Background: Endogenous APP expression in CNS has not been rigorously examined.
Results: We characterized the expression, localization, and stability of endogenous APP and APPsβ using a highly specific antibody.
Conclusion: APP is a neuron-specific protein under basal and neuroinflammatory conditions, and soluble APP is highly stable.
Significance: Our studies clarify important properties of APP, which have direct implications in APP biology and AD pathogenesis.
Keywords: Alzheimer Disease, Amyloid Precursor Protein, Astrocytes, Immunohistochemistry, Neurons
Abstract
APP processing and amyloid-β production play a central role in Alzheimer disease pathogenesis. APP has been considered a ubiquitously expressed protein. In addition to amyloid-β, α- or β-secretase-dependent cleavage of APP also generates soluble secreted APP (APPsα or APPsβ, respectively). Interestingly, APPsβ has been shown to be subject to further cleavage to create an N-APP fragment that binds to the DR6 death receptor and mediates axon pruning and degeneration under trophic factor withdrawal conditions. By performing APP immunocytochemical staining, we found that, unexpectedly, many antibodies yielded nonspecific staining in APP-null samples. Screening of a series of antibodies allowed us to identify a rabbit monoclonal antibody Y188 that is highly specific for APP and prompted us to re-examine the expression, localization, and stability of endogenous APP and APPsβ in wild-type and in APPsβ knock-in mice, respectively. In contrast to earlier studies, we found that APP is specifically expressed in neurons and that its expression cannot be detected in major types of glial cells under basal or neuroinflammatory conditions. Both APPsα and APPsβ are highly stable in the central nervous system (CNS) and do not undergo further cleavage with or without trophic factor support. Our results clarify several key questions with regard to the fundamental properties of APP and offer critical cellular insights into the pathophysiology of APP.
Introduction
Alzheimer disease (AD)2 is the most common cause of dementia in the elderly. Although the exact nature of AD etiology is still controversial, it is clear that amyloidogenic processing of the amyloid precursor protein (APP) generates Aβ peptides, which play a vital role in the pathogenesis of AD.
APP is a type-I transmembrane protein with high abundance in the CNS (1). APP undergoes alternative splicing to generate APP mRNAs encoding proteins of 695, 751, and 770 amino acids (referred to as APP695, APP751, and APP770). It is generally understood that APP695 is predominantly expressed in neurons, whereas APP751 and APP770 isoforms are expressed in most tissues examined, and their expressions are increased in astrocytes and microglia following brain injury (2–4).
APP is subject to cleavage by a number of secretases. The α-secretase or β-secretase cleaves the APP extracellular domain, generating soluble derivatives termed APPsα or APPsβ, respectively. APPsα has been reported to exhibit neurotrophic and synaptogenic activities (reviewed in Ref. 1). Our recent work indicated a potential signaling activity of APPsβ (5). Intriguingly, Nikolaev et al. (6) reported that APPsβ is further cleaved to create a 38-kDa N-terminal fragment (N-APP) found in the conditioned medium of trophic factor deprived axons in dorsal root ganglion cultures. This APPsβ fragment was shown to be a ligand of the death receptor 6 (DR6) and triggers axon pruning and neurodegeneration (6). These findings raise the question of whether N-APP is also produced in the CNS and to what degree it contributes to APPsβ function.
In this study, we first carefully examined a number of APP antibodies using APP-null samples as controls. We found that although most antibodies show excellent specificity for APP on Western blots, only one of them, APP-Y188, a rabbit monoclonal antibody recognizing the YENPTY motif of APP, is able to clearly distinguish the APP knock-out neurons from wild-type neurons on fluorescence immunocytochemistry. We subsequently characterized APP expression in mouse neurons and glial cells in vitro and in vivo, under resting or traumatic brain injury (TBI) conditions, or in the presence of Aβ plaques in AD mouse models. The results demonstrate that APP is only expressed in neurons and is undetectable in most glial cells examined. Furthermore, taking advantage of our APPsβ knock-in mouse model (5) and using a variety of culture conditions, we report that in contrast to Nikolaev et al. (6), APPsβ is highly stable and remains as an intact protein under normal or trophic factor deprivation (TFD) conditions.
EXPERIMENTAL PROCEDURES
Animals Used in This Study
Mice were housed 2–5 per cage with ad libitum access to food and water in a room with a 12-h light/dark cycle in a specific pathogen-free mouse facility. All procedures were performed in accordance with National Institutes of Health guidelines and with the approval of the Baylor College of Medicine Institutional Animal Care and Use Committee (IACUC). All animals used in this study are either C57BL6/J wild-type mice or mice on C57BL6/J background. APP+/− mice were intercrossed to generate APP+/+, APP+/−, and APP−/− offspring (7). APPsβ knock-in (ki) mice have been described previously (5), and APPsβ ki/+ mice were intercrossed to generate wild-type (+/+), APPsβ ki/+, and ki/ki mice. APPsl/Aβ/sl/Aβ mice (8) and PS1M146V/M146V mice (9) were bred together to generate double heterozygous APPsl/Aβ/+: PS1M146V/+ animals, which were intercrossed to produce homozygous APPsl/Aβ/sl/Aβ and PS1M146V/M146V double knock-in mice (APP/hAβ/PS1).
Antibodies
The following APP-related antibodies were tested and used in this study: APP Y188 (rabbit monoclonal, 1:500, Epitomics), APP A8967 (mouse monoclonal, 1:100–1:1000, Sigma), APP ab15272 (rabbit polyclonal, 1:100–1:1000, Abcam), APPc (rabbit polyclonal, 1:100–1:1000, generated in the laboratory, (10)), 22C11 (mouse monoclonal, 1:100–1:1000, Millipore), 4G8 (mouse monoclonal, 1:100–1:1000, Covance), APP SIG-39184 (mouse monoclonal, 1:100–1:1000, Covance), APP SIG-39180 (mouse monoclonal, 1:100–1:1000, Covance), and 5A3/1G7 (mouse monoclonal, 1:100–1:1000, courtesy of Dr. Edward Koo (11)). The following antibodies were also used: anti-V5 tag antibody (mouse monoclonal ab27671, 1:4000, Abcam), anti-DYKDDDDK tag antibody (L5, rat, 1:500, BioLegend), anti-GFAP (mouse monoclonal, MAB360, 1:1000, Millipore), anti-NeuN (mouse monoclonal, MAB377, 1:500, Millipore), anti-CD11b (rat monoclonal, ab8878, 1:100, Abcam), anti-myelin basic protein (MBP) (rat monoclonal, 1:100, Millipore), anti-KDEL (mouse monoclonal, SPA-827, 1:500, StressGen), anti-GM130 (mouse monoclonal, 1:500, 610822, BD Biosciences), anti-EEA1 (mouse monoclonal, 1:500, H00008411-M02, Abnova), anti-M6PR (mouse monoclonal, ab2733, 1:500, Abcam), anti-LAMP1 (rat, 553792, 1:1000, BD Biosciences), and anti-Grp78 (mouse monoclonal, 610979, BD Biosciences).
