Background: Clusterin and COMMD1 interact to down-regulate copper transporters ATP7A and ATP7B.
Results: Clusterin and COMMD1 act independently and under different conditions to target ATP7B degradation via different pathways.
Conclusion: Clusterin and COMMD1 regulate the quality control of ATP7A/ATP7B and directly impact copper homeostasis.
Significance: Clusterin and COMMD1 allelic variations may influence the clinical expression of Menkes and Wilson diseases.
Keywords: ATPases, Copper, Copper Transport, Molecular Chaperone, Protein Degradation, ATP7A, ATP7B, COMMD1, Clusterin, P-type ATPase
Abstract
ATP7A and ATP7B are copper-transporting P1B-type ATPases (Cu-ATPases) that are critical for regulating intracellular copper homeostasis. Mutations in the genes encoding ATP7A and ATP7B lead to copper deficiency and copper toxicity disorders, Menkes and Wilson diseases, respectively. Clusterin and COMMD1 were previously identified as interacting partners of these Cu-ATPases. In this study, we confirmed that clusterin and COMMD1 interact to down-regulate both ATP7A and ATP7B. Overexpression and knockdown of clusterin/COMMD1 decreased and increased, respectively, endogenous levels of ATP7A and ATP7B, consistent with a role in facilitating Cu-ATPase degradation. We demonstrate that whereas the clusterin/ATP7B interaction was enhanced by oxidative stress or mutation of ATP7B, the COMMD1/ATP7B interaction did not change under oxidative stress conditions, and only increased with ATP7B mutations that led to its misfolding. Clusterin and COMMD1 facilitated the degradation of ATP7B containing the same Wilson disease-causing C-terminal mutations via different degradation pathways, clusterin via the lysosomal pathway and COMMD1 via the proteasomal pathway. Furthermore, endogenous ATP7B existed in a complex with clusterin and COMMD1, but these interactions were neither competitive nor cooperative and occurred independently of each other. Together these data indicate that clusterin and COMMD1 represent alternative and independent systems regulating Cu-ATPase quality control, and consequently contributing to the maintenance of copper homeostasis.
Introduction
The copper-transporting P1B-type ATPases (Cu-ATPases),2 ATP7A and ATP7B, are key proteins responsible for regulating copper levels in mammals. They are large transmembrane proteins, with six N-terminal metal-binding domains (MBD). Copper is coordinated by the cysteine residues of the highly conserved GMXCXXC (where X is any amino acid) motif within each MBD (1). These proteins actively transport copper across cellular membranes using the energy derived from ATP hydrolysis (reviewed in Lutsenko et al. (2)). The Cu-ATPases predominantly reside at the trans-Golgi network (TGN) under basal conditions for metallation of copper-dependent enzymes of the secretory pathway. In response to elevated intracellular copper levels, they traffic to vesicles located near the basolateral (ATP7A) (3, 4) or apical (ATP7B) (5–7) membranes where they sequester and mediate export of the excess copper. When intracellular copper levels are restored, the Cu-ATPases recycle back to the TGN. Therefore, the copper status of the cell determines the steady-state localization of the Cu-ATPases (8).
Menkes (MD) and Wilson (WD) diseases are genetically inherited copper metabolism disorders (9) that result from mutation of ATP7A (OMIM 309400) and ATP7B (OMIM 277900), respectively. In MD, a fatal X-linked copper deficiency disorder, the intestinal enterocytes are the primary site of the defect. Defective copper transport across the basolateral membrane of these cells into the portal circulation leads to a systemic copper deficiency with devastating consequences (3, 4, 9, 10). Biochemical abnormalities lead to neurological and developmental defects among many other symptoms (9, 11). WD is an autosomal recessive copper toxicity disorder (9). The ATP7B protein is a key liver protein that regulates the copper status of the body, primarily by regulating biliary copper excretion (12). In WD, copper accumulates to toxic levels in the liver and in 50% of cases, in the brain. Thus, patients may present with either hepatic or neurological symptoms or both (13, 14).
