Abstract
Lipid droplets (LDs) are highly dynamic cell organelles involved in energy homeostasis and membrane trafficking. Here, we review how select pathogens interact with LDs. Several RNA viruses use host LDs at different steps of their life cycle. Some intracellular bacteria and parasites usurp host LDs or encode their own lipid biosynthesis machinery, thus allowing production of LDs independently of their host. Although many mechanistic details of host/pathogen LD interactions are unknown, a picture emerges in which the unique cellular architecture and energy stored in LDs are important in the replication of diverse pathogens.
Keywords: Chlamydia, Hepatitis C Virus, Lipid Droplet, Mycobacteria, Plasmodium, Dengue Virus, Rotavirus
Lipid Droplets: More than Just Fat Storage
Lipid droplets (LDs)3 are cytoplasmic organelles composed of a hydrophobic core of neutral lipids (triglycerides and cholesterol esters) surrounded by a phospholipid monolayer and a growing list of associated proteins (reviewed in Ref. 1). All cells produce LDs, but the LDs vary significantly in size (i.e. <1–100 μm in diameter), triglyceride/cholesterol ester ratio, and protein decoration depending on the cell type.
Several proteins are involved in the generation, maturation, and degradation of LDs. Prominent among these are perilipin, adipocyte differentiation-related protein (ADRP), and TIP47 (tail-interacting protein 47), the founding members of the growing family of LD-associated PAT proteins (reviewed in Ref. 2).
The turnover of LDs is rapid (e.g. 24 h in cultured hepatoma cells), with droplets constantly being produced at the endoplasmic reticulum (ER), trafficked through the cytoplasm by kinesin and dynein motors (3), and degraded by the action of lipases such as adipocyte triglyceride lipase and hormone-sensitive lipase, as well as by macroautophagy (4, 5). Mitochondria are often found in close proximity to LDs, and membrane bridges between the organelles have been described that might facilitate the efflux of fatty acids toward β-oxidation (6). The shielding of LDs through the PAT proteins tightly controls their turnover and the access of lipases (7, 8).
The prevailing model suggests that LDs originate at the ER, with triglycerides and cholesterol esters accumulating between the bilayer of the ER membrane and the cytosolic layer eventually engulfing the lipid content (1). Support for this model comes from the localization of the enzymes required for neutral lipid synthesis such as the diacylglycerol acyltransferases DGAT1 and DGAT2, which are localized mainly in the ER membrane (reviewed in Ref. 9). DGAT2, but not DGAT1, can also traffic onto LDs when cells are exposed to high levels of fatty acids (10). The detailed mechanisms by which proteins are targeted onto the surface of LDs are only emerging; depending on the protein, the process involves diverse mechanisms such as vesicle-mediated trafficking pathways via COPI (coat protein complex I), lateral diffusion within the ER membrane, or yet to be identified shuttling mechanisms. A mature droplet may bud off the membrane or stay attached to ER membranes that often tightly surround LDs (11). LD biology also involves fusion/fission events, but mechanistic details of these processes remain largely unknown (12–14).
Here, we review recent reports that connect LDs with the life cycle of pathogens. We focus on select pathogens to highlight the broad spectrum of adaptation mechanisms that pathogens have evolved to take advantage of unique aspects of LD biology in support of their own replication and persistence in the host.
LDs and Viruses
Hepatitis C Virus
Hepatitis C virus (HCV) is the best defined human pathogen with close ties to host LDs. HCV, a single-copy positive-stranded RNA virus of the Flaviviridae family, is a blood-borne virus that replicates primarily in hepatocytes.
The viral genome is a single positive-stranded RNA. After receptor-mediated endocytosis, the host cell translation machinery translates the viral RNA genome stored in the particles into a single polyprotein precursor (Fig. 1). Host and viral proteases process the viral polyprotein, thereby releasing the three structural proteins (the nucleocapsid core and two envelope proteins, E1 and E2), the viroporin p7, and six nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). Multiprotein RNA replication complexes containing NS3–NS5B proteins replicate the viral RNA within ER-derived structures termed the “membranous web.” After encapsidation of newly synthesized viral RNA, the virus is thought to bud into the ER, thereby acquiring its envelope, and to exit the cell via the exocytotic pathway.
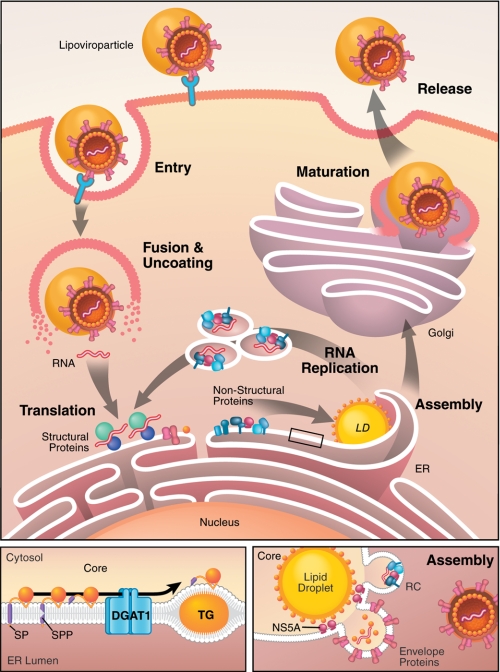
FIGURE 1.
LDs serve as virion assembly platforms during HCV replication. Upper panel, during the HCV life cycle, infectious lipoviroparticles enter hepatocytes through receptor-mediated endocytosis. Upon uncoating and release, the viral RNA is translated at the ER to produce one polyprotein that is cleaved by host and viral proteases, releasing the 10 viral proteins. The viral non-structural proteins (NS3–NS5B) form RNA replication complexes within ER-derived membranous structures termed the membranous web. After encapsidation of newly synthesized viral RNA, viroparticles bud into the ER lumen, mature through interactions with lipoproteins, and exit the cell via the secretory pathway. The lower left panel depicts trafficking of the viral core protein. After translation, the viral nucleocapsid core at the N-terminal end of the polyprotein is released by two subsequent cleavages (signal peptidase (SP) and signal peptide peptidase (SPP)), enabling core to traffic along the ER membrane. Through interaction with DGAT1, core is loaded onto LDs in a DGAT1-dependent manner. TG, triglyceride. The lower right panel shows a model of HCV assembly at LDs. Core is thought to recruit viral RNA replication complexes (RC) to ER membranes close to LDs, where the viral RNA is encapsidated by core.
