Background: Smooth muscle cell (SMC) types are characterized by a growing number of cyto-contractile genes.
Results: A combination of approaches led to the discovery of Leiomodin 1 (Lmod1) as a SMC-restricted gene under control of the serum response factor (SRF)/myocardin (MYOCD) transcriptional switch.
Conclusion: Lmod1 is a new SMC-specific SRF/MYOCD target gene.
Significance: A rationale is established to elucidate LMOD1 function in SM development and disease.
Keywords: Bioinformatics, Smooth Muscle, Transcription Regulation, Transcription Target Genes, Transgenic Mice
Abstract
Smooth muscle cell (SMC) differentiation is defined largely by a number of cell-restricted genes governed directly by the serum response factor (SRF)/myocardin (MYOCD) transcriptional switch. Here, we describe a new SRF/MYOCD-dependent, SMC-restricted gene known as Leiomodin 1 (Lmod1). Conventional and quantitative RT-PCRs indicate that Lmod1 mRNA expression is enriched in SMC-containing tissues of the mouse, whereas its two paralogs, Lmod2 and Lmod3, exhibit abundant expression in skeletal and cardiac muscle with very low levels in SMC-containing tissues. Western blotting and immunostaining of various adult and embryonic mouse tissues further confirm SMC-specific expression of the LMOD1 protein. Comparative genomic analysis of the human LMOD1 and LMOD2 genes with their respective mouse and rat orthologs shows high conservation between the three exons and several noncoding sequences, including the immediate 5′ promoter region. Two conserved CArG boxes are present in both the LMOD1 and LMOD2 promoter regions, although LMOD1 displays much higher promoter activity and is more responsive to SRF/MYOCD stimulation. Gel shift assays demonstrate clear binding between SRF and the two CArG boxes in human LMOD1. Although the CArG boxes in LMOD1 and LMOD2 are similar, only LMOD1 displays SRF or MYOCD-dependent activation. Transgenic mouse studies reveal wild type LMOD1 promoter activity in cardiac and vascular SMC. Such activity is abolished upon mutation of both CArG boxes. Collectively, these data demonstrate that Lmod1 is a new SMC-restricted SRF/MYOCD target gene.
Introduction
SMCs2 invest most hollow organs and much of the vascular system where they provide structural support and regulate the flow of materials through the organs where they reside. Vascular and visceral SMCs are defined by the expression of a number of cell-restricted ion channels as well as signaling, cyto-contractile, and matrix-associated genes whose encoded proteins facilitate the major functions associated with the differentiated SMC phenotype (1). Reduced expression of these SMC differentiation markers results in phenotypic adaptation and is associated with the onset of numerous human diseases such as atherosclerosis, cancer, Alzheimer disease, hypertension, asthma, obstructive bladder, gastrointestinal disease, and reproductive disorders (1–4). Thus, there has been tremendous effort to elucidate the molecular basis for SMC phenotypic adaptation. Although a number of SMC-restricted genes associated with the SMC differentiated phenotype have been identified and characterized, it is likely that there exists additional genes (including nonprotein coding RNAs) whose regulation and function in SMC lineages have yet to be fully described.
The majority of SMC-restricted differentiation genes are regulated directly by a widely expressed transcription factor known as serum response factor (SRF) (5). SRF is a single copy gene whose encoded protein physically binds as a homodimer to a 10-bp cis element known as a CArG box (5–7). SRF is a weak transcription factor, but through its association with over 60 co-factors, it mediates cell- and context-specific programs of gene expression. One such transactivator is myocardin (MYOCD), which was first identified as a cardiac myocyte- and SMC-specific SRF co-factor that stimulates CArG-dependent gene expression in these cell types (8–12). The importance of MYOCD in cardiac myocyte and SMC biology has been demonstrated through genetic inactivation studies (13–15). Although MYOCD appears to be necessary for normal cardiac function (15), it is not sufficient to direct a program of cardiac myocyte differentiation (16). In contrast, ectopic expression of MYOCD is sufficient to impart a functional SMC contractile phenotype (17). Although MYOCD clearly plays a pivotal role in directing CArG-dependent gene expression in SMC, there is a report of MYOCD working in a CArG-independent manner (18). A very recent paper showed that MYOCD interacts with cardiac myocyte-restricted T-box transcription factor 5 in a CArG-independent fashion to specifically activate cardiac muscle genes but not SMC-associated genes (19). Thus, MYOCD has emerged as an important molecular switch for the programs of SMC and cardiac myocyte differentiation through a number of CArG-dependent and CArG-independent mechanisms.
Full disclosure of all CArG-SRF-dependent genes in the genome is incomplete. Several strategies have been used to elucidate the so-called CArGome including computational interrogation of the genome (20) and various wet lab assays such as ChIP-seq (21–27). We recently defined the Ref-Seq CArGome wherein conserved CArG element identification was analyzed across essentially all Ref-Seq genes. These findings revealed 8,252 conserved CArG elements near >5,000 Ref-Seq genes (28). During the course of our work to fully define the human CArGome, we became aware of an obscure family of actin-binding domain containing proteins (29). One of these proteins was initially identified as a 64-kDa autoantigen in an expression screen with sera from patients with Hashimoto's thyroiditis (30). Meanwhile, in an effort to discover genes related to tropomodulins, which represent a family of actin-capping proteins, Conley et al. (31) used a bioinformatics approach and found several expressed sequence tags that appeared to extended the tropomodulin cDNAs. These authors were able to assemble cDNA sequences into two distinct mRNA transcripts, and mRNA dot blotting revealed one of these to be expressed across a wide range of tissues, whereas the other was restricted to heart and skeletal muscle. Because of the apparent enrichment of one transcript in smooth muscle-containing tissues, the gene was formally named leiomodin 1 for smooth (leio) and its similarity to tropomodulins (modin) (official HUGO symbol, LMOD1). LMOD1 was found to be nearly identical to the original 64-kDa autoantigen (30). A second related transcript was found to be enriched in cardiac and skeletal muscle and was named Lmod2 (31).
There is very limited information about the function of LMODs (32), and nothing is known regarding their transcriptional regulation. Here, we extend earlier mRNA expression analysis to mouse tissues and show by Western and immunoblotting the highly restrictive pattern of LMOD1 expression in SMC lineages. In vitro and transgenic mouse studies document the essential requirement for both CArG elements in the expression of LMOD1. In contrast, similarly positioned CArG elements around the LMOD2 gene appear nonfunctional. Together, these data demonstrate LMOD1 as a new, SMC-restricted member of the mammalian CArGome and highlight the importance of validating putative CArG-containing genes with wet lab assays.