Clusterin (apolipoprotein J) recently was shown to interact with both Cu-ATPases (15). The interaction between COMMD1 (copper metabolism MURR1 domain) and ATP7B has been the subject of other studies (16–18). Both proteins target ATP7B, among other proteins (19–26), for degradation and therefore appear to participate in regulating the quality control of ATP7B. Recently, it was demonstrated that COMMD1 also interacts with ATP7A but this interaction led to stabilization of misfolded mutant ATP7A proteins (27).
Clusterin is a 449-amino acid heterodimeric secreted glycoprotein that is ubiquitously expressed and present in most body fluids (28). Although widely recognized as an extracellular chaperone (26, 29), intracellular forms exist as a result of alternative splicing and are localized to the cytosol and nucleus (30). Clusterin has been implicated in pathological conditions in which oxidative stress plays a central role, such as neurodegenerative diseases and cancer progression, as well as aging (31, 32). COMMD1 is a 188-amino acid soluble protein that is expressed in most tissues, with the highest expression in the liver (33). COMMD1 is localized throughout the cytoplasm, in the nucleus, endosomes, and lysosomal compartments (34). The COMMD1 gene was found to be deleted in Bedlington terrier dogs that exhibit canine copper toxicosis, a chronic liver copper overload disease that resembles WD (33). However, more recent data has implicated COMMD1 in a variety of cellular processes and signaling pathways (35). The data in all cases, point to a general role of COMMD1 in controlling protein degradation and stability (35–37).
Clusterin and COMMD1 interact with a vast array of proteins. Clusterin possesses molten globular-like structures that contain putative amphipathic α-helices (38). This structure allows clusterin to interact with the hydrophobic regions of many proteins exposed following stress (24). Hence, clusterin has been implicated in a diverse range of cellular processes including lipid transport, cell differentiation, regulation of apoptosis, and clearance of cellular debris. Clusterin binding to proteins either stabilizes them or facilitates their degradation and this underpins many of the cellular roles with which clusterin has been associated (31, 39). Clusterin functions similarly to the small heat shock proteins with chaperone-like activity, binding to the hydrophobic regions of stressed and misfolded proteins and maintaining them in a state competent for subsequent re-folding by other chaperones (24).
COMMD1 is a member of the COMMD (copper metabolism Murr1 domain) family of proteins that share a highly conserved C-terminal COMM domain (35). All of the COMMD proteins interact with each other via the COMM domains. The N-terminal regions are variable. Due to the instability and aggregation of COMMD1, only the solution structure of the N-terminal domain has been determined (40, 41). This region adopts a novel α-helical fold with two positively charged regions at the surface that may provide sites for interaction with partner proteins (40, 41). A recent study identified specific interactions between COMMD1 and phosphatidylinositols (PtdIns(4,5)P2, PtdIns(4)P, and phosphatidic acid), which may explain the localization of COMMD1 to cell membranes of specific intracellular compartments, and which stabilize large oligomeric complexes formed by COMMD1 (40). Modeling studies and proteolysis data verified the two-domain structure of COMMD1 (40).
We sought to consolidate, compare, and contrast the roles of clusterin and COMMD1 in regulating the quality control of the Cu-ATPases. We confirmed that endogenously expressed ATP7A and ATP7B interact with both COMMD1 and clusterin. However, in contrast to the recent study by Vonk et al. (27), which showed that COMMD1 binding increased the levels of mutant ATP7A proteins, we found that COMMD1 overexpression led to down-regulation of ATP7A and therefore, as for ATP7B, was likely to also facilitate its degradation. This study provides important new insights into the roles and action of clusterin and COMMD1 in regulating the degradation of the Cu-ATPases. Clusterin and COMMD1 interaction with ATP7B occurred independently of each other although the three proteins co-existed in a complex. Although the conditions that promoted clusterin and COMMD1 interaction with the Cu-ATPases were different, and different pathways were targeted, the eventual consequence of these interactions were to facilitate the degradation of misfolded Cu-ATPase molecules. This is important in mediating ATP7A and ATP7B quality control, which in turn is required for the maintenance of normal copper homeostasis and in ensuring cell survival in the context of disease.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Plasmid cDNA constructs encoding wild type (WT) ATP7B, ATP7B (MBD1–6del), and ATP7B (MBD3–5del) were generated previously (42), as were plasmid constructs encoding WT-ATP7A, ATP7Am4–6, and ATP7Am1–6 (43). Clusterin cDNA in pIREShyg1 (Clontech) was kindly provided by Saverio Bettuzzi (University of Parma, Italy). COMMD1 cDNA in pEBB-FLAG was previously described (19), as was the plasmid encoding the COMMD1 shRNA (17) and constructs encoding ATP7B-4193del and Atp7b-tx (6, 44).