Mouse Primary Hippocampal/Cortical Culture
Cerebral hemispheres from postnatal day 0 mice were isolated, and meninges were carefully removed under the stereoscope in dissection medium (Hanks' balanced salt solution 1× with 0.1 m HEPES, 0.6% glucose, 100 units/ml penicillin, and 100 μg/ml streptomycin, Invitrogen). Brain tissue was cut into small pieces with a sharp scissor, and tissue pieces were transferred to a 15-ml tube with 10 ml of dissection medium. After adding 500 μl of 2.5% trypsin (Invitrogen), the brain tissue was then incubated in a 37 °C water bath for 15 min with frequent swirling. 400 μl of soybean trypsin inhibitor (1 mg/ml, Invitrogen) was then added to stop the trypsin activity. The dissection medium was replaced with 2 ml of culture medium (NeurobasalTM medium with 2% B-27 supplements and 0.5 mm l-glutamine, Invitrogen). The tissue was dissociated by passing it through a P1000 tip 10–20 times in the presence of DNase (10 μg/ml, Sigma). Dissociated cells were then transferred to a new 15-ml tube and centrifuged at 1,000 rpm for 5 min, washed with 5 ml of culture medium twice, resuspended with 5 ml of culture medium, and plated onto poly-d-lysine (Sigma) precoated 12-mm glass coverslips/plastic 6-well plate at a density of 10,000 cells/cm2. One day after seeding, the culture medium was changed.
Immunocytochemistry
Cells on coverslips were fixed with 4% paraformaldehyde (PFA) in PBS at 4 °C overnight. Cells were washed with PBS three times and incubated in blocking solution (PBS with 3% BSA, 2% goat serum, and 0.1% Triton X-100) for at least 1 h at room temperature. Cells were then incubated with primary antibody solution (primary antibody diluted in blocking solution) for least 2 h at room temperature or overnight at 4 °C. After washing with PBS three times, cells were incubated with secondary antibody solution for 1 h. Before being mounted onto glass slides, cells were washed with PBS another three times.
Immunohistochemistry
Anesthetized animals were perfused with 4% PFA in PBS, and brains were dissected out for overnight post-fixation in 4% PFA. Brains were dehydrated, embedded in paraffin, and sectioned at 8 μm. For antibody staining, brain sections were then deparaffinized and rehydrated before being boiled in sodium citrate buffer (10 mm, pH 6.0) for 10 min for antigen retrieval. For plaque staining, sections were incubated in 80% formic acid for 5 min at room temperature. Brain sections were rinsed with PBS three times, incubated with blocking solution (PBS with 3% BSA, 2% goat serum, and 0.1% Triton X-100) for at least 1 h, incubated with primary antibody solution (primary antibody diluted in blocking solution) at 4 °C overnight, rinsed with PBS three times, incubated with secondary antibody solution for 1 h, and then rinsed with PBS three times. For fluorescence staining, brain sections were then mounted with DAPI-containing mounting medium (Vector Laboratories). In the case of diaminobenzidine chemical staining, the Elite ABC staining kit (Vector Laboratories) was used according to the user manual.
TBI
TBI was induced as described previously (12). Briefly, mice were anesthetized with isoflurane and intubated to control ventilation. After the mice were placed and secured in a stereotaxic frame, a 3-mm craniotomy was performed over the right parietal cortex. Injury was induced with controlled cortical impact using a voltage-driven impactor (3 m/sec, 1.5-mm deformation, 100 ms, Benchmark stereotaxic impactor, myNeuroLab, St. Louis, MO). The wounds were then sutured closed, and the mice were monitored until full recovery.
Immunoprecipitation
Immunoprecipitation of APPsβ was performed from brains of APPsβ ki/ki mice using rat anti-DYKDDDDK (FLAG) tag antibody conjugated to magnetic beads (catalog number 143.11D, Invitrogen) according to the manufacturer's protocol. Mice were sacrificed under anesthesia induced by isoflurane. Brains were retrieved and homogenized in PBS containing protease inhibitor mixture (catalog number 04693116001, Roche Applied Science), centrifuged at 20,000 × g for 15 min at 4 °C. Pellets were resuspended in PBS containing 1% Triton X-100 and protease inhibitor mixture, rotated for 1 h at 4 °C, and then centrifuged at 20,000 × g for 15 min at 4 °C. Supernatants were used for immunoprecipitation by adding magnetic beads conjugated with anti-DYKDDDDK antibody. This mixture was incubated with rotation at 4 °C for 1 h, washed in ice-cold PBS containing 0.1% Triton X-100 five times, and eluted using 3×FLAG peptide (F4799, Sigma-Aldrich) at 4 °C with rotation for 30 min. Eluted protein was precipitated by adding 4× volume of acetone and incubating overnight at −20 °C. Protein precipitates were centrifuged, air-dried, and rehydrated in water and then mixed with 2× sample loading buffer for SDS-PAGE followed by Western blot analysis.
Immunoprecipitation of full-length APP was performed in similar manner as above from brains of wild-type mice using APPc rabbit polyclonal antibody and protein-G magnetic beads (catalog number 100.07D, Invitrogen). Proteins binding to the beads were eluted by boiling the beads in sample loading buffer and then subjected to SDS-PAGE and Western blot analysis. Normal rabbit IgG was used to control nonspecific bindings to protein-G beads. Brain lysate from APP−/− mice were used to control any nonspecific bindings of APPc antibody.
Purification of APPsβ-V5/His Recombinant Protein
Mouse APPsβ was cloned into the pcDNA3.1-TOPO-V5/His (Invitrogen) mammalian expression vector with the V5/His tag fused to the C terminus of APPsβ. This pcDNA3.1-APPsβ-V5/His vector was transfected into CHO-S cells (Invitrogen) using Lipofectamine 2000 (Invitrogen). Stable clones were selected in serum-containing selection medium (1 mg/ml G418, 10% FBS, and 1× nonessential amino acids in DMEM medium, Invitrogen). After selecting a stable clone with the highest expression/secretion level of APPsβ-V5/His, this clone was further expanded in serum-containing selection medium. When the culture density reached 80% confluency, the medium was replaced with serum-free chemically defined medium (CD-CHO medium containing 8 mm l-glutamine and 10 ml/liter HT supplement, Invitrogen) for another 5 days. The conditioned serum-free medium was collected, centrifuged to remove large cell debris, passed through a 0.45-μm filter, and loaded onto a HisTrap® column (GE HealthCare). The APPsβ-V5/His was then purified by eluting the column with a gradient of imidazole in PBS. The fractions containing APPsβ-V5/His recombinant protein were dialyzed against PBS, filtered with a 0.20-μm filter, aliquoted, and stored in a −80 °C freezer.