EXPERIMENTAL PROCEDURES
Animal Studies
Studies involving RNA and Western blotting were performed on tissues isolated from 3-week-old C57BL/6 mice. The tissues were individually rinsed in PBS, snap frozen in liquid nitrogen, and subsequently processed for total RNA or protein isolation. Srf inducible knock-out mice were generated by crossing a floxed Srf mouse (33) to a tamoxifen-inducible Myh11-Cre mouse (34). Adult male Srf fl/fl/Myh11-Cre+/− mice were treated with tamoxifen (35 μg/g, intraperitoneal) or equal volume of sunflower oil for 5 consecutive days, and tissues were then harvested 3 weeks following the initial injection for expression studies (below). Animal protocols were approved by the University's Institutional Animal Care and Use Committee.
Cell Culture
Rat pulmonary artery SMCs (PAC1) (35), rat aortic SMCs, NIH 3T3, COS-7, C2C12, HEK293, and HeLa cells were cultured in DMEM containing high glucose and 10% FBS without antibiotics or antimycotics. Differentiation of C2C12 cells was induced by replacing existing medium in subconfluent cells with that of DMEM containing high glucose and 2% horse serum for 48 h. Human coronary artery smooth muscle cells (HCASM) were maintained in growth medium 231 or differentiation medium per the manufacturer's instructions (Invitrogen).
RNA Isolation and RT-PCR
Total RNA was extracted from tissues and cell lines using TRIzol (Invitrogen) and purified as per the manufacturer's instructions. cDNA was generated with either the first strand cDNA synthesis kit (GE Healthcare) or the iScript cDNA synthesis kit (Bio-Rad). Differences in mRNA expression were examined initially by conventional RT-PCR. In some instances, mRNA expression levels were quantified using SYBR Green-based real time PCR (MyiQ; Bio-Rad). Information on the primers used to assess the expression of target genes is listed in supplemental Table S1.
Protein Extraction and Western Blotting
To isolate protein from tissues, each tissue sample was manually crushed under liquid nitrogen and homogenized in lysis buffer as described (36). Protein extracts from cultured cells were prepared following brief rinsing in PBS and a 10-min incubation with protein lysis buffer. Protein concentration for all samples was measured using a detergent-compatible colorimetric assay kit supplied by Bio-Rad. Equal amounts of protein samples (6 μg) were separated in 10% SDS-PAGE gels and then transferred onto nitrocellulose membranes. These membranes were blocked in 5% skim milk solution prepared in 1× TBST for 1 h and then probed overnight at 4 °C with antibodies recognizing LMOD1 (Proteintech, 1:2000), α-tubulin (TUBA; Sigma, 1:5000), ACTA2 (Sigma, 1:3000), CNN1 (DAKO, 1:2000), or MYOCD (H300 antibody; Santa Cruz, 1:300). The membranes were subsequently rinsed with TBST and incubated with an appropriate secondary antibody for 1 h at room temperature. Protein expression was detected using SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific). In some experiments, the antibody to LMOD1 was preabsorbed to purified LMOD1 antigen (Proteintech) for 24 h at 4 °C before use in Western blotting.
siRNA, shRNA, and Adenoviral-mediated Gene Transfer Studies
The cells were seeded at ∼70% confluency in 6-well dishes and transfected the next day with either scrambled RNA or siRNA to Lmod1 (Invitrogen) using Lipofectamine 2000 (Invitrogen). Following 6 h of incubation, the cells were rinsed with PBS and then refed with fresh growth medium. Protein was isolated from these cells after 72 h. The cells were similarly transfected with scrambled RNA or a shRNA to SRF as described (37) and target gene expression assessed by extracting total RNA from cultured cell lines for RT-PCR studies. Adenovirus carrying lacZ or human MYOCD constructs were transduced in cells cultured in DMEM containing 2% FBS. 14–16 h later, medium in each of the wells was replaced with DMEM containing 10% FBS. Total RNA or protein was subsequently isolated after 8–10 h.
Immunohistochemistry
LMOD1 protein localization was characterized in wild type or inducible Srf knock-out mouse tissues. Once isolated, the tissues were immediately fixed in 10% neutral-buffered formalin, embedded in paraffin, and sectioned at 5-micron thickness. Following deparaffanization, tissue sections were rehydrated in PBS and then incubated in 3% aqueous hydrogen peroxide. Prior to immunohistochemistry, the tissues were incubated in antigen retrieval buffer (DAKO) and subjected to steam in a pressure cooker for 10 min. Tissue sections were next blocked in serum-free blocking buffer (DAKO) and then incubated overnight at 4 °C with LMOD1 antibody (Proteintech, 1:1000), SRF (Santa Cruz, 1:1000), or rabbit IgG (Dako, 1:1000). After rinsing with PBS, tissue sections were incubated in biotinylated anti-rabbit secondary antibody (Vector Laboratories Inc., 1:1000) for 30 min and then in freshly prepared avidin-biotin complex (Vector Laboratories Inc.) for an additional 30 min. LMOD1 protein localization was then revealed by incubating sections in Vector Red (Vector Laboratories Inc.) for 30 min followed by counterstaining with hematoxylin.
Bioinformatics
Homology between human, mouse, and rat Leiomodin 1 orthologs was determined using Visualization Tools for Alignments (VISTA) (38). Leiomodin 1 genomic sequences were obtained from the University of California at Santa Cruz genome browser (39). Consensus or non-consensus SRF-binding CArG boxes present in the human, mouse, or rat Leiomodin 1 orthologs were identified using the FINDPATTERNS algorithm in the Genetics Computer Group's suite of programs. A similar approach was used to detect all CArG boxes in human, mouse, and rat Leiomodin 2 orthologs. The sequence homology between Leiomodin 1 and Leiomodin 2 CArG boxes was evaluated and graphically represented using WebLogo (40). Detailed standard operating procedures for navigating through each of these computational tools are available upon request.
Generation and Mutagenesis of LMOD1 Promoter
We mapped the transcription start site of mouse Lmod1 by 5′-rapid amplification of cDNA ends and inferred the approximate start site of human LMOD1 based on sequence homology and annotation of cDNA ends in public databases. High quality genomic DNA isolated from HCASM and enzyme-clamped primers (supplemental Table S1) were used to PCR amplify a 540-bp fragment of the human LMOD1 promoter from −291 to +249 bp of the inferred transcription start site. This region contains the two CArG boxes depicted in Fig. 3A. The resulting PCR product was cloned into the XhoI-HindIII site of the pGL3 basic reporter plasmid (Promega). We also PCR-cloned a 922-bp region encompassing two CArG boxes in the human LMOD2 promoter. Mutations in each of the two CArG boxes of LMOD1 were introduced with a QuikChange mutagenesis kit (Stratagene) using primers listed in supplemental Table S1. All of the mutagenesis experiments were confirmed by sequencing each of the plasmids at the Cornell University Life Sciences Core Laboratories Center (Ithaca, NY) to confirm fidelity in nucleotide sequence.
