. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E1–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rusiñol A. E., Cui Z., Chen M. H., Vance J. E. (1994) J. Biol. Chem. 269, 27494–27502 [PubMed] [Google Scholar]
- 12. Stone S. J., Levin M. C., Zhou P., Han J., Walther T. C., Farese R. V., Jr. (2009) J. Biol. Chem. 284, 5352–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuerschner L., Moessinger C., Thiele C. (2008) Traffic 9, 338–352 [DOI] [PubMed] [Google Scholar]
- 14. McFie P. J., Banman S. L., Kary S., Stone S. J. (2011) J. Biol. Chem. 286, 28235–28246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stone S. J., Levin M. C., Farese R. V., Jr. (2006) J. Biol. Chem. 281, 40273–40282 [DOI] [PubMed] [Google Scholar]
- 16. Li L. O., Klett E. L., Coleman R. A. (2010) Biochim. Biophys. Acta 1801, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wendel A. A., Lewin T. M., Coleman R. A. (2009) Biochim. Biophys. Acta 1791, 501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goodman J. M. (2008) J. Biol. Chem. 283, 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H. F. (2005) Cell 122, 85–95 [DOI] [PubMed] [Google Scholar]
- 20. van den Bosch H., de Vet E. C. (1997) Biochim. Biophys. Acta 1348, 35–44 [DOI] [PubMed] [Google Scholar]
- 21. Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. (2007) J. Lipid Res. 48, 837–847 [DOI] [PubMed] [Google Scholar]
- 22. Igal R. A., Wang S., Gonzalez-Baró M., Coleman R. A. (2001) J. Biol. Chem. 276, 42205–42212 [DOI] [PubMed] [Google Scholar]
- 23. Pellon-Maison M., Montanaro M. A., Coleman R. A., Gonzalez-Baró M. R. (2007) Biochim. Biophys. Acta 1771, 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hörl G., Wagner A., Cole L. K., Malli R., Reicher H., Kotzbeck P., Köfeler H., Höfler G., Frank S., Bogner-Strauss J. G., Sattler W., Vance D. E., Steyrer E. (2011) J. Biol. Chem. 286, 17338–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Razani B., Combs T. P., Wang X. B., Frank P. G., Park D. S., Russell R. G., Li M., Tang B., Jelicks L. A., Scherer P. E., Lisanti M. P. (2002) J. Biol. Chem. 277, 8635–8647 [DOI] [PubMed] [Google Scholar]
- 26. Cohen A. W., Razani B., Schubert W., Williams T. M., Wang X. B., Iyengar P., Brasaemle D. L., Scherer P. E., Lisanti M. P. (2004) Diabetes 53, 1261–1270 [DOI] [PubMed] [Google Scholar]
- 27. Wang H., Sreenevasan U., Hu H., Saladino A., Polster B. M., Lund L. M., Gong D. W., Stanley W. C., Sztalryd C. (2011) J. Lipid Res. 52, 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kadereit B., Kumar P., Wang W. J., Miranda D., Snapp E. L., Severina N., Torregroza I., Evans T., Silver D. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W. P., Agarwal A. K., Garg A., Anderson R. G., Goodman J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miranda D. A., Koves T. R., Gross D. A., Chadt A., Al-Hasani H., Cline G. W., Schwartz G. J., Muoio D. M., Silver D. L. (2011) J. Biol. Chem. 286, 42188–42199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fei W., Du X., Yang H. (2011) Trends Endocrinol. Metab. 22, 204–210 [DOI] [PubMed] [Google Scholar]