The following pair of oligonucleotides encoding clusterin-targeted hairpin siRNAs were annealed and cloned into the pSilencer 4.1-CMV puro vector (Ambion). The single-stranded overhangs with BamHI and HindIII restriction enzyme sites are shown in italics; clusterin-targeted sequences are shown in bold: clusterin (forward) 5′-GATCCCCAGAGCTCGCCCTTCTACTTCAAGAGAGTAGAAGGGCGAGCTCTGGTTA-3′, clusterin (reverse) 5′-AGCTTAACCAGAGCTCGCCCTTCTACTCTCTTGAAGTAGAAGGGCGAGCTCTGGG-3′.A hairpin siRNA with limited homology to known sequences in the human, mouse, and rat genomes, cloned into the pSilencer 4.1-CMV puro vector was used alone as a negative control for off-target effects.
Cell Culture and Transfection
Chinese hamster ovary (CHO-K1) cells were cultured at 37 °C as monolayers in Eagle's basal medium (Trace Biosciences) supplemented with 0.2 mm proline, 10% (v/v) fetal calf serum (FCS), 2 mm l-glutamine, 1.2 mm sodium bicarbonate, and 20 mm HEPES. CHO-K1 cells stably transfected with mammalian expression constructs encoding WT or mutant ATP7A/ATP7B were cultured in medium supplemented with 500 μg/ml of G418 (Invitrogen) to maintain transgene expression. Human embryonic kidney 293T cells (HEK293T) were cultured in Dulbecco's modified Eagle's medium (DMEM) (high bicarbonate) (Trace Biosciences) supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, 200 μg/ml of penicillin, and 200 μg/ml of streptomycin. These cells were used for detection and/or co-immunoprecipitation of endogenously or transiently expressed proteins. Human neuroblastoma M17 cells were cultured at 37 °C in Opti-MEM® Reduced Serum Medium (Invitrogen) supplemented with 10% (v/v) FCS, 20 mm HEPES, 28 mm sodium bicarbonate, and 2 mm l-glutamine. Where indicated, the growth medium was supplemented with 200 μm CuCl2, 200 μm bathocuproinedisulfonic acid (Sigma), 200 μm d-penicillamine (Sigma), 2 mm hydrogen peroxide (H2O2) (Sigma), 10 mm diamide (Sigma), 53 μm cycloheximide (CHX) (Sigma), 5 μm MG132 (Merck), 5 μm NH4Cl or 100 μm leupeptin (Sigma), prior to harvesting cells. HEK293T cells were transfected by calcium phosphate precipitation as previously described (45) or using FuGENE® HD (Roche Diagnostics) according to the manufacturer's protocol. The generation of stably transfected CHO-K1 cell lines (WT-ATP7B, MBD1–6del, MBD3–5del, wt-ATP7A, ATP7Am4–6, and ATP7Am1–6) were described previously (42, 43). In this study, “stably” and “transiently” expressed proteins refer to proteins expressed from integrated and nonintegrated plasmids, respectively.
Western Blotting and Immunoprecipitation
To prepare protein samples, cells were washed in 2 ml of phosphate-buffered saline, lysed with buffer containing 50 mm HEPES, pH 7.5, 0.5% (w/v) Nonidet P-40, 0.5% (w/v) Triton X-100, 50 mm sodium chloride, 10% (w/v) glycerol, 1 mm dithiothreitol, 1 mm EDTA, and one half of a complete mini EDTA-free protease inhibitor tablet (Roche Diagnostics) and scraped from the bottom of the flask using a rubber cell scraper (BD Biosciences). The cell suspension was transferred to a 1.7-ml Eppendorf tube on ice for 5 min. The lysate was centrifuged at 10,000 × g for 10 min at 4 °C to pellet the cell debris and the supernatant was transferred into a new tube.