The HCV life cycle is closely tied to the lipid metabolism in the infected cells (Fig. 1) (15). Strikingly, infectious virions are bound to lipoproteins circulating in the blood of infected people (16). These so-called “lipoviroparticles” enter host cells by receptor-mediated contact of viral and lipoprotein components. Several HCV receptors have been identified, including the tetraspanin protein CD81, scavenger receptor class B type I, the tight-junction proteins claudin-1 and occludin, and the LDL receptor. Interestingly, none of these is exclusively expressed on the surface of hepatocytes or other target cells (reviewed in Ref. 17).
With the discovery of the long-sought infectious cell culture system for HCV (HCVcc) in 2005 (18–20), cytosolic LDs rapidly emerged as putative viral assembly sites (21). In HCVcc-infected cells, the nucleocapsid protein core and components of the viral RNA replication complex (NS5A and NS3 and the viral RNA) localize to LDs. Electron micrographs revealed viral budding-like structures in ER membranes surrounding LDs (21). The current view is that LDs may serve as assembly “platforms” for HCV, with the viral core protein trafficking to the LD surface and recruiting viral RNA replication complexes to ER membranes in close proximity to the LD surface for encapsidation of newly synthesized viral RNA. Why HCV assembly requires LDs remains an open question, but mechanistic insight into the localization of core to LDs has been gained.
Core is the first viral protein translated from the viral RNA and is released from the polyprotein by two subsequent cleavage events, the first mediated by the signal peptidase and the second by the signal peptide peptidase. These events generate a 179-amino acid mature protein that is believed to migrate via lateral diffusion into ER subcompartments and onto the surface of LDs (22, 23). Mutations in the second cleavage site restrict the localization of core to ER membranes, as mutated core retains the C-terminal membrane-spanning domain that hinders association with the phospholipid monolayer of LDs (23). The hydrophobic LD-binding domain of core lies in its so-called D2 domain (amino acids 119–179) and is composed of an amphipathic helix-turn-helix motif termed a proline knot motif, which has strong similarities to the LD-binding domain of both the GB virus-B core protein and the plant protein oleosin (22, 24, 25). The tight association of the core protein with LDs is thought to disturb normal LD biology: it dissociates ADRP from LDs and causes microtubule-dependent clustering of core-coated droplets around the nucleus (26). Core is palmitoylated at Cys-172; mutation of this residue does not affect LD association of core but is known to severely impair virus production through a mechanism that is not yet understood (27).
A host factor involved in localizing core to LDs is the triglyceride-synthesizing enzyme DGAT1 (28). DGAT1 and DGAT2 enzymes catalyze the same enzymatic reaction, the acylation of diacylglycerol, the final and only committed step in triglyceride biosynthesis (reviewed in Ref. 9). However, only DGAT1 is critical in HCV infection. The core protein physically interacts with DGAT1 at the ER, and this interaction and active DGAT1 triglyceride synthesis are required for core to access LDs (28). These findings support a model in which core access to LDs is not random but geared toward DGAT1-generated LDs. As localization of core to LDs is a prerequisite for successful HCV particle assembly, pharmacologically suppressing DGAT1 activity inhibits HCV replication at the assembly step. However, the overall number of LDs is unaffected by DGAT1 inhibition because of the activity of DGAT2.
Core also causes an accumulation of droplets (steatosis), a phenotype frequently observed in infected patients (reviewed in Ref. 29). The severity of clinical steatosis is linked to polymorphisms in the LD-binding domain of core, which connects LD binding and steatogenic properties of core (30). Steatosis is more frequent in HCV genotype 3 strains, and the genotype 3a core protein induces the formation of large LDs in cultured cells by diminishing the expression of PTEN (phosphatase and tensin homolog) and IRS-1 (insulin receptor substrate 1) (31). Core expression also increases lipogenesis through activation/induction of SREBP-1c (sterol regulatory element-binding protein 1c), peroxisome proliferator-activated receptor γ, and retinoid X receptor α (32, 33) and is associated with inhibition of the microsomal triglyceride transfer protein (34), a key protein in VLDL assembly (reviewed in Ref. 35).
The role of LDs has widened from core to some nonstructural proteins of HCV. NS2, NS3, and NS5A proteins are also found at LDs in the context of an ongoing infection (36–40), but only the localization of NS5A, a phosphoprotein that is thought to recruit viral RNA for encapsidation, has been more closely studied so far. Several mutations were identified in NS5A that disrupt trafficking of the protein to LDs; these mutations also impair assembly and release of infectious virions (21, 41, 42). Phosphorylation of Ser-457 in the C terminus of NS5A may determine when the viral life cycle switches from RNA replication to packaging (43). This C-terminal region of NS5A also interacts with apolipoprotein E (44, 45), an interaction that might facilitate egress of viral particles via VLDL secretion. This pathway is important in the maturation of HCV particles to highly infectious lipoviroparticles that circulate in the bloodstream and represents another potential avenue for therapeutic intervention. Indeed, inhibition of VLDL secretion impairs HCVcc maturation and release (46–48). As the lipoprotein secretion pathway is unique to hepatocytes, the dependence of HCV infection on this pathway may partially explain why hepatocytes are the natural targets for productive HCV infection.
Dengue Virus
Dengue virus (DENV) is a single-copy positive-stranded RNA virus that, like HCV, belongs to the Flaviviridae family. The life cycle of DENV is similar to that of HCV, but the unique aspects of DENV include the involvement of a mosquito vector (Aedes aegypti), infection of different host cells (mainly immune cells), and a slightly different genetic repertoire, resulting in differences in the life cycle (reviewed in Ref. 49).