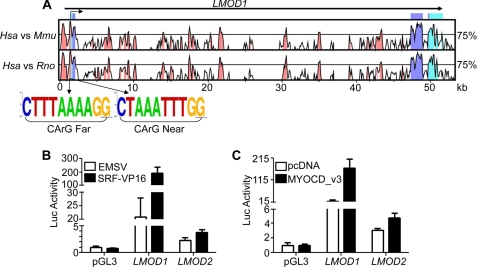
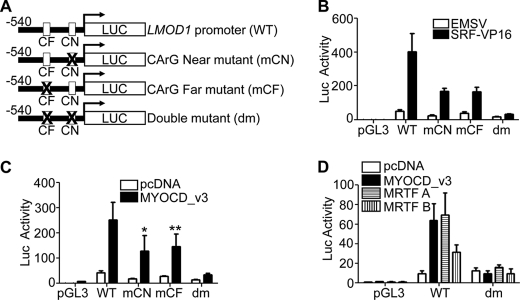
FIGURE 3.
Functional analysis of human LMOD1 and LMOD2 promoters. A, VISTA plot indicating nucleotide sequence homology between human (Hsa), mouse (Mmu) and rat (Rno) LMOD1 over a 50-kb genomic interval. The x axis represents the human base sequence, and the y axis indicates the percentage of homology between Hsa versus Mmu (top plot) and Hsa versus Rno (bottom plot). The bent arrow at the top represents the inferred transcription start site. Light teal peaks represent untranslated regions; dark blue peaks represent protein-coding exons; and pink peaks are the conserved nonprotein coding sequences. Sequence logos of two CArG-like elements near LMOD1 are shown below the VISTA plots and illustrate the conservation of each CArG box across six vertebrate species. B and C, LMOD1 and LMOD2 promoter activity in PAC1 SMCs transfected either with SRF-VP16 (B) or MYOCD_v3 (C). The promoter activity reported here and below is a ratio between luciferase and Renilla normalized to that of the pGL3 basic plasmid set to 1. Similar SRF/MYOCD-mediated promoter activity was observed in at least three independent studies.
Luciferase Reporter Assays
Human LMOD1 and LMOD2 promoter-related luciferase assays were carried out in COS-7 and PAC1 SMCs. The cells were plated at ∼70% confluency in 24-well dishes and co-transfected with indicated plasmids the next day using the calcium phosphate co-precipitation method (41). All of the cells were co-transfected with a Renilla reporter gene that served as an internal control. Following a 12–14-h incubation, medium in each of the wells was replaced with DMEM containing 10% FBS. The cells were then lysed 24 h later and prepared for luminometry as specified by the manufacturer (Promega). All of the transfections were done in quadruplicate, and each experiment was repeated at least three times. The data were analyzed and graphically represented using GraphPad Prism 5.0. The data are reported as the means ± standard deviation.
Electrophoretic Mobility Shift Assay
SRF protein was generated in vitro using a plasmid carrying the entire human SRF coding sequence as a template in an in vitro transcription/translation assay (TnT Quick PCR for DNA; Promega). Recombinant SRF protein was incubated with unlabeled or 32P-labeled oligonucleotides (supplemental Table S1) for 20 min at room temperature as described (42). Controls included incubation of the labeled oligonucleotides plus SRF with either an SRF specific antibody (Santa Cruz) or a MEF2A antibody (Santa Cruz) or 100-fold molar excess of unlabeled, mutant oligonucleotides for 20 min at room temperature. Nucleoprotein complexes were resolved in a 5% nondenaturing polyacrylamide gel, vacuum-dried, transferred onto blotting paper, and exposed to Kodak Biomax XAR film at −80 °C for varying lengths of time.
Transgenic Mouse Studies
Human LMOD1 promoter constructs containing intact SRF-binding CArG boxes or mutated CArG boxes were independently cloned into a SalI-HindIII site immediately upstream of a nuclear lacZ reporter gene (36). Each of these plasmids was then linearized and independently microinjected into fertilized mouse oocytes through services provided by Cyagen Biosciences (Guangzhou, China). Embryonic day 12.5 mouse embryos were genotyped, fixed, stained with lacZ, and “cleared” as described (36). We examined a total of 25 and 10 independent transgenic founders carrying either wild type or CArG mutant transgenes, respectively.
Statistical Analysis
Where deemed necessary, a two-way analysis of variance followed by Bonferroni's post-hoc test for individual significance was done using GraphPad Prism 5.0.
RESULTS
Leiomodin 1 Is Expressed Primarily in Adult and Embryonic SMC-rich Tissues
The expression profile of LMOD1 is incomplete. Thus, we first assessed the mRNA and protein expression of LMOD1 across a broad spectrum of adult mouse tissues as well as embryonic mice. Conventional RT-PCR data indicate that Lmod1 mRNA is expressed in SMC-enriched tissues such as aorta, bladder, colon, intestine, stomach, and uterus (Fig. 1A). Little to no Lmod1 mRNA expression is detected in the brain, liver, skeletal muscle, and spleen. The expression of Lmod1 mRNA is very similar to Cnn1 (Fig. 1A), a known SMC-restricted gene (43). Quantitative RT-PCR reveals highest levels of Lmod1 mRNA in aorta, bladder, and other SMC-enriched tissues; comparatively less Lmod1 mRNA is seen in brain, heart, liver, and skeletal muscle (Fig. 1B). In contrast to Lmod1, expression of Lmod2 and Lmod3 mRNA is restricted largely to cardiac and skeletal muscle (Fig. 1A).
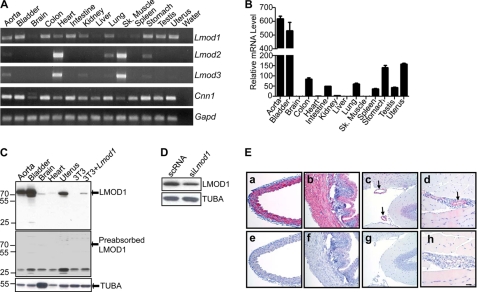
FIGURE 1.