For co-immunoprecipitations, protein G-coupled magnetic beads (Dynabeads Protein G, Invitrogen) were used according to the recommended protocols. Briefly, ∼3 mg of total cell protein was applied to Dynabeads prepared with immobilized antibody, sheep anti-ATP7B (NC36, ammonium sulfate precipitated) (42) (∼50 μg), sheep anti-ATP7A (R17-BX, affinity purified) (46) (∼3 μg), rabbit anti-COMMD1 (34) (∼3 μg), or preimmune serum. Protein complexes were eluted with 1 m glycine, pH 2.0, neutralized in 1 m Tris-HCl, pH 8.0, and analyzed by immunoblotting together with ∼50 μg of total protein extracts. For sequential co-immunoprecipitation, ∼3 mg of total cell protein was applied to Dynabeads prepared with sheep anti-ATP7B (NC36) antibody (∼50 μg) or preimmune serum for the negative control. Protein complexes were eluted with 1 m glycine, pH 2.0, and neutralized in 1 m Tris-HCl, pH 8.0. This eluate was applied to Dynabeads prepared with rabbit anti-COMMD1 antibody (∼3 μg) or preimmune serum for the negative control. Protein complexes were eluted as above and analyzed by immunoblotting together with ∼50 μg of total protein extracts. In all experiments, ∼4% of the total protein extract is represented in lanes labeled “lysate,” whereas the proteins evident in the lanes labeled “IP” are derived from ∼46% of the total protein extract.
For SDS-PAGE, protein samples were prepared with 4× loading dye (10% (w/v) glycerol, 20% (w/v) SDS, 1.5 m Tris-HCl, pH 6.8, 1.8 m bromphenol blue, dH2O) and 10% (v/v) 2-mercaptoethanol (Sigma) in a 4:1:1 ratio of sample:10% (v/v) SDS:loading dye, respectively. Samples were fractionated by SDS-PAGE (6–15% (v/v) acrylamide). Proteins were transferred to nitrocellulose membranes (GE Healthcare) using the XCellIITM Blot Module and the Xcell SurelockTM Mini-cell system (Invitrogen). The membranes were blocked with Tris-buffered saline (10 mm Tris, pH 8.0, and 150 mm NaCl) containing 5% (w/v) skim milk powder (Diploma) for 1 h at room temperature. The membranes were immunoblotted with the following antibodies; sheep anti-ATP7B (42) (diluted 1:1,000), rabbit anti-(mouse)Atp7b (44) (diluted 1:1,000), sheep anti-ATP7A (46) (diluted 1:1,000), goat anti-clusterin (diluted 1:1,000) (Sigma or Santa Cruz), or rabbit anti-COMMD1 (34) (diluted 1:1,000). For protein loading controls either mouse monoclonal anti-β-actin (diluted 1:5,000) (Sigma) or rabbit anti-β-tubulin antibodies (diluted 1:5,000) (Abcam) were used. Secondary antibodies included horseradish peroxidase (HRP)-conjugated sheep anti-rabbit IgG (Millipore), rabbit anti-goat IgG (Sigma), and sheep anti-mouse IgG (Dako) (diluted 1:10,000). Proteins were detected using Immobilon Western HRP Substrate Peroxide Solution (Millipore) and images were captured using the Luminescent Image Analyzer LAS-3000 (Fujifilm).
Statistical Analysis
Unless otherwise indicated, data are reported as mean ± S.E., where n = at least 3. Statistical analysis was carried out using Student's t test.
RESULTS
ATP7B, Clusterin, and COMMD1 Exist in a Complex
Co-immunoprecipitation experiments using HEK293T cells (Fig. 1, A and B) verified that endogenous ATP7B interacts with both clusterin and COMMD1 in mammalian cells as previously reported (15–17). The anti-ATP7B antibody immunoprecipitated ATP7B, clusterin, and COMMD1, whereas preimmune serum did not precipitate these proteins (Fig. 1, A and B). Endogenous ATP7A also interacted with COMMD1 in HEK293T cells (Fig. 1C) and in M17 neuroblastoma cells (data not shown). Previous studies demonstrated that endogenously expressed clusterin and COMMD1 interacted in the PC-3 (human prostate cancer) cell line (23). We verified that this interaction also occurs with the endogenously expressed proteins in HEK293T cells (Fig. 1D). The anti-COMMD1 antibody (34) immunoprecipitated both COMMD1 and clusterin, whereas preimmune serum did not precipitate either protein (Fig. 1D).