After the bite of an infected mosquito, DENV initially infects Langerhans cells in the skin, which, after recognizing the pathogen, move to the nearest lymph node. Progeny virions then infect other white blood cells such as monocytes and macrophages. The virus enters target cells through receptor-mediated endocytosis by binding to the C-type lectin DC-SIGN, the mannose receptor, and CLEC5A (50, 51). After release into the cytoplasm, the single-copy positive-strand viral RNA is replicated in ER-derived membranous structures. As in HCV, the viral RNA encodes a single polyprotein that is cleaved into three structural proteins (capsid C, prM (membrane) protein, and envelope protein E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (52). Newly synthesized RNA is encapsidated by the C protein, whereas prM/E heterodimers located in the ER form trimers that induce a curved surface that guides virion budding (53, 54). Immature viral particles are thought to be transported through the Golgi, where they mature before exiting the cell via the exocytotic pathway (55).
The DENV C protein, similar to the HCV core protein, localizes to LDs in human and insect cells and also causes a 2-fold increase in LD content in infected baby hamster kidney cells (56). In contrast, in human hepatoma cells, which harbor LDs in the uninfected state, DENV infection up-regulates autophagy, leading to LD (especially triglyceride) degradation (57). Interestingly, the viral protein NS3 interacts with host fatty acid synthase and recruits this enzyme to sites of viral RNA replication (58). Like core, the DENV C protein is released from the polyprotein by two subsequent peptidase cleavages, one by signal peptidase and one by the viral NS3/NS2B protease (59). The second cleavage by NS3/NS2B completely removes the C protein membrane-spanning anchor. Intriguingly and in contrast to the HCV core protein, the DENV C protein localizes to the LD surface even if the second cleavage site is mutated, suggesting a different route of LD translocation than core (56). Nevertheless, the association of the C protein with LDs is critical for viral replication, as mutations in hydrophobic residues located within the α2 helix of the C protein result in mislocalization of the C protein and impaired viral replication (56). In addition, treatment of infected cells with the fatty acid synthase inhibitor C75, which blocks generation of all LDs, attenuates viral replication at the step of assembly and release of infectious virions (56).
These data uncover unique ties of two different Flaviviridae family members with LDs and suggest that infectious particle production and release of other members of this virus family may also be directly linked to LDs. An interesting aspect is that the two viruses infect different host cells: HCV infects primarily hepatocytes, and DENV replicates in mosquito and human monocytes and macrophages. Although these cells underlie different metabolic regulations, both cell types have a unique propensity for excessive fat storage, which may be an aspect exploited by viral infection.
Rotavirus
Rotaviruses (RVs) are double-stranded non-enveloped RNA viruses that belong to the family Reoviridae (from respiratory enteric orphan viruses) (reviewed in Refs. 60 and 61). RVs infect enterocytes of the villi of the small intestine and are a major cause of acute gastroenteritis in children worldwide (62). They contain a 18,555-nucleotide RNA genome of 11 segments encoding six structural (VP1–VP4, VP6, and VP7) and six nonstructural (NSP1–NSP6) proteins. Infectious virions are so-called triple-layered particles (TLPs) containing four major capsid proteins (VP2, VP4, VP6, and VP7) and two minor proteins (VP1 and VP3). After endocytotic entry of the virus, the outer layer is removed within the endocytic vesicles. The resulting double-layered particles (DLPs) transcribe the viral RNA into mRNA, which is released into the cytosol of infected cells (Fig. 2).
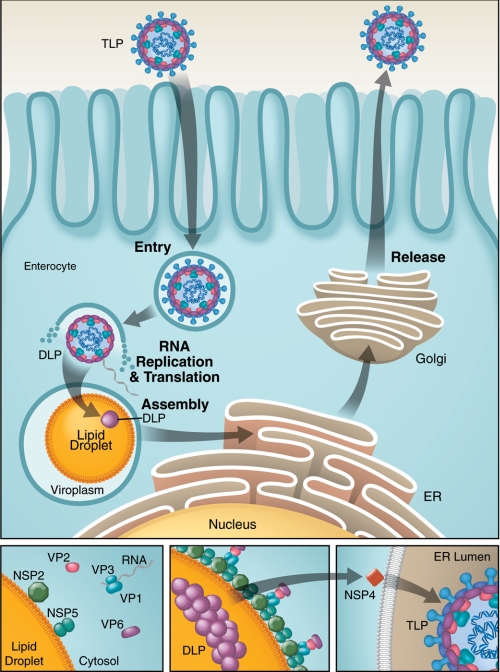
FIGURE 2.
Viroplasms (virion factories) associate with LDs in RV-infected cells. Upper panel, during the RV life cycle, infectious virions (TLPs) enter enterocytes through endocytosis, whereby the outer layer is removed within endocytic vesicles. DLPs actively transcribe the viral RNA into mRNA, which is released and serves as a template for translation and viral RNA replication. Viral RNA replication and the first steps of viral assembly occur in inclusion bodies called viroplasms, colocalizing with cellular LDs, which might serve as an assembly platform. Two viral proteins, NSP2 and NSP5, bind to LDs to form viroplasm-like structures. These viroplasm-like structures then associate with precore complexes (VP1, VP3, and viral RNA), VP2, and VP6 at the surface of LDs (highlighted in the lower panels). Through a yet to be defined mechanism, viroplasms form, and DLPs assemble and traffic to the ER, where they interact with NSP4 and are processed to become TLPs, which are released through the exocytotic pathway.
The viral mRNA serves as a template for viral protein translation and dsRNA replication. Early stages of viral assembly and viral RNA replication take place in virus-induced inclusion bodies called viroplasms, from which partially assembled DLPs are released to acquire the outer layer from the rough ER. Mature viruses are thought to be released through the exocytotic pathway.
Viroplasms, which contain active RNA replication complexes, colocalize with the LD-binding proteins perilipin and ADRP in infected Caco-2 cells. A similar colocalization was observed with the lipophilic dye Nile red, which is frequently used to visualize LDs in cells (63). Furthermore, viral double-stranded RNA, the NSP5 protein (a component of viroplasms of infected cells), and the LD-binding protein ADRP are all present in low density fractions isolated from infected cells, indicating a tight association of viroplasms with LDs (63). LDs seem to be recruited to viroplasms in a time-dependent manner after the initial infection. In uninfected cells expressing only the two nonstructural proteins NSP2 (an NTPase involved in RNA packaging) and NSP5, a process that induces viroplasm-like structures, these structures also associate with LDs (63). Indeed, NSP5 and perilipin colocalize in transfected MA104 kidney cells, as shown by FRET (63). The knockdown of NSP5 in infected cells blocks the recruitment of LDs to viroplasms (63), which points to a direct role of NSP5 in the relocalization of LDs and associated proteins. Additionally, the viral NSP4 protein, an enterotoxin, interacts with caveolin-1 (64), which localizes to LDs and functions in LD biogenesis and degradation (65).