Leiomodin 1 mRNA and protein expression analysis. A, RT-PCR analysis of Lmod1, Lmod2, and Lmod3 in the indicated adult mouse tissues. Note the similar mRNA expression pattern between Lmod1 and Cnn1, a well known SMC-restricted gene (43). B, quantitative RT-PCR of Lmod1 mRNA in the same tissues as A. Expression levels are displayed as normalized fold increases over brain (set to 1). C, Western blot of LMOD1 protein expression in mouse tissues and in 3T3 cells transfected either with an empty control plasmid or an expression plasmid carrying Lmod1 (top panel). LMOD1 protein expression following preabsorption of the LMOD1 antibody with LMOD1 antigen (middle panel). D, endogenous LMOD1 protein expression in PAC1 SMCs following siRNA knockdown of LMOD1. E, immunohistochemistry of LMOD1 protein in aorta (panels a and e), bladder (panels b and f), brain (panels c and g), and skeletal muscle (panels d and h) using either the LMOD1 antibody (panels a–d) or a nonimmune, isotype-matched IgG control (panels e–h). The bar in panel h is 30 μm for all panels. The data shown in all of the panels are representative of three independent experiments. Arrows in panels c and d indicate LMOD1 positive blood vessels.
We next used a commercially available antibody to LMOD1 in Western blots and immunohistochemistry. We found a protein of ∼70 kDa to be highly expressed in the aorta, bladder and uterus with barely detectable levels seen in brain and heart (Fig. 1C). Three lines of evidence support the authenticity of the 70-kDa band being LMOD1. First, 3T3 cells transfected with an LMOD1 expression plasmid show the clear presence of a protein band that co-migrates with that seen in various adult tissues (Fig. 1C, top panel). Second, preincubation of the LMOD1 antibody with purified LMOD1 antigen blocked the detection of the 70-kDa protein (Fig. 1C, middle panel). Interestingly, this experiment showed the emergence of an immunoreactive band of ∼25 kDa; the nature of this protein product is presently unclear, but the fact that its presence appears higher in brain than in aorta would suggest it is not a form of LMOD1. Finally, transfection of PAC1 SMC with siRNA to Lmod1 results in attenuated expression of the 70-kDa protein (Fig. 1D). To determine whether LMOD1 expression is modulated under conditions promoting SMC differentiation, we examined steady-state LMOD1 protein levels in HCASM in growth or differentiation-inducing medium. The results reveal a consistent (n = 4 independent studies) elevation in LMOD1 protein upon HCASM differentiation (supplemental Fig. S1).
Having demonstrated the specificity of the LMOD1 antibody and the presence of LMOD1 protein in whole tissue/cell extracts, we next turned to the question of where in tissues LMOD1 protein resides. Immunohistochemistry of adult aorta (Fig. 1E, panel a), bladder (Fig. 1E, panel b), brain (Fig. 1E, panel c), and skeletal muscle (Fig. 1E, panel d) demonstrates a SMC-specific pattern of LMOD1 protein expression. Further, analysis of LMOD1 in the developing mouse shows early expression of LMOD1 in heart (Fig. 2A) that persists up to embryonic day (E) 12.5 (below) but then is virtually undetectable at E13.5 (Fig. 2D) and E15.5 (Fig. 2F). Vascular and visceral SMC expression of LMOD1 is easily detected at E13.5 and E15.5 (Fig. 2) similar to other SMC-restricted genes (43, 44). We also note LMOD1 protein expression in the neural tube (data not shown). Taken together, mRNA and protein expression data firmly establish LMOD1 as a new SMC-restricted gene associated with the SMC differentiated phenotype.
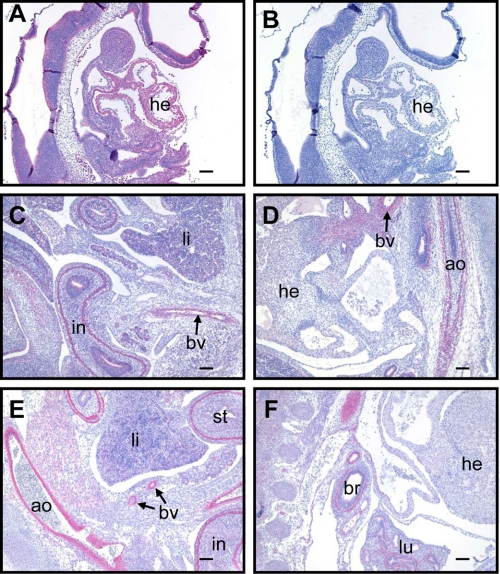
FIGURE 2.
LMOD1 protein expression in developing mouse embryos. Immunohistochemistry of LMOD1 protein expression (red stain) in sagittal sections of E9.5 (A), E13.5 (C and D), and E15.5 (E and F) mouse embryos. An isotype-matched IgG control antibody shows no staining of an E9.5 embryo (B). Similar lack of staining was seen with the IgG control applied to E13.5 and E15.5 embryo sections (not shown). ao, aorta; br, bronchiole; bv, blood vessel; he, heart; in, intestine; li, liver; lu, lung; st, stomach. The bars in A and B represent 100 μm, and the bars in C–F represent 500 μm.
The LMOD1 Gene Harbors Two Functional CArG Elements
Most SMC-restricted genes contain one or more SRF-binding CArG elements present within 4 kb of the transcription start site (7). To ascertain whether CArG elements exist in or around the Leoimodin 1 gene, we performed comparative sequence analysis between human, mouse, and rat orthologs using various bioinformatics tools. A VISTA plot shows that the ∼50-kb human LMOD1 gene comprises three highly conserved exons and several conserved noncoding sequences, including the immediate 5′ promoter region (Fig. 3A). The FINDPATTERNS algorithm in Genetics Computer Group software revealed many CArG elements in and around the LMOD1 gene. Two of these CArG boxes, located just upstream of a mapped transcription start site, are completely conserved in multiple vertebrate species (Fig. 3A). We also found similar CArG elements near the LMOD2 promoter (supplemental Fig. S2); no conserved CArG elements exist around the LMOD3 gene. Based on their relative position to the annotated transcription start site and previous work from the Olson lab (45), the LMOD1 CArG elements are referred to as CArG Far and CArG Near (Fig. 3A). To determine whether the CArG boxes in LMOD1 and LMOD2 are functionally active, we PCR-cloned each promoter into a luciferase reporter plasmid and tested the response of each promoter to expression plasmids carrying either SRF-VP16 or MYOCD_v3 (long, SMC isoform) (46) in cultured cells. Despite the presence of conserved CArG elements in LMOD2, this promoter is weakly active in PAC1 SMC (Fig. 3, B and C) and COS-7 cells (data not shown) as compared with LMOD1. Moreover, the LMOD2 promoter is only weakly responsive to SRF-VP16 or MYOCD_v3 overexpression (Fig. 3, B and C, and data not shown). On the other hand, SRF-VP16 and MYOCD_v3 each stimulate LMOD1 promoter activity 10-fold or greater (Fig. 3, B and C). These data suggest that LMOD1 expression is under direct control of SRF/MYOCD.