FIGURE 1.
ATP7B, clusterin, and COMMD1 exist in a complex together. Co-immunoprecipitation of endogenous (A) ATP7B and clusterin, (B) ATP7B and COMMD1, (C) ATP7A and COMMD1, and (D) clusterin and COMMD1 from HEK293T cells. Dynabeads (Protein G) were bound with the antibody as indicated (IP Ab). Proteins were co-immunoprecipitated (IP) from whole cell lysates and then fractionated and immunoblotted with anti-ATP7A, anti-ATP7B, anti-clusterin, and anti-COMMD1 antibodies (WB Ab). Preimmune serum was used as a negative control. E, sequential co-immunoprecipitation of ATP7B, clusterin, and COMMD1. Dynabeads bound with anti-ATP7B antibody (IP Ab) were used to co-immunoprecipitate ATP7B and any interacting proteins from HEK293T cell lysates. Once eluted, the protein samples were re-precipitated with the anti-COMMD1 antibody (IP Ab). Proteins were again eluted, then fractionated and immunoblotted with anti-ATP7B, anti-clusterin, and anti-COMMD1 antibodies (WB Ab). Preimmune serum was used as a negative control. WB, Western blot.
To explore the possibility that ATP7B, clusterin, and COMMD1 exist in a complex, sequential co-immunoprecipitation experiments were conducted in HEK293T cells (Fig. 1E). We first immunoprecipitated ATP7B from HEK293T cell lysates using Dynabeads bound with the anti-ATP7B antibody. Immunoprecipitated proteins were eluted without denaturing and subsequently re-precipitated with the anti-COMMD1 antibody. Immunoprecipitated proteins were then fractionated and immunoblotted with anti-ATP7B, anti-clusterin, and anti-COMMD1 antibodies, which showed that ATP7B, clusterin, and COMMD1 could all be detected (Fig. 1E). This method was used previously to identify proteins that exist in complexes (19). This data suggests that the binding of both clusterin and COMMD1 to ATP7B is not mutually exclusive and the three proteins are able to co-exist in a complex.
Oxidative Stress Does Not Influence the Interaction between ATP7B and COMMD1
Previous observations revealed that oxidative stress increased the interaction between clusterin and ATP7B (15), and that both clusterin and COMMD1 act to facilitate the degradation of ATP7B (15, 17). To determine whether the interaction between ATP7B and COMMD1 also can be modulated by oxidative stress, HEK293T cells were treated with CuCl2 (200 μm), bathocuproinedisulfonic acid/d-penicillamine (200 μm), diamide (10 mm), or H2O2 (2 mm). We previously showed that these conditions effectively increase oxidative stress levels in HEK293T cells by demonstrating an increase in the levels of hemoxygenase-1, a commonly used marker of oxidative stress (15). Co-immunoprecipitation experiments were carried out using the anti-ATP7B antibody (Fig. 2A). The anti-ATP7B antibody immunoprecipitated ATP7B and COMMD1 as expected. However, there was no difference in the amount of COMMD1 that co-precipitated when cells were exposed to copper or the other oxidants, or were depleted of copper, compared with untreated cells. Similar results were obtained when the anti-ATP7A antibody was used to co-immunoprecipitate ATP7A and COMMD1 under the same oxidative stress-inducing conditions (Fig. 2B). These results are in contrast to our previous data, which demonstrated that clusterin interacted to a greater extent with ATP7A and ATP7B when cells were treated with the same oxidizing agents (15). The data presented here suggests that COMMD1 interaction with the Cu-ATPases occurs in response to different cellular signals compared with clusterin.
FIGURE 2.