RESULTS
Identification of a Specific Antibody for APP Immunostaining
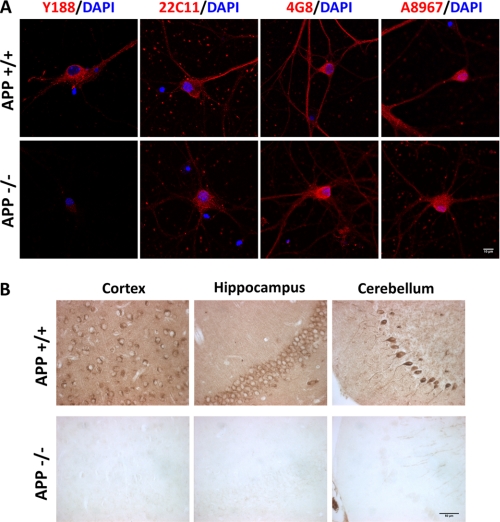
A variety of APP antibodies are available either commercially or from academic research laboratories. Although these antibodies have been extensively used for biochemical and immunohistochemical assays, we found that when performing immunocytochemical staining and comparing wild-type with APP-null neuronal cultures, most of the antibody stainings were indistinguishable between the two cultures, suggesting nonspecific staining for endogenous APP. We therefore tested a selection of APP antibodies from different origins and against different epitopes on fixed mouse primary hippocampal cultures (Table 1 and Fig. 1A) and identified a rabbit monoclonal antibody (Y188), raised against the C-terminal sequences of APP, that displayed strong staining in wild-type neurons but almost no staining in APP KO neurons. Y188 antibody staining is also highly specific in paraffin-fixed brain sections (Fig. 1B). In addition, Western blot analysis showed that only the full-length APP, but not α- or β-secretase cleaved C-terminal fragments, is produced in cultured neurons at physiological levels (data not shown). As such, the Y188 antibody recognizes full-length APP translated from all mRNA isoforms (herein referred to as APP or FL-APP). Although different experimental conditions may give rise to different results, our data call for the importance of using APP-deficient samples as negative controls for APP expression studies.
TABLE 1.
Summary of APP antibodies and their specificity
General characteristics of the APP antibodies and their specificity tested by immunocytochemical staining of 14 DIV wild type and APP null primary hippocampal cultures fixed with 4% PFA are shown. The origins of the antibodies can be found under “Experimental Procedures.” ICC, immunocytochemistry; mAb, monoclonal antibody; pAb, polyclonal antibody; aa, amino acids.
| Antibody | Species | Epitope | ICC specificity |
|---|---|---|---|
| Y188 | Rabbit mAb | YENPTY motif | High |
| A8967 | Rabbit pAb | 46–60 aa | No |
| Ab15272 | Rabbit pAb | 44–62 aa | No |
| APPc | Rabbit pAb | C terminus | No |
| 22C11 | Mouse mAb | 66–81 aa | No |
| 4G8 | Mouse mAb | Aβ 17–24 aa | No |
| APP 39184 | Mouse mAb | 61–77 aa | No |
| APP 39180 | Mouse mAb | 573–596 aa | No |
| 5A3/1G7 | Mouse mAb | N terminus | Low |
FIGURE 1.
Specificity of the Y188 antibody in vitro and in vivo. A, representative immunocytochemical staining images of 14 DIV wild-type (APP+/+) and APP KO (APP−/−) primary hippocampal cultures stained with Y188, 22C11, 4G8, and A8967 antibodies. Blue, DAPI; Red, APP. Scale bar, 10 μm. B, immunohistochemical staining of wild-type (APP+/+) and APP KO (APP−/−) cortex, hippocampus, and cerebellum sections using the Y188 antibody. Scale bar, 50 μm.
APP Is Specifically Expressed in Neurons but Not in Major Glial Cells
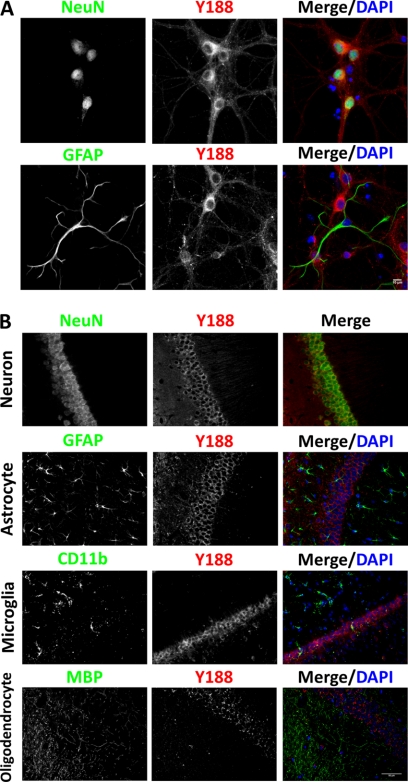
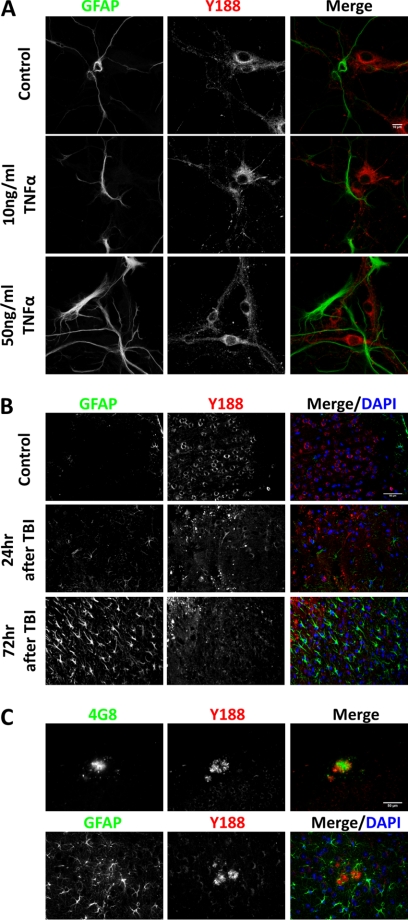
We subsequently examined endogenous APP expression and localization using the Y188 antibody. Unexpectedly, we found that in primary hippocampal neurons cultured 14 days in vitro (DIV), we could only detect APP in NeuN-positive neurons, but not in GFAP-positive astrocytes (Fig. 2A). Another two major CNS glial cell types, microglia and oligodendrocytes, were not present in our in vitro culture system (data not shown). Therefore, we performed APP immunohistochemical staining on adult mouse hippocampal sections. Consistent with the neuronal culture result, the NeuN-positive CA1 neurons abundantly express APP, but nearby GFAP-positive astrocytes, CD11b-positive microglia, and MBP-positive oligodendrocytes were devoid of APP expression (Fig. 2B). Because APP has been shown to be induced in activated astrocytes under neuroinflammation or brain injury conditions (2–4), we further investigated possible astrocytic APP expression using several conditions known to lead to astrocyte activation. We first treated primary hippocampal cultures with 10 or 50 ng/ml human TNFα, concentrations known to trigger neuroinflammation in vitro (13) (Fig. 3A). When compared with vehicle controls, GFAP immunoreactivity increased dramatically in a dose-dependent manner in TNFα-treated groups, and astrocytes displayed hypertrophic morphology, suggesting activation. However, similar to vehicle-treated controls, APP expression was not detected in the reactive astrocytes (Fig. 3A). Next we tested APP expression in vivo in response to TBI using the controlled cortical impact protocol (Fig. 3B). When compared with uninjured controls where APP staining is observed within neuronal cell bodies (Fig. 3B, Control), 24 h after TBI, we observed the presence of punctate APP staining at the injury site, presumably in the swelling axons as reported previously (14, 15). Although there are limited GFAP-positive cells at this stage, profound reactive astrocytosis can be seen 72 h after TBI (Fig. 3B). Consistent with the in vitro culture experiment, APP expression is absent in GFAP-positive reactive astrocytes in brain tissue after TBI. It is interesting to note that the induction of GFAP immunoreactivity at this stage is correlated with diminished APP punctates, suggesting that these reactive astrocytes play an active role in clearing APP deposits. Lastly, we examined APP expression in a homozygous knock-in mouse model of AD expressing APP with a humanized Aβ sequence and both APP and PS1 familial AD mutations (16). In this model, due to physiological expression levels of human mutations, amyloid pathology can be detected after 14 months of age. Consistent with the previous observations in human AD brains (17–19), Aβ (4G8) and APP (Y188) co-immunostaining showed profound APP immunoreactivity surrounding the core of amyloid plaques (Fig. 3C), likely due to its accumulation in dystrophic neurites. However, although abundant GFAP-positive reactive astrocytes can be observed in the vicinity of amyloid plaques, these astrocytes are devoid of APP expression (Fig. 3C). Taken together, we conclude that endogenous APP expression is extremely low or absent in astrocytes under physiological or pathological conditions induced by TBI or AD pathology.