The interactions of RV proteins with LDs are functionally relevant, as interference with LD biogenesis with triacsin C, an inhibitor of long-chain fatty acyl-CoA synthetase, inhibited viral RNA replication, viroplasm formation, and virus-induced cell death in cultured cells (63). Inducing LD fragmentation through treatment with isoproterenol and isobutylmethylxanthine, which both elevate cAMP levels and activate lipolysis, also inhibited RV replication (63). These data point to a unique role of LDs in RV replication and associated pathogenesis and might provide new opportunities for therapeutic intervention.
Other Viruses
Another member of the Reoviridae family, the orthoreoviruses, are connected with LDs. Orthoreoviruses infect vertebrates (including humans) without causing disease symptoms. In cultured cells, orthoreoviral infection is used to study virally induced apoptosis. The orthoreovirus outer capsid protein μ1 was recently shown to localize to membranes of the ER, mitochondria, and LDs via a C-terminal amphipathic helix that also mediates cell death (66). Intriguingly, besides localizing to the ER and LDs, the HCV core protein is also found at the outer mitochondrial membrane, although the relevance of this interaction in the context of infectious HCV particle production remains to be determined (22, 67). As the HCV core protein also induces apoptosis via the induction of ER stress and mitochondrial dysfunctions (68, 69), the possibility exists that the localization to LDs may represent a unique strategy to control viral protein-induced stress reactions and cell death during viral infection.
Other viruses with connections to LDs include the hepatitis B virus, which, through expression of the HBx protein, causes lipid accumulation by up-regulating the liver X receptor and its lipogenic target genes SREBP-1, FAS, and peroxisome proliferator-activated receptor γ (70, 71). Human adenovirus 36, the serotype frequently associated with obesity, decreases fatty acid oxidation and increases lipogenesis in cultured cells by inducing the expression of Cidec/FSP27 (fat-specific protein 27) (72). Human polyomavirus BK agnoprotein, a protein of unknown function, colocalizes with LDs through an amphipathic helix located in the center of the protein (73).
LDs and Bacteria
Chlamydia trachomatis
C. trachomatis is an obligate intracellular pathogen that is sexually transmitted and causes diseases of the urogenital tract through infection of epithelial mucosal cells. Different strains also infect the epithelium of the eye, causing trachoma and potential blindness.
Host LDs appear critical for C. trachomatis to capture triglycerides and cholesterol esters for bacterial growth during active propagation. The bacterial life cycle can be divided in two phases, an environmentally stable, metabolically inactive, inert phase (elementary body) and a vegetative, metabolically active phase (reticulate bodies (RBs)) (74). RBs replicate in inclusions, which are membrane-bound parasitophorous vacuoles that are mostly inaccessible to the host cell trafficking machinery and are devoid of most endosomal, lysosomal, or Golgi proteins (Fig. 3) (75). Nevertheless, trafficking of host lipids to the inclusions might be essential for the bacterial replication (76). Indeed, during replication, Chlamydia acquires host-derived glycerophospholipids, sphingolipids, and cholesterol through rerouting of exocytotic vesicles but also via non-vesicle transport mechanisms (77, 78). During active replication, LDs rich in cholesterol esters accumulate at the cytosolic side of the bacterial inclusions (79). Three bacterial proteins are translocated into the host cytosol and target vacuole-associated LDs (79). These proteins are now termed LD-associated proteins: Lda1 (CT156), Lda2 (CT163), and Lda3 (CT473).
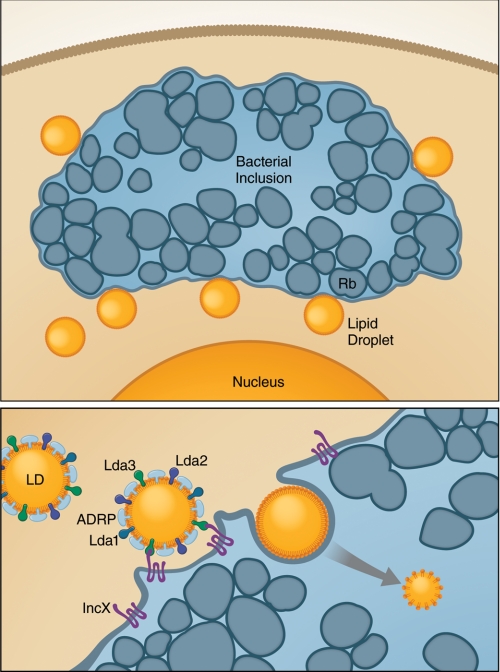
FIGURE 3.
During Chlamydia replication, host LDs are actively transported into bacterial inclusions. Upper panel, Chlamydia RBs replicate within membrane-bound parasitophorous vacuoles. Bacterial Lda proteins are translocated into the host cytosol and bind to vacuole-associated LDs. Lda3 at the surface of LDs interacts with the bacterial protein IncX to transport cytosolic LDs into the inclusion. The LDs in the bacterial inclusions lack host ADRP, which may be required to enable degradation and utilization of host lipids (lower panel).
The association of the bacterial Lda proteins with LDs is key for their cytotoxic effects. Indeed, yeast strains devoid of LDs are protected from Lda2 cytotoxicity (79). Interference with neutral lipid biosynthesis by treatment with triacsin C negatively affects the stability of the bacterial Lda proteins as well as their cytotoxic effects (79). In triacsin C-treated cells, bacterial inclusions are small, and bacterial growth is inhibited (79). The bacterial Lda3 proteins associate with cytosolic LDs and, together with the bacterial protein IncX, cause translocation of cytosolic LDs into bacterial inclusions (Fig. 3) (80). During this process, host ADRP is displaced from the surface of translocating LDs, a process that might be required to render lipids stored in the droplets accessible for degradation (80).