Oxidative stress has no affect on the interaction between the Cu-ATPases and COMMD1. Co-immunoprecipitation of endogenous (A, i) ATP7B and COMMD1 and (B, i) ATP7A and COMMD1 from HEK293T cells that were untreated or treated with CuCl2 (200 μm, 24 h), bathocuproinedisulfonic acid (BCS)/d-penicillamine (D-PEN) (200 μm 72 h), diamide (10 mm, 15 min), or H2O2 (2 mm, 20 min). Cell lysates and co-immunoprecipitated (IP) proteins were fractionated and immunoblotted with anti-ATP7B, anti-ATP7A, anti-COMMD1, or anti-β-actin antibodies (WB Ab). A (ii) and B (ii) densitometric analysis of co-precipitated COMMD1 relative to ATP7B or ATP7A, respectively, expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different, *, p < 0.05. WB, Western blot.
COMMD1 Binds to Misfolded ATP7B Protein
Our data above showed that the ATP7B and COMMD1 interaction is not influenced by oxidative stress. Previously de Bie et al. (17) showed that more COMMD1 interacted with the ATP7B N terminus that contained several WD-causing mutations than with the WT N terminus. We sought to determine: 1) if COMMD1 interacts with regions of ATP7B outside of the N terminus, and 2) if the interaction is enhanced by mutations that affect the misfolding of ATP7B in the context of the full-length protein. ATP7B variants truncated at the N terminus were previously generated and shown to localize to the endoplasmic reticulum of the cell, indicative of unstable proteins (15, 42) (Fig. 3A). Retention in the endoplasmic reticulum is common for unstable ATP7B proteins, which can often regain function and correct localization at the TGN when cultured at 30 °C (6, 42, 47, 48). Previous immunoblot analysis of CHO-K1 cells stably expressing two different truncated ATP7B proteins in which MBDs 1–6 (MBD1–6del) or 3–5 (MBD3–5del) were deleted (Fig. 3A), verified that these truncations caused temperature-sensitive instability (15).
FIGURE 3.
Increased binding of COMMD1 to unstable ATP7B mutants MBD1–6del and MBD3–5del. A, schematic representation of the N terminus of WT-ATP7B and truncated mutants MBD1–6del and MBD3–5del. B, co-immunoprecipitation (IP) of endogenous COMMD1 and stably expressed WT-ATP7B, MBD1–6del, or MBD3–5del from CHO-K1 cells cultured at either 37 or 30 °C. Cell lysates and co-immunoprecipitated proteins were fractionated and immunoblotted with anti-ATP7B, anti-COMMD1, and anti-β-actin antibodies (WB Ab). Densitometric analysis of co-immunoprecipitated COMMD1 relative to immunoprecipitated ATP7B is shown, expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different, *, p < 0.05. WB, Western blot.
To establish if ATP7B instability affected its interaction with COMMD1, we investigated COMMD1 binding to these truncated ATP7B mutants after cells were cultured at either 30 or 37 °C (Fig. 3B). The levels of WT-ATP7B and the two mutants increased at 30 °C, when compared with their levels at 37 °C. In addition, the amount of COMMD1 that co-precipitated with both ATP7B variants was markedly increased at 37 °C, when the proteins were unstable, compared with the amount that co-precipitated at 30 °C when the proteins were more stable. This data shows that more COMMD1 binds to unstable, misfolded ATP7B proteins than to the stable proteins. Importantly, we show for the first time that COMMD1 interaction with ATP7B is not restricted to the ATP7B N terminus as originally reported (16), but also recognizes sites elsewhere in the molecule. Like clusterin, it may recognize some common feature(s) of misfolded proteins.
COMMD1 and Clusterin Interact Independently with ATP7B
To determine whether the clusterin and COMMD1 interaction with ATP7B was competitive or cooperative, co-immunoprecipitation experiments with endogenous ATP7B were carried out following manipulation of either clusterin (Fig. 4, A and B) or COMMD1 levels (Fig. 4C). HEK293T cells were transfected with plasmids containing either the full-length clusterin cDNA (Fig. 4A) or clusterin shRNA (Fig. 4B), then ATP7B was immunoprecipitated using the anti-ATP7B antibody as described above. Similar levels of COMMD1 co-precipitated with ATP7B in both experiments, indicating that changes in clusterin levels did not affect the extent of the interaction between ATP7B and COMMD1 (Fig. 4, A and B). Similarly, when COMMD1 levels were altered by overexpression or knockdown, the amount of clusterin that co-precipitated with ATP7B was not appreciably altered (Fig. 4C). These findings suggested that clusterin and COMMD1 interact with ATP7B independently of each other and that the interactions are neither competitive nor cooperative. These results support the earlier observation that the three proteins can co-exist in a complex (Fig. 1E).