FIGURE 2.
Endogenous APP is a neuron-specific protein in vitro and in vivo. A, co-staining of NeuN (upper) or GFAP (lower) with Y188 antibodies of 14 DIV primary hippocampal cultures showed that APP is only expressed in NeuN-positive, but not GFAP-positive, cells. Blue, DAPI; Green, NeuN or GFAP; Red, Y188. Scale bar, 10 μm. B, co-staining of adult mouse hippocampus sections using Y188 and neuron marker NeuN, astrocyte marker GFAP, microglia marker CD11b, or oligodendrocyte marker MBP revealed that although all NeuN-positive neurons express APP, APP expression in major types of glial cells is below detection. Blue, DAPI; Green, NeuN, GFAP, CD11b, and MBP; Red, Y188. Scale bar, 50 μm.
FIGURE 3.
APP expression is not induced in activated astrocytes in vitro and in vivo. A, 12 DIV primary hippocampal cultures were treated with PBS (Control), 10 ng/ml TNFα, and 50 ng/ml TNFα followed by immunocytochemical staining for APP. Blue, DAPI; Green, GFAP; Red, Y188. Scale bar, 10 μm. B, controlled cortical impact-induced TBI was performed on adult mouse brain. APP immunostaining was carried out in uninjured controls (Control), 24 or 72 h after TBI. Blue, DAPI; Green, GFAP; Red, Y188. Scale bar, 50 μm. C, immunostaining of 20 month-old APP/hAβ/PS1 brains. Upper panel, APP detected by Y188 (red) was found around 4G8-positive Aβ plaques (green). Lower panel, GFAP-positive reactive astrocytes (green) do not express APP (red). Scale bar, 50 μm.
Subcellular Localization of APP in Neurons
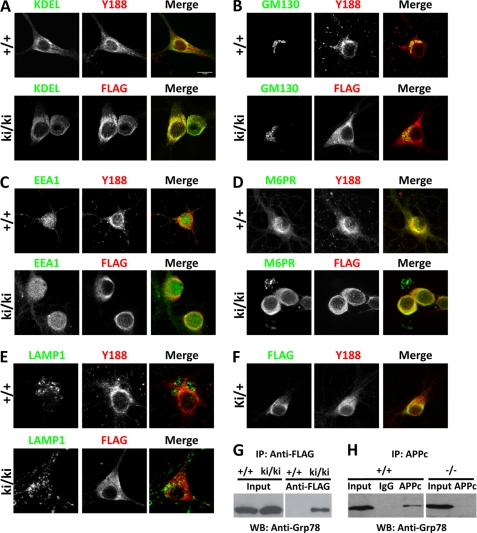
APP has been reported to be localized to the plasma membrane (20), the ER and endosomes (21), and the Golgi apparatus (22, 23) and accumulated in the lysosomes (24). Because these studies were performed using overexpression systems or employing antibodies that may result in nonspecific staining, we thought to re-evaluate the subcellular localization of endogenous APP in primary wild-type neuronal cultures using the Y188 antibody and co-staining with a panel of organelle markers, including ER (KDEL), Golgi complex (GM130), early endosomes (EEA1), late endosomes (M6PR), and lysosomes (LAMP1) (Fig. 4, A–E, +/+). We found that APP is largely localized to the Golgi complex and late endosomes. Its localization is limited in the ER and early endosomes and almost completely absent in lysosomes.
FIGURE 4.
Subcellular localization of FL-APP and APPsβ. A–E, 14 DIV hippocampal neurons cultured from wild-type (+/+) or homozygous APPsβ knock-in (ki/ki) mice were co-stained with APP (red) and organelle markers (green). A, ER (anti-KDEL); B, Golgi (GM130); C, early endosome (EEA1); D, late endosome (M6PR); E, lysosome (LAMP1). APP and APPsβ were recognized by Y188 or anti-FLAG antibody, respectively. F, co-staining of FL-APP and APPsβ in APPsβ ki/+ neurons. Green, FLAG; Red, Y188. G, interaction between Grp78 and the APP soluble domain. Wild-type (+/+) or homozygous APPsβ knock-in (ki/ki) brain lysates were immunoprecipitated (IP) using the anti-FLAG antibody conjugated to magnetic beads followed by elution with 3×FLAG peptide and Western blotting (WB) using an antibody against Grp78. Input, total lysates. H, interaction between Grp78 and FL-APP. Wild-type (+/+) or APP-null (−/−) brain lysates were immunoprecipitated using the APPc antibody or rabbit IgG plus protein-G beads followed by Western blotting using an antibody against Grp78.
We previously created a strain of FLAG-tagged APPsβ ki mice by inserting the FLAG tag and stop codon immediately after the BACE1 cleavage site (5). The resulting knock-in allele expresses truncated APPsβ fused with the FLAG tag and lacks the transmembrane and the intracellular domains. We reported that the APPsβ is expressed and secreted similar to wild-type APP (5). Because APP is known to undergo intracellular processing, to understand the cellular dynamics of APP and APPsβ, we performed similar subcellular localization studies in homozygous APPsβ knock-in neurons using an anti-FLAG antibody and compared these with that of FL-APP (Fig. 4, A–E, ki/ki). We found that APPsβ (FLAG) and FL-APP (Y188) were localized in distinct subcellular compartments: the highest levels of APPsβ were found in the ER instead of the Golgi. This distinct subcellular localization is further evidenced by co-staining of APPsβ heterozygous (ki/+) neurons with Y188 and FLAG, which allows direct comparison of endogenous APPsβ and FL-APP in the same neuron (Fig. 4F). Nevertheless, similar to FL-APP, APPsβ is significantly localized to the late endosomes (Fig. 4D) and is absent in the lysosomes (Fig. 4E). Overall, these results indicate that the transmembrane and the intracellular sequences of APP are required for trafficking from the ER to Golgi, whereas the APP extracellular domain is sufficient for its localization to endosomal compartments.