Mycobacterium tuberculosis and Mycobacterium leprae
M. tuberculosis is another obligate intracellular bacterium that infects macrophages in the respiratory tract. M. tuberculosis is transmitted via aerosol droplets. Once in the respiratory tract, it infects primarily alveolar macrophages. Host defense mechanisms trap the bacteria in granulomas, which are infected macrophages surrounded by “foamy” lipid-laden macrophages, mononuclear phagocytes, and lymphocytes inside a fibrous layer of endothelial cells (81, 82). In this dormant state, the bacteria can be latent for decades until activated by a weakened immune system.
M. tuberculosis utilizes triglycerides as its main energy source. These triglycerides are synthesized by the bacterial Tgs1 (triacylglycerol synthase 1) protein (83, 84). Host-derived fatty acids are imported and used for the synthesis of triglycerides that are deposited in bacterial LDs (85). M. tuberculosis also encodes its own lipase (LipY), a homolog of hormone-sensitive lipases that might release stored fatty acids in times of energy shortage but may also degrade host triglycerides for utilization by the pathogen (86). In addition, M. tuberculosis infection causes a lipid accumulation in infected and uninfected bystander macrophages just before their necrotic cell death, a process that leads to the release of lipids into their surroundings (87).
Mycobacterium bovis infection, which causes bovine tuberculosis, also induces foamy macrophages. In a murine model of tuberculosis, M. bovis infection triggers a dose- and time-dependent increase in LDs that is dependent on TLR2 (Toll-like receptor 2) signaling, but not TLR4 signaling (88). Toll-like receptors play a fundamental role in pathogen recognition and the activation of innate immunity. The function of LD induction caused by M. bovis infection is not clear at this point, but it may be part of the host innate immune response or may have evolved as a defense mechanism of the bacteria against the immune attack.
M. leprae is the causative agent of leprosy, a granulomatous disease of the mucosa of the respiratory tract and the peripheral nerves. This uncultivable pathogen has an exceptionally long generation time (up to 13 days), which makes deciphering its life cycle difficult. M. leprae is also transmitted via aerial droplets. After phagocytotic entry into target cells and evasion of host defense responses, the bacteria replicate inside intracellular vesicles.
M. leprae has long been known to cause a foamy phenotype in dermal lesions (89, 90). Infected macrophages express high levels of ADRP, which localizes to the bacterium-containing phagosome (91, 92). Schwann cells also exhibit a relocalization of ADRP-positive LDs to bacterium-containing phagosomes in cultured cells and in nerve biopsies of infected patients (92). This relocalization of LDs depends on PI3K signaling and requires cytoskeletal rearrangements (92). Blocking LD movement by interfering with cytoskeleton function decreases bacterial survival in infected cells. Again, infection-induced formation of LDs requires innate immune response signaling, but in contrast to M. bovis infection, it is TLR6 (not TLR2)-dependent in M. leprae infection (93). Blocking LD formation enabled infected cells to kill the invading bacteria, supporting a model in which induced LD biogenesis is part of the bacterial immune evasion strategy required for persistent infection (93).
LDs and Parasites
Plasmodium falciparum is a parasitic protist that causes the most virulent form of malaria. P. falciparum is transmitted to humans by the bite of an infected anopheline mosquito. The released sporozoites infect hepatocytes and undergo asexual replication that culminates in the release of tens of thousands of merozoites. These merozoites enter erythrocytes, where they multiply again. The clinical features of malaria infection, high fever and chills, are caused by the synchronous rupture of infected erythrocytes. Terminally differentiated erythrocytes lack internal organelles, the machinery for de novo protein and lipid synthesis, and trafficking machineries. The main component of mature erythrocytes is hemoglobin, which is degraded and used as an amino acid source by P. falciparum.
Infected erythrocytes display a dramatic increase in phospholipid and triglyceride content, but not in cholesterol esters (94, 95). Interestingly, to produce triglyceride-rich LDs, the parasite encodes its own DGAT enzyme (a homolog to membrane-bound O-acyltransferases), which is expressed specifically in the intra-erythrocyte stage and is essential for the proliferation of the parasite (95–97).
However, P. falciparum lacks the capability to degrade triglycerides to produce energy. Newly synthesized LDs accumulate in the food vacuole and are involved in the detoxification of heme (Fig. 4) (98). These results are in line with recent findings that suggest that LDs can store and thereby “neutralize” hydrophobic proteins that have the tendency to form toxic aggregates (99, 100). They suggest that, in addition to proteins, also specific metabolites such as heme could be detoxified using LDs as an intracellular “dumping” ground.
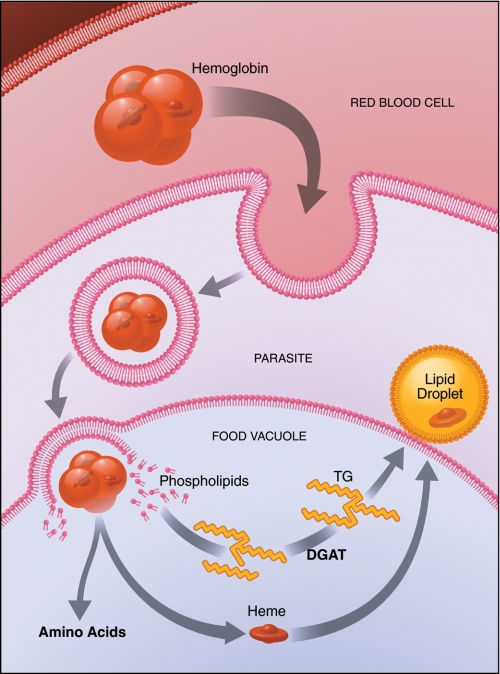
FIGURE 4.
P. falciparum in infected erythrocytes produces LDs for heme detoxification. P. falciparum infects mature erythrocytes and replicates on a diet of hemoglobin, which is degraded and used as an amino acid source. The parasite accumulates LDs inside infected erythrocytes that may be used to detoxify the released heme. TG, triglyceride.
Concluding Remarks
New details of LD biology are emerging rapidly. The findings that viruses, bacteria, and parasites hijack these organelles to support their own life cycle are intriguing and point to new previously unrecognized aspects of LD biology.