SRF Binds the Two CArG Elements in LMOD1
Both CArG elements in LMOD1 deviate from the consensus (CCW6GG) sequence by 1 bp. To determine whether SRF binds the two CArG-like elements found in LMOD1, we performed a gel shift assay. Consistent with a prior report (42), SRF binds the consensus CArG element located in the first intron of CNN1 resulting in a prominent CArG-SRF nucleoprotein complex (Fig. 4). Although weaker in intensity, similar nucleoprotein complexes are seen between SRF and each of the CArG elements found in LMOD1. A clear supershift in each of the nucleoprotein complexes is seen when the nucleoprotein complex is incubated with an antibody to SRF. In contrast, no supershift is detected with an antibody to MEF2A. The addition of unlabeled wild type oligonucleotides results in a loss in the CArG-SRF nucleoprotein complex, whereas no such loss is seen when unlabeled mutant oligonucleotides are added (Fig. 4). Collectively, these in vitro results reveal functional CArG elements in the LMOD1 promoter.
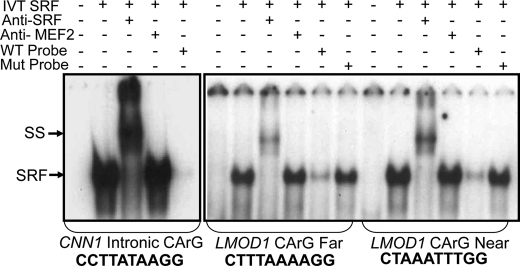
FIGURE 4.
SRF binding to LMOD1 CArG elements. 32P-Labeled double-stranded oligonucleotides containing CArG elements from CNN1 and LMOD1 promoter CArG Far and CArG Near were incubated with the in vitro translated (IVT) SRF in the presence of SRF or MEF2A antibody or a 100-fold molar excess of unlabeled wild type (WT) or mutant (Mut) oligonucleotide. SRF-CArG nucleoprotein complex (SRF) and supershift (SS) are indicated with arrows. Exposure times for each EMSA were 11 h (for CNN1) or 64 h (for LMOD1).
SRF/MYOCD-dependent Transactivation of the LMOD1 Promoter in Vitro Requires Intact CArG Boxes
To assess the importance of the two CArG-like elements in mediating LMOD1 promoter activity, we mutated each (Fig. 5A) and examined accompanying changes in SRF-VP16- and MYOCD_v3-mediated promoter activity in cultured cells. Although SRF-VP16 induces WT LMOD1 promoter activity, this activity is reduced upon mutating CArG near (mCN) or CArG far (mCF). In addition, SRF-VP16-mediated promoter activity is nearly abolished after mutating both CArG boxes (Fig. 5B, dm). Similar attenuated LMOD1 promoter activity is noted following transfection with MYOCD_v3, MRTF-A and MRTF-B (Fig. 5, C and D). These findings show that SRF and members of the MYOCD family mediate LMOD1 promoter activity in a CArG-dependent fashion.
FIGURE 5.
Functional analysis of LMOD1 CArG elements in vitro. A, schematic of the wild type (WT) LMOD1 promoter and various CArG mutants. B and C, each indicated LMOD1 promoter construct was transfected into PAC-1 SMCs in the presence of SRF-VP16 (B) or MYOCD_v3 (C), and luciferase activity was measured. D, COS-7 cells were similarly transfected with WT or double CArG mutant LMOD1 promoter and either MYOCD_v3, MRTF-A, or MRTF-B. The results shown were reproduced in one independent experiment. *, p < 0.001; **, p < 0.01.
SRF and MYOCD Direct Endogenous Leiomodin 1 Expression
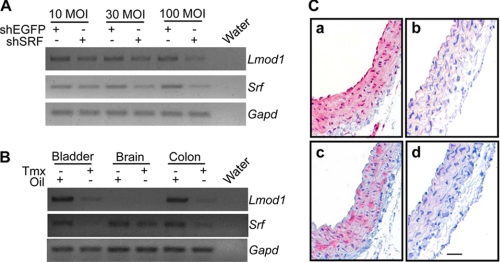
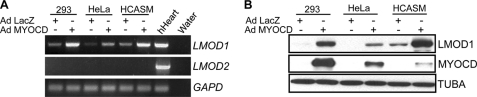
Using three complementary approaches, we next examined the ability of SRF to effect endogenous Leiomodin 1 expression. First, we transduced PAC-1 SMCs with shSRF for 72 h and then assessed mRNA expression of Lmod1 by RT-PCR. shSRF represses endogenous Lmod1 mRNA expression in a dose-dependent manner (Fig. 6A). Consistent with promoter data above, shSRF reduces Lmod1 mRNA in differentiated C2C12 cells. Interestingly, although there appears to be weak activation of the LMOD2 promoter by SRF/MYOCD (Fig. 3), shSRF had no observable effect on the endogenous expression of Lmod2 (supplemental Fig. S3). In the next series of experiments, we analyzed Lmod1 mRNA expression in tissues isolated from mice wherein a floxed Srf gene (33) is inducibly deleted with a tamoxifen-responsive Cre driver mouse (34). Lmod1 mRNA expression is severely attenuated in such SMC-rich tissues as the bladder and colon upon Srf reduction (Fig. 6B); no detectable changes in Lmod1 expression are observed in the brain consistent with earlier data (Fig. 1A versus Fig. 6B). Using the same conditional Srf knock-out mouse, we extended these mRNA expression studies by analyzing LMOD1 protein expression in the aorta. Consistent with the mRNA expression data in bladder and colon, tamoxifen treatment effectively reduces SRF protein in the aorta (Fig. 6C, panel a versus panel b). Importantly, LMOD1 protein expression is reduced in the aorta following Cre-mediated excision of the Srf allele (Fig. 6C, panel c versus panel d). Finally, to investigate whether MYOCD induces endogenous human LMOD1 expression, we transduced different human cultured cells with an adenovirus carrying human MYOCD for 24 h and then examined endogenous LMOD1 mRNA expression by RT-PCR. MYOCD induces LMOD1 mRNA expression in cultured HCASM as well as non-SMC types (Fig. 7A). Consistent with the weak activation of LMOD2 promoter activity (Fig. 3C), ectopic MYOCD did not elicit detectable LMOD2 mRNA transcripts (Fig. 7A). Western blotting shows clear increases in endogenous LMOD1 protein upon ectopic MYOCD expression (Fig. 7B). Collectively, these results offer firm evidence for the requirement of SRF and MYOCD in the expression of endogenous LMOD1.
FIGURE 6.