In an effort to search for potential binding partners for soluble APP, we took advantage of the FLAG tag fused at the end of the APPsβ sequence in APPsβ knock-in mice and performed immunoprecipitation of APPsβ brain lysates using magnetic beads conjugated with anti-FLAG antibodies followed by elution with 3×FLAG peptide. Preliminary proteomic analysis of the elution mix identified Grp78 (glucose-regulated protein 78 kDa), an ER chaperone protein (data not shown). Because we observed a large pool of APPsβ in the ER, we performed Western blot analysis to further confirm the specific binding between Grp78 and APPs by Western blotting the elutes with an anti-Grp78 antibody (Fig. 4G). Indeed, Grp78 can be detected in the APPsβ ki/ki samples, but not the wild-type controls lacking the FLAG sequence. In addition, we also confirmed the endogenous interaction between FL-APP and Grp78, which was previously reported in an in vitro HEK293 co-transfection experiment (25) and also identified in recent proteomic studies (26, 27). An APP C-terminal antibody, APPc, can pull down Grp78 from wild-type brain lysate but not from IgG control or from APP-null samples (Fig. 4H). Taken together, our results support a physiological interaction of APP and Grp78 through the soluble APP ectodomain, suggesting that this interaction may facilitate the folding and trafficking of APP.
APPsβ Is Highly Stable under Normal and TFD Conditions
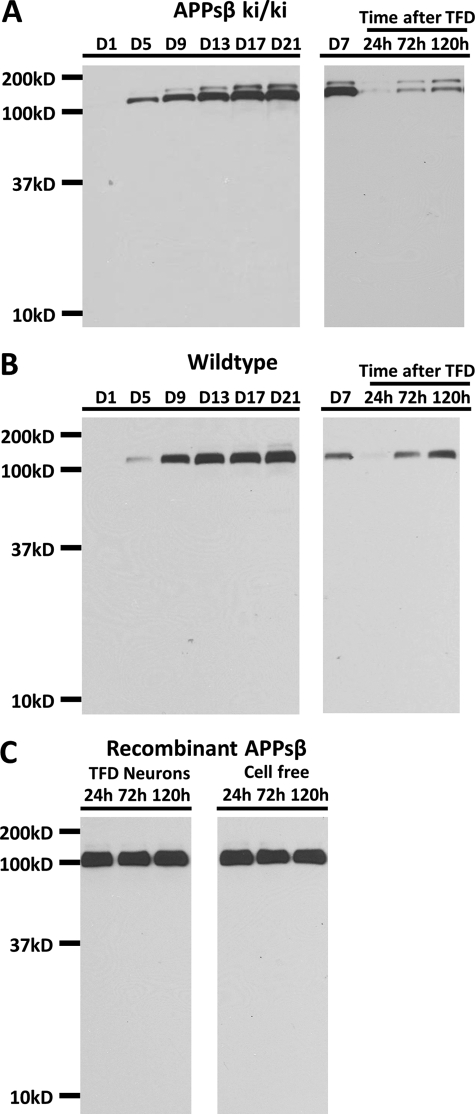
Nikolaev et al. (6) showed that in dorsal root ganglion culture, APPsβ is further cleaved upon TFD to produce a truncated product named N-APP, which serves as a DR6 ligand to mediate axon pruning and degeneration (6). Generation of the APPsβ knock-in mice and the demonstration of the normal secretion of the APPsβ protein afford us with a unique opportunity to examine this cleavage event in the absence of APPsα. We thus investigated the stability and metabolism of APPsβ in the APPsβ ki/ki primary neuronal cultures while using wild-type cultures for total APPs (APPsα and APPsβ). We collected 5% of the conditioned medium from 1, 5, 9, 13, 17, and 21 DIV cultures and replaced it with the same amount of fresh medium to maintain the total culture volume. Western blot analysis of the conditioned medium using the 22C11 antibody, which detects APPsβ and total APPs in APPsβ ki/ki (Fig. 5A, left), and wild-type mice (Fig. 5B, left), respectively, showed that the sole band detected was ∼100 kDa, representing intact APPs. No lower molecular weight product resembling N-APP was detectable.
FIGURE 5.
APPsβ is a highly stable protein. A and B, Western blot analysis using the 22C11 antibody on conditioned medium collected from APPsβ ki/ki (A) or wild-type (B) cultures under normal neuronal culture conditions (day 1, 5, 9, 13, 17, and 21), at day 7 immediately before TFD, or 24, 72, or 120 h after TFD, showing the intact nature of APPs recovered from residual surviving neurons. C, anti-V5 Western blot of fresh medium containing APPsβ-V5/His recombinant protein incubated with TFD-treated neurons or cell-free culture for 24 h, 72 h or 120 h.
We next washed the APPsβ and wild-type neurons with fresh medium multiple times at 7 DIV, which depleted all the trophic factors secreted from the neurons, and then we replaced the medium with fresh medium for 24, 72, or 120 h (Fig. 5, A and B, right). Only a small amount of neurons survived from the severe TFD, which accumulated overtime along with increased APPs. However, no APP cleavage products could be detected at any time following the TFD. To rule out the possibility that there was not enough APPs as substrate, we added purified V5/His-tagged APPsβ recombinant protein into the fresh medium in wild-type cultures immediately after TFD to test the existence of any enzymatic activity induced by TFD. Western blot analysis using the anti-V5 antibody showed that there was no difference between APPsβ exposed to TFD-treated neurons or fresh cell-free medium and that recombinant APPsβ protein is stable under both conditions (Fig. 5C). Taken together, we conclude that APPs is highly stable under normal or TFD conditions in the CNS.
DISCUSSION
In this study, we first tested a panel of APP antibodies to distinguish wild-type versus APP KO neurons by immunocytochemistry and identified a highly specific and potent APP antibody, Y188, which yields specific staining in paraffin-fixed brain sections. Taking advantage of this antibody, we show that only neurons, but not astrocytes, microglia, nor oligodendrocytes, express APP under basal or stressed conditions. Endogenous APP is mainly localized to the Golgi, whereas APPsβ expressed in APPsβ knock-in neurons is found to be enriched in the ER where it binds with the ER chaperone Grp78. Furthermore, biochemical analyses of total soluble APP or APPsβ expressed in wild-type or APPsβ knock-in neurons, respectively, allow the conclusion that secreted APP extracellular proteins are highly stable in regular or trophic factor deprivation conditions.