We suspect that the findings highlighted here only scratch the surface of this rich new area of investigation into LD biology and potential use in therapies. Viperin (for virus inhibitory protein, ER-associated, interferon-inducible), an intriguing interferon-induced antiviral cellular factor with characteristic LD localization, has activity against a broad range of viruses, including HCV and DENV, but also human CMV, influenza virus, and West Nile virus, suggesting that LDs may play a role in the life cycle of these and many other pathogens.
Acknowledgments
We thank John Carroll and Giovanni Maki for help in preparing the figures and Gary Howard for editorial support.
This work was supported, in whole or in part, by National Institutes of Health Grants R56 AI085056 and P30 DK026743 (to the UCSF Liver Center). This work was also supported by the Gladstone Institutes and the Hellman Family Foundation. This is the second article in the Thematic Minireview Series on the Lipid Droplet, a Dynamic Organelle of Biomedical and Commercial Importance.
- LD
- lipid droplet
- ADRP
- adipocyte differentiation-related protein
- ER
- endoplasmic reticulum
- HCV
- hepatitis C virus
- HCVcc
- HCV cell culture system
- DENV
- Dengue virus
- RV
- rotavirus
- TLP
- triple-layered particle
- DLP
- double-layered particle
- RB
- reticulate body.
REFERENCES
- 1. Farese R. V., Jr., Walther T. C. (2009) Cell 139, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bickel P. E., Tansey J. T., Welte M. A. (2009) Biochim. Biophys. Acta 1791, 419–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Welte M. A., Cermelli S., Griner J., Viera A., Guo Y., Kim D. H., Gindhart J. G., Gross S. P. (2005) Curr. Biol. 15, 1266–1275 [DOI] [PubMed] [Google Scholar]
- 4. Zechner R., Kienesberger P. C., Haemmerle G., Zimmermann R., Lass A. (2009) J. Lipid Res. 50, 3–21 [DOI] [PubMed] [Google Scholar]
- 5. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009) Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jägerström S., Polesie S., Wickström Y., Johansson B. R., Schröder H. D., Højlund K., Boström P. (2009) Cell Biol. Int. 33, 934–940 [DOI] [PubMed] [Google Scholar]
- 7. Brasaemle D. L. (2007) J. Lipid Res. 48, 2547–2559 [DOI] [PubMed] [Google Scholar]
- 8. Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. (2007) J. Lipid Res. 48, 2751–2761 [DOI] [PubMed] [Google Scholar]
- 9. Yen C. L., Stone S. J., Koliwad S., Harris C., Farese R. V., Jr. (2008) J. Lipid Res. 49, 2283–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuerschner L., Moessinger C., Thiele C. (2008) Traffic 9, 338–352 [DOI] [PubMed] [Google Scholar]
- 11. Robenek H., Hofnagel O., Buers I., Robenek M. J., Troyer D., Severs N. J. (2006) J. Cell Sci. 119, 4215–4224 [DOI] [PubMed] [Google Scholar]
- 12. Boström P., Andersson L., Rutberg M., Perman J., Lidberg U., Johansson B. R., Fernandez-Rodriguez J., Ericson J., Nilsson T., Borén J., Olofsson S. O. (2007) Nat. Cell Biol. 9, 1286–1293 [DOI] [PubMed] [Google Scholar]
- 13. Murphy S., Martin S., Parton R. G. (2010) PLoS ONE 5, e15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marcinkiewicz A., Gauthier D., Garcia A., Brasaemle D. L. (2006) J. Biol. Chem. 281, 11901–11909 [DOI] [PubMed] [Google Scholar]
- 15. Herker E., Ott M. (2011) Trends Endocrinol. Metab. 22, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomssen R., Bonk S., Propfe C., Heermann K. H., Köchel H. G., Uy A. (1992) Med. Microbiol. Immunol. 181, 293–300 [DOI] [PubMed] [Google Scholar]
- 17. Sabahi A. (2009) Virol. J. 6, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., Rice C. M. (2005) Science 309, 623–626 [DOI] [PubMed] [Google Scholar]
- 20. Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 22. Hope R. G., McLauchlan J. (2000) J. Gen. Virol. 81, 1913–1925 [DOI] [PubMed] [Google Scholar]
- 23. McLauchlan J., Lemberg M. K., Hope G., Martoglio B. (2002) EMBO J. 21, 3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hope R. G., Murphy D. J., McLauchlan J. (2002) J. Biol. Chem. 277, 4261–4270 [DOI] [PubMed] [Google Scholar]
- 25. Boulant S., Montserret R., Hope R. G., Ratinier M., Targett-Adams P., Lavergne J. P., Penin F., McLauchlan J. (2006) J. Biol. Chem. 281, 22236–22247 [DOI] [PubMed] [Google Scholar]
- 26. Boulant S., Douglas M. W., Moody L., Budkowska A., Targett-Adams P., McLauchlan J. (2008) Traffic 9, 1268–1282 [DOI] [PubMed] [Google Scholar]
- 27. Majeau N., Fromentin R., Savard C., Duval M., Tremblay M. J., Leclerc D. (2009) J. Biol. Chem. 284, 33915–33925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herker E., Harris C., Hernandez C., Carpentier A., Kaehlcke K., Rosenberg A. R., Farese R. V., Jr., Ott M. (2010) Nat. Med. 16, 1295–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Syed G. H., Amako Y., Siddiqui A. (2010) Trends Endocrinol. Metab. 21, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jhaveri R., McHutchison J., Patel K., Qiang G., Diehl A. M. (2008) J. Infect. Dis. 197, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clement S., Peyrou M., Sanchez-Pareja A., Bourgoin L., Ramadori P., Suter D., Vinciguerra M., Guilloux K., Pascarella S., Rubbia-Brandt L., Negro F., Foti M. (2011) Hepatology 54, 38–49 [DOI] [PubMed] [Google Scholar]