- 32. Magré J., Delépine M., Khallouf E., Gedde-Dahl T., Jr., Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A., Verloes A., d'Abronzo F. H., Seemanova E., Assan R., Baudic N., Bourut C., Czernichow P., Huet F., Grigorescu F., de Kerdanet M., Lacombe D., Labrune P., Lanza M., Loret H., Matsuda F., Navarro J., Nivelon-Chevalier A., Polak M., Robert J. J., Tric P., Tubiana-Rufi N., Vigouroux C., Weissenbach J., Savasta S., Maassen J. A., Trygstad O., Bogalho P., Freitas P., Medina J. L., Bonnicci F., Joffe B. I., Loyson G., Panz V. R., Raal F. J., O'Rahilly S., Stephenson T., Kahn C. R., Lathrop M., Capeau J. (2001) Nat. Genet. 28, 365–370 [DOI] [PubMed] [Google Scholar]
- 33. Cui X., Wang Y., Tang Y., Liu Y., Zhao L., Deng J., Xu G., Peng X., Ju S., Liu G., Yang H. (2011) Hum. Mol. Genet. 20, 3022–3030 [DOI] [PubMed] [Google Scholar]
- 34. Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A. J., Wenk M. R., Parton R. G., Yang H. (2008) J. Cell Biol. 180, 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tian Y., Bi J., Shui G., Liu Z., Xiang Y., Liu Y., Wenk M. R., Yang H., Huang X. (2011) PLoS Genet. 7, e1001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fei W., Li H., Shui G., Kapterian T. S., Bielby C., Du X., Brown A. J., Li P., Wenk M. R., Liu P., Yang H. (2011) J. Lipid Res. 52, 2136–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Binns D., Lee S., Hilton C. L., Jiang Q. X., Goodman J. M. (2010) Biochemistry 49, 10747–10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abell B. M., Holbrook L. A., Abenes M., Murphy D. J., Hills M. J., Moloney M. M. (1997) Plant Cell 9, 1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang A. H. (1996) Plant Physiol. 110, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wolins N. E., Skinner J. R., Schoenfish M. J., Tzekov A., Bensch K. G., Bickel P. E. (2003) J. Biol. Chem. 278, 37713–37721 [DOI] [PubMed] [Google Scholar]
- 41. Wolins N. E., Quaynor B. K., Skinner J. R., Schoenfish M. J., Tzekov A., Bickel P. E. (2005) J. Biol. Chem. 280, 19146–19155 [DOI] [PubMed] [Google Scholar]
- 42. Skinner J. R., Shew T. M., Schwartz D. M., Tzekov A., Lepus C. M., Abumrad N. A., Wolins N. E. (2009) J. Biol. Chem. 284, 30941–30948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., Goodman J. M. (2011) J. Cell Biol. 192, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., Schneiter R. (2011) J. Cell Sci. 124, 2424–2437 [DOI] [PubMed] [Google Scholar]
- 45. Kohlwein S. D. (2010) J. Biol. Chem. 285, 15663–15667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fei W., Wang H., Fu X., Bielby C., Yang H. (2009) Biochem. J. 424, 61–67 [DOI] [PubMed] [Google Scholar]
- 47. Beller M., Sztalryd C., Southall N., Bell M., Jäckle H., Auld D. S., Oliver B. (2008) PLoS Biol. 6, e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. (2008) Nature 453, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soni K. G., Mardones G. A., Sougrat R., Smirnova E., Jackson C. L., Bonifacino J. S. (2009) J. Cell Sci. 122, 1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brasaemle D. L., Subramanian V., Garcia A., Marcinkiewicz A., Rothenberg A. (2009) Mol. Cell. Biochem. 326, 15–21 [DOI] [PubMed] [Google Scholar]
- 51. Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., Brasaemle D. L. (2004) J. Biol. Chem. 279, 42062–42071 [DOI] [PubMed] [Google Scholar]
- 52. Yamaguchi T., Omatsu N., Matsushita S., Osumi T. (2004) J. Biol. Chem. 279, 30490–30497 [DOI] [PubMed] [Google Scholar]
- 53. Granneman J. G., Moore H. P., Granneman R. L., Greenberg A. S., Obin M. S., Zhu Z. (2007) J. Biol. Chem. 282, 5726–5735 [DOI] [PubMed] [Google Scholar]