LMOD1 expression in SRF-deficient cells and tissues. RT-PCR analysis of Lmod1 and Srf mRNA either in PAC-1 SMC transduced with shEGFP or shSRF for 72 h (A) or indicated tissues from adult mice carrying homofloxed Srf alleles and the tamoxifen (Tmx)-inducible Myh11-Cre driver (34) (B). Note the reduction in both Srf and Lmod1 mRNA in tissues derived from mice treated with Tmx (to activate Cre recombinase and effect excision of the Srf gene) versus the vehicle control (Oil). The latter results were extended by comparing protein expression of SRF (C, panels a and b) versus LMOD1 (C, panels c and d) in similar mice treated either with Tmx (C, panels b and d) or sunflower oil (C, panels a and c). The bar in panel d of C is 30 μm for all panels. The images in A and B were inverted in Adobe Photoshop so as to better indicate the bands in each gel.
FIGURE 7.
LMOD1 mRNA and protein expression in human cells overexpressing MYOCD. A, the indicated human cell lines were transduced with equal amounts of adenovirus carrying MYOCD or LacZ and endogenous LMOD1 and LMOD2 mRNA expression assessed by RT-PCR. B, same experiment as in A only LMOD1 and MYOCD protein expression were determined using Western blotting. This result was reproduced in an independent experiment.
In Vivo LMOD1 Promoter Activity in Cardiac and Vascular SMC Requires Intact CArG Elements
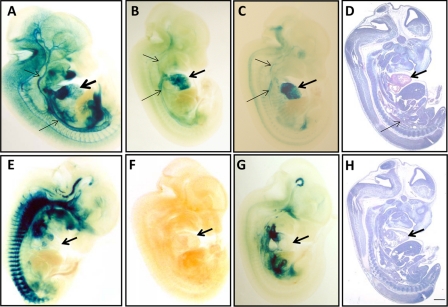
To establish whether LMOD1 promoter activity mirrors endogenous protein expression during development and whether such activity is CArG-dependent, we evaluated LMOD1 promoter activity in E12.5 mice. Transgenic mice carrying the wild type LMOD1 promoter display cardiac muscle activity (Fig. 8, A–C), a finding consistent with endogenous LMOD1 protein expression (Fig. 8D). Vascular SMC LMOD1 promoter activity is seen in some of these transgenic mice as well (Fig. 8, A–C). In contrast, LMOD1 transgenic mice carrying mutations in both CArG elements show either no promoter activity whatsoever (6 of 10 founders; Fig. 8F) or ectopic activity exhibiting little, if any, cardiac muscle and vascular SMC activity (Fig. 8, E and G). Thus, we conclude that the human LMOD1 promoter displays cardiovascular activity in transgenic mouse embryos in a CArG-dependent manner.
FIGURE 8.
LMOD1 promoter activity in transgenic mice. Sagittal E12.5 day mouse embryos stained either with β-galactosidase to assess LMOD1 promoter activity (A–C and E–G) or an antibody to LMOD1 (D) or control IgG (H). Shown are three WT LMOD1 promoter mice (A–C) and three double CArG mutant LMOD1 promoter mice (E–G). The thick arrows indicate the heart, and the thinner arrows point to aorta and vessels of head. The bar in H is 1 mm for all panels. Eight of 25 wild type founders displayed promoter activity in cardiac muscle, and five of 25 showed promoter activity in vascular tissue. In contrast none of the 10 CArG mutant founders showed LMOD1 promoter activity in cardiac muscle or vascular tissue.
DISCUSSION
The genome is punctuated with more than 1 million regulatory elements binding ubiquitous or cell-specific transcription factors that collectively drive proper spatiotemporal patterns of gene expression across some 250 cell types comprising the human body. We recently reported conserved SRF-binding CArG elements adjacent to ∼20% of Ref-seq genes, including all previously validated CArG-containing genes (28). Most muscle-restricted genes harbor at least two closely spaced CArG elements that are critical for gene expression and normal muscle homeostasis. Here, we report on the functionality of two closely spaced CArG elements in an ill-defined gene called Leiomodin 1 (LMOD1) (31). Gel shift and luciferase assays verify SRF binding and activity over the two LMOD1 CArG elements. Moreover, the potent SRF coactivator, MYOCD, is shown to transactivate the LMOD1 promoter and induce the endogenous transcript and protein in cells that otherwise exhibit very low levels of MYOCD expression. Genetic inactivation studies further demonstrate the requirement for SRF in normal LMOD1 expression. Importantly, similar to several other SMC-restricted promoters, we have demonstrated that the cardiovascular-restricted expression of LMOD1 during embryonic development is recapitulated in transgenic mice carrying the human LMOD1 promoter. This pattern of promoter activity is shown to be strictly dependent on intact CArG elements. The adult SMC-specific pattern of LMOD1 and its dependence on SRF/MYOCD for normal expression establishes LMOD1 as a new SMC-restricted CArG-containing gene.
Leiomodins comprise three paralogous genes encoding highly similar proteins that are thought to play a role in actin cytoskeleton assembly (47). LMOD3 has not been examined in any context to date. We show here that expression of Lmod3 mRNA is highly enriched in both cardiac and skeletal muscle. This striated muscle-restricted pattern of expression is virtually identical to that of Lmod2 (31). In contrast to these two Lmod family members, Lmod1 mRNA appears to be expressed widely across many tissues. However, extensive analysis of LMOD1 protein expression demonstrates a SMC-specific pattern of expression which is in keeping with an earlier, noncomprehensive report (29). Moreover, we demonstrate clear endogenous LMOD1 protein as well as transgenic promoter activity in the embryonic mouse heart. Thus, Lmod1 joins a growing list of adult SMC-specific genes (e.g. Tagln1, Cnn1, Acta2, Csrp1, Fhl2, and Kcnmb1) exhibiting embryonic cardiac myocyte expression. It is unclear whether expression of these genes and their encoded proteins serves an important physiological role in the embryonic heart or is merely a manifestation of the activity of SRF/MYOCD in the absence of suppressors (microRNAs?) that only emerge later in cardiac development.