- 32. Kim K. H., Hong S. P., Kim K., Park M. J., Kim K. J., Cheong J. (2007) Biochem. Biophys. Res. Commun. 355, 883–888 [DOI] [PubMed] [Google Scholar]
- 33. Tsutsumi T., Suzuki T., Shimoike T., Suzuki R., Moriya K., Shintani Y., Fujie H., Matsuura Y., Koike K., Miyamura T. (2002) Hepatology 35, 937–946 [DOI] [PubMed] [Google Scholar]
- 34. Perlemuter G., Sabile A., Letteron P., Vona G., Topilco A., Chrétien Y., Koike K., Pessayre D., Chapman J., Barba G., Bréchot C. (2002) FASEB J. 16, 185–194 [DOI] [PubMed] [Google Scholar]
- 35. Roingeard P., Hourioux C. (2008) J. Viral Hepat. 15, 157–164 [DOI] [PubMed] [Google Scholar]
- 36. Jirasko V., Montserret R., Lee J. Y., Gouttenoire J., Moradpour D., Penin F., Bartenschlager R. (2010) PLoS Pathog. 6, e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stapleford K. A., Lindenbach B. D. (2011) J. Virol. 85, 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma Y., Anantpadma M., Timpe J. M., Shanmugam S., Singh S. M., Lemon S. M., Yi M. (2011) J. Virol. 85, 86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma Y., Yates J., Liang Y., Lemon S. M., Yi M. (2008) J. Virol. 82, 7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mousseau G., Kota S., Takahashi V., Frick D. N., Strosberg A. D. (2011) J. Gen. Virol. 92, 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Appel N., Zayas M., Miller S., Krijnse-Locker J., Schaller T., Friebe P., Kallis S., Engel U., Bartenschlager R. (2008) PLoS Pathog. 4, e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Masaki T., Suzuki R., Murakami K., Aizaki H., Ishii K., Murayama A., Date T., Matsuura Y., Miyamura T., Wakita T., Suzuki T. (2008) J. Virol. 82, 7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tellinghuisen T. L., Foss K. L., Treadaway J. (2008) PLoS Pathog. 4, e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cun W., Jiang J., Luo G. (2010) J. Virol. 84, 11532–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Benga W. J., Krieger S. E., Dimitrova M., Zeisel M. B., Parnot M., Lupberger J., Hildt E., Luo G., McLauchlan J., Baumert T. F., Schuster C. (2010) Hepatology 51, 43–53 [DOI] [PubMed] [Google Scholar]
- 46. Huang H., Sun F., Owen D. M., Li W., Chen Y., Gale M., Jr., Ye J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gastaminza P., Cheng G., Wieland S., Zhong J., Liao W., Chisari F. V. (2008) J. Virol. 82, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chang K. S., Jiang J., Cai Z., Luo G. (2007) J. Virol. 81, 13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodenhuis-Zybert I. A., Wilschut J., Smit J. M. (2010) Cell. Mol. Life Sci. 67, 2773–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller J. L., deWet B. J., Martinez-Pomares L., Radcliffe C. M., Dwek R. A., Rudd P. M., Gordon S. (2008) PLoS Pathog. 4, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van der Schaar H. M., Rust M. J., Chen C., van der Ende-Metselaar H., Wilschut J., Zhuang X., Smit J. M. (2008) PLoS Pathog. 4, e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Clyde K., Kyle J. L., Harris E. (2006) J. Virol. 80, 11418–11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kuhn R. J., Zhang W., Rossmann M. G., Pletnev S. V., Corver J., Lenches E., Jones C. T., Mukhopadhyay S., Chipman P. R., Strauss E. G., Baker T. S., Strauss J. H. (2002) Cell 108, 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y., Zhang W., Ogata S., Clements D., Strauss J. H., Baker T. S., Kuhn R. J., Rossmann M. G. (2004) Structure 12, 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zybert I. A., van der Ende-Metselaar H., Wilschut J., Smit J. M. (2008) J. Gen. Virol. 89, 3047–3051 [DOI] [PubMed] [Google Scholar]
- 56. Samsa M. M., Mondotte J. A., Iglesias N. G., Assunção-Miranda I., Barbosa-Lima G., Da Poian A. T., Bozza P. T., Gamarnik A. V. (2009) PLoS Pathog. 5, e1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heaton N. S., Randall G. (2010) Cell Host Microbe 8, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Heaton N. S., Perera R., Berger K. L., Khadka S., Lacount D. J., Kuhn R. J., Randall G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Niyomrattanakit P., Winoyanuwattikun P., Chanprapaph S., Angsuthanasombat C., Panyim S., Katzenmeier G. (2004) J. Virol. 78, 13708–13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patton J. T., Silvestri L. S., Tortorici M. A., Vasquez-Del Carpio R., Taraporewala Z. F. (2006) Curr. Top. Microbiol. Immunol. 309, 169–187 [DOI] [PubMed] [Google Scholar]
- 61. Pesavento J. B., Crawford S. E., Estes M. K., Prasad B. V. (2006) Curr. Top. Microbiol. Immunol. 309, 189–219 [DOI] [PubMed] [Google Scholar]
- 62. Parashar U. D., Gibson C. J., Bresse J. S., Glass R. I. (2006) Emerg. Infect. Dis. 12, 304–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheung W., Gill M., Esposito A., Kaminski C. F., Courousse N., Chwetzoff S., Trugnan G., Keshavan N., Lever A., Desselberger U. (2010) J. Virol. 84, 6782–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parr R. D., Storey S. M., Mitchell D. M., McIntosh A. L., Zhou M., Mir K. D., Ball J. M. (2006) J. Virol. 80, 2842–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cohen A. W., Razani B., Schubert W., Williams T. M., Wang X. B., Iyengar P., Brasaemle D. L., Scherer P. E., Lisanti M. P. (2004) Diabetes 53, 1261–1270 [DOI] [PubMed] [Google Scholar]