- 54. Granneman J. G., Moore H. P., Krishnamoorthy R., Rathod M. (2009) J. Biol. Chem. 284, 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamaguchi T., Omatsu N., Morimoto E., Nakashima H., Ueno K., Tanaka T., Satouchi K., Hirose F., Osumi T. (2007) J. Lipid Res. 48, 1078–1089 [DOI] [PubMed] [Google Scholar]
- 56. Wang H., Bell M., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., Gong D., Brasaemle D., Sztalryd C. (2011) J. Biol. Chem. 286, 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gruber A., Cornaciu I., Lass A., Schweiger M., Poeschl M., Eder C., Kumari M., Schoiswohl G., Wolinski H., Kohlwein S. D., Zechner R., Zimmermann R., Oberer M. (2010) J. Biol. Chem. 285, 12289–12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. (2006) Cell Metab. 3, 309–319 [DOI] [PubMed] [Google Scholar]
- 59. Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., Zimmermann R. (2006) J. Biol. Chem. 281, 40236–40241 [DOI] [PubMed] [Google Scholar]
- 60. Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. (2003) J. Cell Biol. 161, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russell D., Gong D., Londos C., Yamaguchi T., Holm C., Rizzo M. A., Brasaemle D., Sztalryd C. (2009) J. Biol. Chem. 284, 32116–32125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. (2002) J. Biol. Chem. 277, 44507–44512 [DOI] [PubMed] [Google Scholar]
- 63. Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H. W., Schmidt-Supprian M., Vance D. E., Mann M., Farese R. V., Jr., Walther T. C. (2011) Cell Metab. 14, 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xie M., Smith J. L., Ding Z., Zhang D., Cornell R. B. (2004) J. Biol. Chem. 279, 28817–28825 [DOI] [PubMed] [Google Scholar]
- 65. Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. (2011) J. Biol. Chem. 286, 21330–21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nagayama M., Shimizu K., Taira T., Uchida T., Gohara K. (2010) FEBS Lett. 584, 86–92 [DOI] [PubMed] [Google Scholar]
- 67. Nagayama M., Uchida T., Gohara K. (2007) J. Lipid Res. 48, 9–18 [DOI] [PubMed] [Google Scholar]
- 68. Boström P., Andersson L., Rutberg M., Perman J., Lidberg U., Johansson B. R., Fernandez-Rodriguez J., Ericson J., Nilsson T., Borén J., Olofsson S. O. (2007) Nat. Cell Biol. 9, 1286–1293 [DOI] [PubMed] [Google Scholar]
- 69. Nishino N., Tamori Y., Tateya S., Kawaguchi T., Shibakusa T., Mizunoya W., Inoue K., Kitazawa R., Kitazawa S., Matsuki Y., Hiramatsu R., Masubuchi S., Omachi A., Kimura K., Saito M., Amo T., Ohta S., Yamaguchi T., Osumi T., Cheng J., Fujimoto T., Nakao H., Nakao K., Aiba A., Okamura H., Fushiki T., Kasuga M. (2008) J. Clin. Invest. 118, 2808–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Toh S. Y., Gong J., Du G., Li J. Z., Yang S., Ye J., Yao H., Zhang Y., Xue B., Li Q., Yang H., Wen Z., Li P. (2008) PLoS ONE 3, e2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Matsusue K., Kusakabe T., Noguchi T., Takiguchi S., Suzuki T., Yamano S., Gonzalez F. J. (2008) Cell Metab. 7, 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu K., Zhou S., Kim J. Y., Tillison K., Majors D., Rearick D., Lee J. H., Fernandez-Boyanapalli R. F., Barricklow K., Houston M. S., Smas C. M. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E1395–E1413 [DOI] [PubMed] [Google Scholar]
- 73. Puri V., Konda S., Ranjit S., Aouadi M., Chawla A., Chouinard M., Chakladar A., Czech M. P. (2007) J. Biol. Chem. 282, 34213–34218 [DOI] [PubMed] [Google Scholar]