SRF directly regulates scores of CArG-containing actin cytoskeleton genes (48, 49). In the absence of SRF, cells and tissues fail to organize an actin cytoskeleton necessary for normal cellular homeostasis (50). Conditional deletion of SRF in heart and vascular SMC results in severe defects in cardiac sarcomerogenesis and trabeculation as well as reduced vascular SMC recruitment to the dorsal aorta (33, 51). In this context, tropomodulin 1, a member of a family of proteins related to leiomodins, is involved with the capping of actin filaments at the slowly growing (pointed) ends (52). Genetic loss of Tmod1 results in embryonic death at day 10 in the mouse because of perturbations in cardiac and yolk sac vessel development (53). These defects could be rescued upon cardiac-specific expression of TMOD1, suggesting the vascular phenotype was secondary to the cardiac pathology (54). Interestingly, Tmod1 is the only Tmod family member with conserved CArG elements in its locus (data not shown). Whether Tmod1 is reduced in the Srf knock-out and thus contributing to the observed cardiovascular phenotypes in this model is presently unknown. Similar to Tmod1, Lmod2 expression is enriched in cardiac myocytes. In vitro knockdown studies reveal a role for LMOD2 in normal cardiac sarcomere assembly (32). Further, we report here the existence of two conserved CArG elements in LMOD2 whose sequence and position relative to the transcription start site are similar to the two CArG elements in LMOD1. Although there is some detectable increase in LMOD2 promoter activity upon co-transfection with SRF or MYOCD, we deem such increases biologically inert because no changes in endogenous LMOD2 are seen with loss of SRF or gain of MYOCD. These results indicate that the presence of conserved CArG elements in or around a gene locus may not always portend SRF dependence, thus highlighting the need to carefully validate hypothetical CArG-dependent genes using multiple, complementary assays.
The only reported function of LMOD1 relates to its association with tropomyosin in an in vitro binding assay (55). Attempts to define a function of LMOD1 in cultured SMC using gain- and loss-of-function studies have, as of this date, been uninformative. Thus, insight into the precise function of LMOD1 awaits further analysis, including genetic inactivation and/or gain-of-function transgenic mouse studies. Given our findings that expression of Lmod1 overlaps with other SMC-restricted genes and that its regulation occurs through the SRF/MYOCD axis, we speculate that LMOD1 function may be linked to SMC contractile activity or actin cytoskeletal homeostasis.
In conclusion, we introduce a new member of the SMC-restricted CArGome: Lmod1. LMOD1 is shown to exhibit an embryonic and adult expression pattern similar to that of many other SRF-dependent SMC cyto-contractile proteins. The challenge moving forward will be defining functions related to LMOD1 through gene targeting and perhaps gain-of-function studies as has been reported for the related family member, TMOD1 (53, 56).
Supplementary Material
Acknowledgments
We thank Sarah Cowan, Xiaochun Long, Orazio Slivano, Xun Tang, and Qian Zhou for assistance throughout these studies.
This work was supported, in whole or in part, by National Institutes of Health Grants HL091168 and HL62572.

This article contains supplemental Table S1 and Figs. S1–S3.
- SMC
- smooth muscle cell
- SRF
- serum response factor
- MYOCD
- myocardin
- HCASM
- human coronary artery smooth muscle cells
- En
- embryonic day n
- sh
- short hairpin.
REFERENCES
- 1. Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 2. Chow N., Bell R. D., Deane R., Streb J. W., Chen J., Brooks A., Van Nostrand W., Miano J. M., Zlokovic B. V. (2007) Serum response factor and myocardin mediate arterial hypercontractility and cerebral blood flow dysregulation in Alzheimer's phenotype. Proc. Natl. Acad. Sci. U.S.A. 104, 823–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirota J. A., Nguyen T. T., Schaafsma D., Sharma P., Tran T. (2009) Airway smooth muscle in asthma. Phenotype plasticity and function. Pulm. Pharmacol. Ther. 22, 370–378 [DOI] [PubMed] [Google Scholar]
- 4. Beamish J. A., He P., Kottke-Marchant K., Marchant R. E. (2010) Molecular regulation of contractile smooth muscle cell phenotype. Implications for vascular tissue engineering. Tissue Eng. Part B Rev. 16, 467–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Norman C., Runswick M., Pollock R., Treisman R. (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55, 989–1003 [DOI] [PubMed] [Google Scholar]
- 6. Minty A., Kedes L. (1986) Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells. Presence of an evolutionarily conserved repeated motif. Mol. Cell Biol. 6, 2125–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miano J. M. (2003) Serum response factor. Toggling between disparate programs of gene expression. J. Mol. Cell Cardiol. 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 8. Wang D. Z., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 9. Chen J., Kitchen C. M., Streb J. W., Miano J. M. (2002) Myocardin. A component of a molecular switch for smooth muscle differentiation. J. Mol. Cell Cardiol. 34, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 10. Du K., Ip H. S., Li J., Chen M., Dandre F., Yu W., Lu M. M., Owens G. K., Parmacek M. S. (2003) Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell Biol. 23, 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshida T., Sinha S., Dandre F., Wamhoff B. R., Hoofnagle M. H., Kremer B. E., Wang D. Z., Olson E. N., Owens G. K. (2003) Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ. Res. 92, 856–864 [DOI] [PubMed] [Google Scholar]
- 12. Wang Z., Wang D. Z., Pipes G. C., Olson E. N. (2003) Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. U.S.A. 100, 7129–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li S., Wang D. Z., Richardson J. A., Olson E. N. (2003) The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. U.S.A. 100, 9366–9370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang J., Cheng L., Li J., Chen M., Zhou D., Lu M. M., Proweller A., Epstein J. A., Parmacek M. S. (2008) Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J. Clin. Invest. 118, 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang J., Lu M. M., Cheng L., Yuan L. J., Zhu X., Stout A. L., Chen M., Li J., Parmacek M. S. (2009) Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc. Natl. Acad. Sci. U.S.A. 106, 18734–18739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Long X., Bell R. D., Gerthoffer W. T., Zlokovic B. V., Miano J. M. (2008) Myocardin is sufficient for a smooth muscle-like contractile phenotype. Arterioscler. Thromb. Vasc. Biol. 28, 1505–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qiu P., Ritchie R. P., Fu Z., Cao D., Cumming J., Miano J. M., Wang D. Z., Li H. J., Li L. (2005) Myocardin enhances Smad3-mediated transforming growth factor-β1 signaling in a CArG box-independent manner. Smad-binding element is an important cis element for SM22α transcription in vivo. Circ. Res. 97, 983–991 [DOI] [PubMed] [Google Scholar]