- 66. Coffey C. M., Sheh A., Kim I. S., Chandran K., Nibert M. L., Parker J. S. (2006) J. Virol. 80, 8422–8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schwer B., Ren S., Pietschmann T., Kartenbeck J., Kaehlcke K., Bartenschlager R., Yen T. S., Ott M. (2004) J. Virol. 78, 7958–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Korenaga M., Wang T., Li Y., Showalter L. A., Chan T., Sun J., Weinman S. A. (2005) J. Biol. Chem. 280, 37481–37488 [DOI] [PubMed] [Google Scholar]
- 69. Benali-Furet N. L., Chami M., Houel L., De Giorgi F., Vernejoul F., Lagorce D., Buscail L., Bartenschlager R., Ichas F., Rizzuto R., Paterlini-Bréchot P. (2005) Oncogene 24, 4921–4933 [DOI] [PubMed] [Google Scholar]
- 70. Kim K. H., Shin H. J., Kim K., Choi H. M., Rhee S. H., Moon H. B., Kim H. H., Yang U. S., Yu D. Y., Cheong J. (2007) Gastroenterology 132, 1955–1967 [DOI] [PubMed] [Google Scholar]
- 71. Na T. Y., Shin Y. K., Roh K. J., Kang S. A., Hong I., Oh S. J., Seong J. K., Park C. K., Choi Y. L., Lee M. O. (2009) Hepatology 49, 1122–1131 [DOI] [PubMed] [Google Scholar]
- 72. Wang Z. Q., Yu Y., Zhang X. H., Floyd E. Z., Cefalu W. T. (2010) Int. J. Obes. 34, 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Unterstab G., Gosert R., Leuenberger D., Lorentz P., Rinaldo C. H., Hirsch H. H. (2010) Virology 399, 322–331 [DOI] [PubMed] [Google Scholar]
- 74. Belland R., Ojcius D. M., Byrne G. I. (2004) Nat. Rev. Microbiol. 2, 530–531 [DOI] [PubMed] [Google Scholar]
- 75. Fields K. A., Hackstadt T. (2002) Annu. Rev. Cell Dev. Biol. 18, 221–245 [DOI] [PubMed] [Google Scholar]
- 76. Van Ooij C., Kalman L., Van Ijzendoorn S., Nishijima M., Hanada K., Mostov K., Engel J. N. (2000) Cell. Microbiol. 2, 627–637 [DOI] [PubMed] [Google Scholar]
- 77. Hackstadt T., Rockey D. D., Heinzen R. A., Scidmore M. A. (1996) EMBO J. 15, 964–977 [PMC free article] [PubMed] [Google Scholar]
- 78. Carabeo R. A., Mead D. J., Hackstadt T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kumar Y., Cocchiaro J., Valdivia R. H. (2006) Curr. Biol. 16, 1646–1651 [DOI] [PubMed] [Google Scholar]
- 80. Cocchiaro J. L., Kumar Y., Fischer E. R., Hackstadt T., Valdivia R. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zahrt T. C. (2003) Microbes Infect. 5, 159–167 [DOI] [PubMed] [Google Scholar]
- 82. Zhang Y. (2004) Front. Biosci. 9, 1136–1156 [DOI] [PubMed] [Google Scholar]
- 83. Daniel J., Deb C., Dubey V. S., Sirakova T. D., Abomoelak B., Morbidoni H. R., Kolattukudy P. E. (2004) J. Bacteriol. 186, 5017–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sirakova T. D., Dubey V. S., Deb C., Daniel J., Korotkova T. A., Abomoelak B., Kolattukudy P. E. (2006) Microbiology 152, 2717–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Daniel J., Maamar H., Deb C., Sirakova T. D., Kolattukudy P. E. (2011) PLoS Pathog. 7, e1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Deb C., Daniel J., Sirakova T. D., Abomoelak B., Dubey V. S., Kolattukudy P. E. (2006) J. Biol. Chem. 281, 3866–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim M. J., Wainwright H. C., Locketz M., Bekker L. G., Walther G. B., Dittrich C., Visser A., Wang W., Hsu F. F., Wiehart U., Tsenova L., Kaplan G., Russell D. G. (2010) EMBO Mol. Med. 2, 258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. D'Avila H., Melo R. C., Parreira G. G., Werneck-Barroso E., Castro-Faria-Neto H. C., Bozza P. T. (2006) J. Immunol. 176, 3087–3097 [DOI] [PubMed] [Google Scholar]
- 89. Virchow R. L. K. (1863) Die krankhaften Geschwülste: dreissig Vorlesungen, gehalten während des Wintersemesters 1862–1863 an der Universität zu Berlin, Hirschwald, Berlin [Google Scholar]
- 90. Scollard D. M., Joyce M. P., Gillis T. P. (2006) Clin. Infect. Dis. 43, e19–e22 [DOI] [PubMed] [Google Scholar]
- 91. Tanigawa K., Suzuki K., Nakamura K., Akama T., Kawashima A., Wu H., Hayashi M., Takahashi S., Ikuyama S., Ito T., Ishii N. (2008) FEMS Microbiol. Lett. 289, 72–79 [DOI] [PubMed] [Google Scholar]
- 92. Mattos K. A., Lara F. A., Oliveira V. G., Rodrigues L. S., D'Avila H., Melo R. C., Manso P. P., Sarno E. N., Bozza P. T., Pessolani M. C. (2011) Cell. Microbiol. 13, 259–273 [DOI] [PubMed] [Google Scholar]
- 93. Mattos K. A., Oliveira V. G., D'Avila H., Rodrigues L. S., Pinheiro R. O., Sarno E. N., Pessolani M. C., Bozza P. T. (2011) J. Immunol. 187, 2548–2558 [DOI] [PubMed] [Google Scholar]
- 94. Holz G. G., Jr. (1977) Bull. World Health Organ. 55, 237–248 [PMC free article] [PubMed] [Google Scholar]
- 95. Palacpac N. M., Hiramine Y., Mi-ichi F., Torii M., Kita K., Hiramatsu R., Horii T., Mitamura T. (2004) J. Cell Sci. 117, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 96. Vielemeyer O., McIntosh M. T., Joiner K. A., Coppens I. (2004) Mol. Biochem. Parasitol. 135, 197–209 [DOI] [PubMed] [Google Scholar]
- 97. Palacpac N. M., Hiramine Y., Seto S., Hiramatsu R., Horii T., Mitamura T. (2004) Biochem. Biophys. Res. Commun. 321, 1062–1068 [DOI] [PubMed] [Google Scholar]
- 98. Jackson K. E., Klonis N., Ferguson D. J., Adisa A., Dogovski C., Tilley L. (2004) Mol. Microbiol. 54, 109–122 [DOI] [PubMed] [Google Scholar]
- 99. Cermelli S., Guo Y., Gross S. P., Welte M. A. (2006) Curr. Biol. 16, 1783–1795 [DOI] [PubMed] [Google Scholar]
- 100. Welte M. A. (2007) Trends Cell Biol. 17, 363–369 [DOI] [PubMed] [Google Scholar]