Neuron-specific Expression of APP and Its Implications in TBI and AD
In performing APP immunostaining using APP-null samples as controls, we found that surprisingly, many well established antibodies failed to distinguish between APP KO and wild-type neurons, indicating nonspecific staining under the conditions we employ. Although we cannot exclude the possibility that protocols used by other investigators may result in more specific signal, our result calls for the necessity of using APP knock-out mice as controls in all immunostaining experiments. It also prompted us to re-evaluate some of the fundamental properties of APP.
It has been well accepted that neurons serve as the primary source of APP in the CNS. However, APP, in particular the 751/770 isoforms, have also been reported to be expressed in astrocytes (2, 3, 28–32), One study even stated that the total amount of astroglial APP is 90% higher than neurons (4). In addition, there are numerous studies reporting increased APP expression in reactive astrocytes induced by different insults, such as lesions or injury of the hippocampus (4, 33, 34), heterotopic transplantation (35), cerebral artery occlusion (36), needle stab injury (14), cuprizone intoxication (37), kainic acid-induced neurotoxic damage (38), and exposure to Aβ peptides (39). Increased APP expression in astrocytes was also suggested to contribute to Aβ plaque deposition and AD pathogenesis (4, 40). We show here that under basal conditions, we could not detect astroglial APP staining in primary cultures or in brain sections. APP expression remained undetectable in astrocytes when these cells were activated by TNFα treatment in vitro or upon traumatic brain injury in vivo. The punctate APP staining following TBI is likely due to its accumulation in the damaged neuronal axons (14, 15). Interestingly, axonal APP accumulation is transient, and most of the staining disappeared within 72 h of the TBI, when significant reactive astrocytosis takes place. Although correlative, this finding indicates that the activated astrocytes may play an active role in clearing APP deposits and possibly damaged axons. On the other hand, the transient and restricted nature of APP accumulation suggests that it is secondary to axonal damage rather than serving a primary role in the TBI response. In this regard, it is worth noting that TBI has been associated with increased risk of dementia (41). However, a consistent relationship between TBI and amyloid pathology has been difficult to establish in mouse models (41). Our APP expression analysis argues against a major effect of TBI in inducing Aβ pathology through astrocytic APP. Previous publications suggest that APP-independent processes, such as Tau modulation or neuroinflammation, are likely to play important roles in TBI-induced dementia (42, 43).
Examination of APP expression in an APP/hAβ/PS1 knock-in AD mouse model detected strong APP staining surrounding Aβ plaques, presumably originating from dystrophic neurites (44–46). However, and consistent with its neuron-specific expression, no APP can be detected in reactive astrocytes in the proximity of Aβ deposits. Because reactive astrocytosis and APP aggregates co-exist, the activated astrocytes do not appear to be effective in clearing APP deposits and dystrophic neurites. The reason for the apparently distinct actions of astroglia in the TBI and the AD model is not clear and may be due to differences in the nature of the insults (acute versus chronic). Nevertheless, these studies, combined, provide strong evidence that astrocytes express negligible amounts of APP under basal or stress/injury conditions and that neurons provide most, if not all, APP and, by extension, Aβ peptides.
Trafficking and Stability of APPs
Grp78, also referred to as heat shock 70-kDa protein 5 (HSPA5), is a key ER chaperone involved in protein folding, protein quality control, and the regulation of ER stress and the unfolded protein response (47). Grp78 was previously found to interact with APP in co-transfected HEK293 cells (25) and also from an APP interactome study (26). Overexpression of Grp78 together with APP in cells has been shown to decrease Aβ production, perhaps due to inhibition of APP maturation (25, 48). Furthermore, a polymorphism allele of the human Grp78 promoter, which results in greater responses to ER stress, was reported to decrease the risk of AD (49). We, for the first time, validated this interaction in an endogenous model in vivo and identified the APP N-terminal domain as its interaction site. It is tempting to speculate that this interaction indirectly contributes to Aβ production and AD pathogenesis through regulating APP maturation and trafficking and that molecules that can modulate the APPs-Grp78 interaction provide a potential therapeutic target.
An intriguing study reported that secreted APPsβ is cleaved upon TFD and that an N-terminal fragment generated from this cleavage (N-APP) serves as a DR6 ligand to trigger axonal degeneration (6). The creation of the APPsβ knock-in mice offers a unique opportunity to examine this cleavage event in the absence of APPsα and compare it with total soluble APP produced in wild-type neurons (5). Using regular neuronal cultures or trophic factor-depleted cultures, and by co-incubating the recombinant APPsβ with TFD-treated cells, we demonstrate that APPsβ and total APPs are both highly stable, and no further cleavage could be identified. Although intrinsic differences between primary cortical and dorsal root ganglion cultures or differences in the culture conditions could explain the different results found by us or by Nikolaev et al. (6), our results using CNS cultures nevertheless argue against a significant role of the N-APP/DR6 pathway in APP biological function or pathogenic processes.
In this study, we employed a highly specific APP antibody and an APPsβ knock-in mouse model and reinvestigated some of the fundamental biological properties of APP and APPs in vitro and in vivo. Our data demonstrate that endogenously produced APP is a neuron-specific protein not expressed in major types of glial cells. APP ectodomain binds to Grp78 in the ER, whereas the membrane and intracellular sequences of APP are required for its localization to the Golgi apparatus. Both neuron-secreted and recombinant APPsβ are highly stable and remain intact upon trophic factor deprivation. We conclude that neuronal APP is the origin of Aβ in both basal and brain injury conditions and that the APP ectodomain mediates downstream effects as a stable and intact protein.
Acknowledgments
We thank members of the Zheng laboratory for helpful and stimulating discussions and Nadia Aithmitti and Xiaoli Chen for technical support. We thank Dr. Matthew Rasband for providing the MBP antibody and the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (IDDRC) Administrative and Confocal Cores for their assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 NS38660 (to C. S. R.) and R01 AG032051, AG033467, and NS076117 (to H. Z.).
- AD
- Alzheimer disease
- Aβ
- amyloid-β
- hAβ
- human Aβ
- APP
- amyloid precursor protein
- APPs
- soluble APP
- FL-APP
- full-length APP
- TFD
- trophic factor deprivation
- TBI
- traumatic brain injury
- GFAP
- glial fibrillary acidic protein
- MBP
- myelin basic protein
- ER
- endoplasmic reticulum
- ki
- knock-in
- DIV
- days in vitro.