- 19. Wang C., Cao D., Wang Q., Wang D. Z. (2011) Synergistic activation of cardiac genes by myocardin and Tbx5. PLoS One 6, e24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Q., Chen G., Streb J. W., Long X., Yang Y., Stoeckert C. J., Jr., Miano J. M. (2006) Defining the mammalian CArGome. Genome Res. 16, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Selvaraj A., Prywes R. (2004) Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Philippar U., Schratt G., Dieterich C., Müller J. M., Galgóczy P., Engel F. B., Keating M. T., Gertler F., Schüle R., Vingron M., Nordheim A. (2004) The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell 16, 867–880 [DOI] [PubMed] [Google Scholar]
- 23. Zhang S. X., Gras E. G., Wycuff D. R., Marriot S. J., Kadeer N., Yu W., Olson E. N., Garry D. J., Parmacek M. S., Schwartz R. J. (2005) Identification of direct serum-response factor gene targets during Me2SO-induced P19 cardiac cell differentiation. J. Biol. Chem. 280, 19115–19126 [DOI] [PubMed] [Google Scholar]
- 24. Balza R. O., Jr., Misra R. P. (2006) Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J. Biol. Chem. 281, 6498–6510 [DOI] [PubMed] [Google Scholar]
- 25. Valouev A., Johnson D. S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R. M., Sidow A. (2008) Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Meth. 5, 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cooper S. J., Trinklein N. D., Nguyen L., Myers R. M. (2007) Serum response factor binding sites differ in three human cell types. Genome Res. 17, 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benson C. C., Zhou Q., Long X., Miano J. M. (2011) Identifying functional single nucleotide polymorphisms in the human CArGome. Physiol. Genomics 43, 1038–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Conley C. A. (2001) Leiomodin and tropomodulin in smooth muscle. Am. J. Physiol. Cell Physiol. 280, C1645–C1656 [DOI] [PubMed] [Google Scholar]
- 30. Dong Q., Ludgate M., Vassart G. (1991) Cloning and sequencing of a novel 64-kDa autoantigen recognized by patients with autoimmune thyroid disease. J. Clin. Endocrinol. Metab. 72, 1375–1381 [DOI] [PubMed] [Google Scholar]
- 31. Conley C. A., Fritz-Six K. L., Almenar-Queralt A., Fowler V. M. (2001) Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics 73, 127–139 [DOI] [PubMed] [Google Scholar]
- 32. Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D. B., Rebowski G., Lappalainen P., Pollard T. D., Dominguez R. (2008) Leiomodin is an actin filament nucleator in muscle cells. Science 320, 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miano J. M., Ramanan N., Georger M. A., de Mesy-Bentley K. L., Emerson R. L., Balza R. O., Jr., Xiao Q., Weiler H., Ginty D. D., Misra R. P. (2004) Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. U.S.A. 101, 17132–17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wirth A., Benyo Z., Lukasova M., Leutgeb B., Wettschureck N., Gorbey S., Orsy P., Horvath B., Maser-Gluth C., Greiner E., Lemmer B., Schutz G., Gutkind J. S., Offermanns S. (2008) G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 14, 64–68 [DOI] [PubMed] [Google Scholar]
- 35. Rothman A., Kulik T. J., Taubman M. B., Berk B. C., Smith C. W., Nadal-Ginard B. (1992) Development and characterization of a cloned rat pulmonary arterial smooth muscle cell line that maintains differentiated properties through multiple subcultures. Circulation 86, 1977–1986 [DOI] [PubMed] [Google Scholar]
- 36. Long X., Tharp D. L., Georger M. A., Slivano O. J., Lee M. Y., Wamhoff B. R., Bowles D. K., Miano J. M. (2009) The smooth muscle cell-restricted KCNMB1 ion channel subunit is a direct transcriptional target of serum response factor and myocardin. J. Biol. Chem. 284, 33671–33682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Streb J. W., Miano J. M. (2005) AKAP12α, an atypical serum response factor-dependent target gene. J. Biol. Chem. 280, 4125–4134 [DOI] [PubMed] [Google Scholar]
- 38. Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., Pachter L. S., Dubchak I. (2000) VISTA. Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 [DOI] [PubMed] [Google Scholar]
- 39. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo. A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Graham F. L., Van der Eb A. J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52, 456–467 [DOI] [PubMed] [Google Scholar]
- 42. Miano J. M., Carlson M. J., Spencer J. A., Misra R. P. (2000) Serum response factor-dependent regulation of the smooth muscle calponin gene. J. Biol. Chem. 275, 9814–9822 [DOI] [PubMed] [Google Scholar]
- 43. Miano J. M., Olson E. N. (1996) Expression of the smooth muscle cell calponin gene marks the early cardiac and smooth muscle cell lineages during mouse embryogenesis. J. Biol. Chem. 271, 7095–7103 [DOI] [PubMed] [Google Scholar]
- 44. Li L., Miano J. M., Cserjesi P., Olson E. N. (1996) SM22α, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ. Res. 78, 188–195 [DOI] [PubMed] [Google Scholar]
- 45. Li L., Liu Z. C., Mercer B., Overbeek P., Olson E. N. (1997) Evidence for serum response factor-mediated regulatory networks governing SM22α transcription in smooth, skeletal, and cardiac muscle cells. Dev. Biol. 187, 311–321 [DOI] [PubMed] [Google Scholar]
- 46. Imamura M., Long X., Nanda V., Miano J. M. (2010) Expression and functional activity of four myocardin isoforms. Gene 464, 1–10 [DOI] [PubMed] [Google Scholar]
- 47. Qualmann B., Kessels M. M. (2009) New players in actin polymerization–WH2-domain-containing actin nucleators. Trends Cell Biol. 19, 276–285 [DOI] [PubMed] [Google Scholar]
- 48. Miano J. M., Long X., Fujiwara K. (2007) Serum response factor. Master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 292, C70–C81 [DOI] [PubMed] [Google Scholar]
- 49. Olson E. N., Nordheim A. (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schratt G., Philippar U., Berger J., Schwarz H., Heidenreich O., Nordheim A. J. Cell Biol. 156, 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Niu Z., Iyer D., Conway S. J., Martin J. F., Ivey K., Srivastava D., Nordheim A., Schwartz R. J. (2008) Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc. Natl. Acad. Sci. U.S.A. 105, 17824–17829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kostyukova A. S. (2008) Biochemistry 73, 1467–1472 [DOI] [PubMed] [Google Scholar]
- 53. Chu X., Chen J., Reedy M. C., Vera C., Sung K. L., Sung L. A. (2003) E-Tmod capping of actin filaments at the slow-growing end is required to establish mouse embryonic circulation. Am. J. Physiol. Heart Circ. Physiol. 284, H1827–H1838 [DOI] [PubMed] [Google Scholar]
- 54. McKeown C. R., Nowak R. B., Moyer J., Sussman M. A., Fowler V. M. (2008) Tropomodulin1 is required in the heart but not the yolk sac for mouse embryonic development. Circ. Res. 103, 1241–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kostyukova A. S. (2007) Leiomodin/tropomyosin interactions are isoform specific. Arch. Biochem. Biophys. 465, 227–230 [DOI] [PubMed] [Google Scholar]
- 56. Sussman M. A., Welch S., Cambon N., Klevitsky R., Hewett T. E., Price R., Witt S. A., Kimball T. R. (1998) Myofibril degeneration caused by tropomodulin overexpression leads to dilated cardiomyopathy in juvenile mice. J. Clin. Invest. 101, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.