REFERENCES
- 1. Zheng H., Koo E. H. (2011) Biology and pathophysiology of the amyloid precursor protein. Mol. Neurodegener. 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chauvet N., Apert C., Dumoulin A., Epelbaum J., Alonso G. (1997) Mab22C11 antibody to amyloid precursor protein recognizes a protein associated with specific astroglial cells of the rat central nervous system characterized by their capacity to support axonal outgrowth. J. Comp. Neurol. 377, 550–564 [PubMed] [Google Scholar]
- 3. LeBlanc A. C., Papadopoulos M., Bélair C., Chu W., Crosato M., Powell J., Goodyer C. G. (1997) Processing of amyloid precursor protein in human primary neuron and astrocyte cultures. J. Neurochem. 68, 1183–1190 [DOI] [PubMed] [Google Scholar]
- 4. Rohan de Silva H. A., Jen A., Wickenden C., Jen L. S., Wilkinson S. L., Patel A. J. (1997) Cell-specific expression of β-amyloid precursor protein isoform mRNAs and proteins in neurons and astrocytes. Brain Res. Mol. Brain Res. 47, 147–156 [DOI] [PubMed] [Google Scholar]
- 5. Li H., Wang B., Wang Z., Guo Q., Tabuchi K., Hammer R. E., Südhof T. C., Zheng H. (2010) Soluble amyloid precursor protein (APP) regulates transthyretin and Klotho gene expression without rescuing the essential function of APP. Proc. Natl. Acad. Sci. U.S.A. 107, 17362–17367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nikolaev A., McLaughlin T., O'Leary D. D., Tessier-Lavigne M. (2009) APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Zheng H., Jiang M., Trumbauer M. E., Sirinathsinghji D. J., Hopkins R., Smith D. W., Heavens R. P., Dawson G. R., Boyce S., Conner M. W., Stevens K. A., Slunt H. H., Sisoda S. S., Chen H. Y., Van der Ploeg L. H. (1995) β-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell 81, 525–531 [DOI] [PubMed] [Google Scholar]
- 8. Köhler C., Ebert U., Baumann K., Schröder H. (2005) Alzheimer disease-like neuropathology of gene-targeted APP-SLxPS1mut mice expressing the amyloid precursor protein at endogenous levels. Neurobiol. Dis. 20, 528–540 [DOI] [PubMed] [Google Scholar]
- 9. Guo Q., Fu W., Sopher B. L., Miller M. W., Ware C. B., Martin G. M., Mattson M. P. (1999) Increased vulnerability of hippocampal neurons to excitotoxic necrosis in presenilin-1 mutant knock-in mice. Nat. Med. 5, 101–106 [DOI] [PubMed] [Google Scholar]
- 10. Xia X., Qian S., Soriano S., Wu Y., Fletcher A.M., Wang X. J., Koo E. H., Wu X., Zheng H. (2001) Loss of presenilin 1 is associated with enhanced β-catenin signaling and skin tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 10863–10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu D. C., Rabizadeh S., Chandra S., Shayya R. F., Ellerby L. M., Ye X., Salvesen G. S., Koo E. H., Bredesen D. E. (2000) A second cytotoxic proteolytic peptide derived from amyloid β-protein precursor. Nat. Med. 6, 397–404 [DOI] [PubMed] [Google Scholar]
- 12. Bitner B. R., Brink D. C., Mathew L. C., Pautler R. G., Robertson C. S. (2010) Impact of arginase II on CBF in experimental cortical impact injury in mice using MRI. J. Cereb. Blood Flow. Metab. 30, 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shim D. J., Yang L., Reed J. G., Noebels J. L., Chiao P. J., Zheng H. (2011) Disruption of the NF-κB/IκBα autoinhibitory loop improves cognitive performance and promotes hyperexcitability of hippocampal neurons. Mol. Neurodegener. 6, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Otsuka N., Tomonaga M., Ikeda K. (1991) Rapid appearance of β-amyloid precursor protein immunoreactivity in damaged axons and reactive glial cells in rat brain following needle stab injury. Brain Res. 568, 335–338 [DOI] [PubMed] [Google Scholar]
- 15. Pierce J. E., Trojanowski J. Q., Graham D. I., Smith D. H., McIntosh T. K. (1996) Immunohistochemical characterization of alterations in the distribution of amyloid precursor proteins and β-amyloid peptide after experimental brain injury in the rat. J. Neurosci. 16, 1083–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H., Wang Z., Wang B., Guo Q., Dolios G., Tabuchi K., Hammer R. E., Südhof T. C., Wang R., Zheng H. (2010) Genetic dissection of the amyloid precursor protein in developmental function and amyloid pathogenesis. J. Biol. Chem. 285, 30598–30605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palmert M. R., Podlisny M. B., Witker D. S., Oltersdorf T., Younkin L. H., Selkoe D. J., Younkin S. G. (1988) Antisera to an amino-terminal peptide detect the amyloid protein precursor of Alzheimer disease and recognize senile plaques. Biochem. Biophys. Res. Commun. 156, 432–437 [DOI] [PubMed] [Google Scholar]
- 18. Palmert M. R., Podlisny M. B., Golde T. E., Cohen M. L., Kovacs D. M., Tanzi R. E., Gusella J. F., Whitehouse P. J., Witker D. S., Oltersdorf T., (1989) The β-amyloid protein precursor: mRNAs, membrane-associated forms, and soluble derivatives. Prog. Clin. Biol. Res. 317, 971–984 [PubMed] [Google Scholar]
- 19. Hyman B. T., Tanzi R. E., Marzloff K., Barbour R., Schenk D. (1992) Kunitz protease inhibitor-containing amyloid β-protein precursor immunoreactivity in Alzheimer disease. J. Neuropathol. Exp. Neurol. 51, 76–83 [DOI] [PubMed] [Google Scholar]
- 20. Shivers B. D., Hilbich C., Multhaup G., Salbaum M., Beyreuther K., Seeburg P. H. (1988) Alzheimer disease amyloidogenic glycoprotein: expression pattern in rat brain suggests a role in cell contact. EMBO J. 7, 1365–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaden D., Voigt P., Munter L. M., Bobowski K. D., Schaefer M., Multhaup G. (2009) Subcellular localization and dimerization of APLP1 are strikingly different from APP and APLP2. J. Cell Sci. 122, 368–377 [DOI] [PubMed] [Google Scholar]
- 22. Palacios G., Palacios J. M., Mengod G., Frey P. (1992) β-amyloid precursor protein localization in the Golgi apparatus in neurons and oligodendrocytes. An immunocytochemical structural and ultrastructural study in normal and axotomized neurons. Brain Res. Mol. Brain Res. 15, 195–206 [DOI] [PubMed] [Google Scholar]
- 23. Caporaso G. L., Takei K., Gandy S. E., Matteoli M., Mundigl O., Greengard P., De Camilli P. (1994) Morphologic and biochemical analysis of the intracellular trafficking of the Alzheimer β/A4 amyloid precursor protein. J. Neurosci. 14, 3122–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benowitz L. I., Rodriguez W., Paskevich P., Mufson E. J., Schenk D., Neve R. L. (1989) The amyloid precursor protein is concentrated in neuronal lysosomes in normal and Alzheimer disease subjects. Exp. Neurol. 106, 237–250 [DOI] [PubMed] [Google Scholar]
- 25. Yang Y., Turner R. S., Gaut J. R. (1998) The chaperone BiP/GRP78 binds to amyloid precursor protein and decreases Aβ40 and Aβ42 secretion. J. Biol. Chem. 273, 25552–25555 [DOI] [PubMed] [Google Scholar]
- 26. Bai Y., Markham K., Chen F., Weerasekera R., Watts J., Horne P., Wakutani Y., Bagshaw R., Mathews P. M., Fraser P. E., Westaway D., St George-Hyslop P., Schmitt-Ulms G. (2008) The in vivo brain interactome of the amyloid precursor protein. Mol. Cell. Proteomics 7, 15–34 [DOI] [PubMed] [Google Scholar]
- 27. Norstrom E. M., Zhang C., Tanzi R., Sisodia S. S. (2010) Identification of NEEP21 as a β-amyloid precursor protein-interacting protein in vivo that modulates amyloidogenic processing in vitro. J. Neurosci. 30, 15677–15685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yasuoka K., Hirata K., Kuraoka A., He J. W., Kawabuchi M. (2004) Expression of amyloid precursor protein-like molecule in astroglial cells of the subventricular zone and rostral migratory stream of the adult rat forebrain. J. Anat. 205, 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forloni G., Demicheli F., Giorgi S., Bendotti C., Angeretti N. (1992) Expression of amyloid precursor protein mRNAs in endothelial, neuronal, and glial cells: modulation by interleukin-1. Brain Res. Mol. Brain Res. 16, 128–134 [DOI] [PubMed] [Google Scholar]
- 30. Berkenbosch F., Refolo L. M., Friedrich V. L., Jr., Casper D., Blum M., Robakis N. K. (1990) The Alzheimer amyloid precursor protein is produced by type-I astrocytes in primary cultures of rat neuroglia. J. Neurosci. Res. 25, 431–440 [DOI] [PubMed] [Google Scholar]
- 31. Young M. J., Lee R. K., Jhaveri S., Wurtman R. J. (1999) Intracellular and cell surface distribution of amyloid precursor protein in cortical astrocytes. Brain Res. Bull. 50, 27–32 [DOI] [PubMed] [Google Scholar]
- 32. Tran M. D. (2011) P2 receptor stimulation induces amyloid precursor protein production and secretion in rat cortical astrocytes. Neurosci. Lett. 492, 155–159 [DOI] [PubMed] [Google Scholar]
- 33. Siman R., Card J. P., Nelson R. B., Davis L. G. (1989) Expression of β-amyloid precursor protein in reactive astrocytes following neuronal damage. Neuron 3, 275–285 [DOI] [PubMed] [Google Scholar]
- 34. Palacios G., Mengod G., Tortosa A., Ferrer I., Palacios J. M. (1995) Increased β-amyloid precursor protein expression in astrocytes in the gerbil hippocampus following ischemia: association with proliferation of astrocytes. Eur. J. Neurosci. 7, 501–510 [DOI] [PubMed] [Google Scholar]
- 35. Martins R. N., Taddei K., Kendall C., Evin G., Bates K. A., Harvey A. R. (2001) Altered expression of apolipoprotein E, amyloid precursor protein, and presenilin-1 is associated with chronic reactive gliosis in rat cortical tissue. Neuroscience 106, 557–569 [DOI] [PubMed] [Google Scholar]
- 36. Nihashi T., Inao S., Kajita Y., Kawai T., Sugimoto T., Niwa M., Kabeya R., Hata N., Hayashi S., Yoshida J. (2001) Expression and distribution of β-amyloid precursor protein and β-amyloid peptide in reactive astrocytes after transient middle cerebral artery occlusion. Acta Neurochir. (Wien.) 143, 287–295 [DOI] [PubMed] [Google Scholar]
- 37. Clarner T., Buschmann J. P., Beyer C., Kipp M. (2011) Glial amyloid precursor protein expression is restricted to astrocytes in an experimental toxic model of multiple sclerosis. J. Mol. Neurosci. 43, 268–274 [DOI] [PubMed] [Google Scholar]
- 38. Solà C., García-Ladona F. J., Mengod G., Probst A., Frey P., Palacios J. M. (1993) Increased levels of the Kunitz protease inhibitor-containing β-APP mRNAs in rat brain following neurotoxic damage. Brain Res. Mol. Brain Res. 17, 41–52 [DOI] [PubMed] [Google Scholar]
- 39. Moreno-Flores M. T., Salinero O., Wandosell F. (1998) βA amyloid peptide (25–35) induced APP expression in cultured astrocytes. J. Neurosci. Res. 52, 661–671 [DOI] [PubMed] [Google Scholar]
- 40. Matsui T., Ingelsson M., Fukumoto H., Ramasamy K., Kowa H., Frosch M. P., Irizarry M. C., Hyman B. T. (2007) Expression of APP pathway mRNAs and proteins in Alzheimer disease. Brain Res. 1161, 116–123 [DOI] [PubMed] [Google Scholar]
- 41. Szczygielski J., Mautes A., Steudel W. I., Falkai P., Bayer T. A., Wirths O. (2005) Traumatic brain injury: cause or risk of Alzheimer disease? A review of experimental studies. J. Neural. Transm. 112, 1547–1564 [DOI] [PubMed] [Google Scholar]
- 42. Tran H. T., LaFerla F. M., Holtzman D. M., Brody D. L. (2011) Controlled cortical impact traumatic brain injury in 3×Tg-AD mice causes acute intra-axonal amyloid-β accumulation and independently accelerates the development of Tau abnormalities. J. Neurosci. 31, 9513–9525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zink B. J., Szmydynger-Chodobska J., Chodobski A. (2010) Emerging concepts in the pathophysiology of traumatic brain injury. Psychiatr. Clin. North Am. 33, 741–756 [DOI] [PubMed] [Google Scholar]
- 44. Shoji M., Hirai S., Yamaguchi H., Harigaya Y., Kawarabayashi T. (1990) Amyloid β-protein precursor accumulates in dystrophic neurites of senile plaques in Alzheimer-type dementia. Brain Res. 512, 164–168 [DOI] [PubMed] [Google Scholar]
- 45. Cras P., Kawai M., Lowery D., Gonzalez-DeWhitt P., Greenberg B., Perry G. (1991) Senile plaque neurites in Alzheimer disease accumulate amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 88, 7552–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cummings B. J., Su J. H., Geddes J. W., Van Nostrand W. E., Wagner S. L., Cunningham D. D., Cotman C. W. (1992) Aggregation of the amyloid precursor protein within degenerating neurons and dystrophic neurites in Alzheimer disease. Neuroscience 48, 763–777 [DOI] [PubMed] [Google Scholar]
- 47. Ni M., Zhang Y., Lee A. S. (2011) Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signaling and therapeutic targeting. Biochem. J. 434, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoshino T., Nakaya T., Araki W., Suzuki K., Suzuki T., Mizushima T. (2007) Endoplasmic reticulum chaperones inhibit the production of amyloid-β-peptides. Biochem. J. 402, 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hsu W. C., Wang H. K., Lee L. C., Fung H. C., Lin J. C., Hsu H. P., Wu Y. R., Ro L. S., Hu F. J., Chang Y. T., Lee-Chen G. J., Chen C. M. (2008) Promoter polymorphisms modulating HSPA5 expression may increase susceptibility to Taiwanese Alzheimer disease. J. Neural. Transm. 115, 1537–1543 [DOI] [PubMed] [Google Scholar]