FIGURE 4.
COMMD1 and clusterin interact independently with ATP7B. Co-immunoprecipitation (IP) of endogenous ATP7B and COMMD1 from HEK293T cells transiently transfected with a clusterin cDNA construct (A, i) and a clusterin shRNA plasmid construct (B, i). Cell lysates and co-immunoprecipitated proteins were fractionated and immunoblotted with anti-ATP7B, anti-clusterin, anti-COMMD1, and anti-β-actin antibodies (WB Ab). Preimmune serum was used as a negative control. C, (i) co-immunoprecipitation of endogenous ATP7B and clusterin from HEK293T cells transiently transfected with either a COMMD1 shRNA plasmid construct or a COMMD1-FLAG cDNA construct. Cell lysates and co-immunoprecipitated proteins were fractionated and immunoblotted with anti-ATP7B, anti-clusterin, anti-COMMD1, and anti-β-tubulin antibodies (WB Ab). Preimmune serum was used as a negative control. Densitometric analysis of co-immunoprecipitated COMMD1 ((A, ii) and (B, ii)) and clusterin (C, ii) relative to immunoprecipitated ATP7B is shown, expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different, *, p < 0.05. WB, Western blot.
Alteration of Clusterin and COMMD1 Levels Affect Cu-ATPase Levels
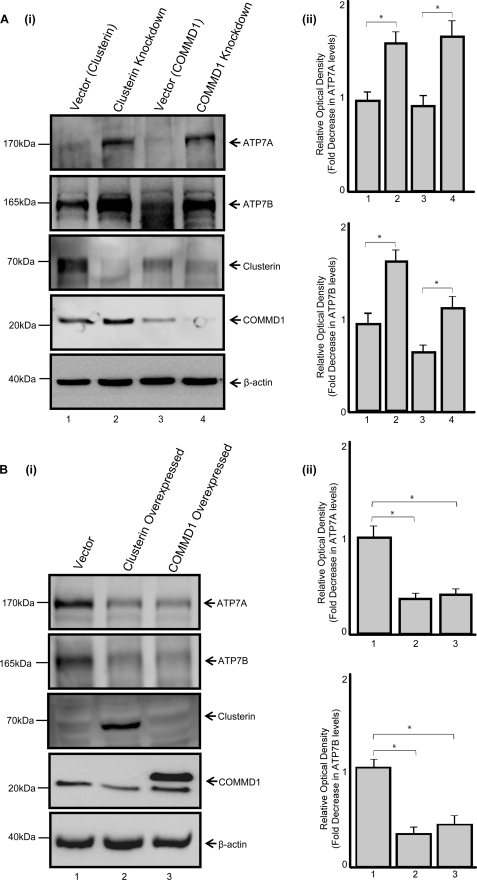
We sought to further demonstrate that clusterin and COMMD1 acted similarly but independently to regulate ATP7A and ATP7B levels. The effect of altering clusterin or COMMD1 levels on the abundance of endogenous ATP7A and ATP7B in the cell were investigated. HEK293T cells were transfected with plasmids encoding either clusterin or COMMD1 shRNA, to knockdown clusterin or COMMD1, respectively (Fig. 5A). When the levels of either of these proteins were reduced, the amount of ATP7A and ATP7B proteins was significantly increased compared with the empty vector control (Fig. 5A). Conversely, when either clusterin or COMMD1 were exogenously overexpressed, the levels of ATP7A and ATP7B were significantly decreased (Fig. 5B).
FIGURE 5.
Clusterin and COMMD1 affect the levels of endogenous ATP7A and ATP7B. A, (i) HEK293T cells were transiently transfected with an empty vector or plasmids encoding clusterin or COMMD1 shRNAs. Proteins were fractionated and immunoblotted with anti-ATP7A, anti-ATP7B, anti-clusterin, anti-COMMD1, and anti-β-actin antibodies. B, (i) HEK293T cells were transiently transfected with an empty vector or either clusterin or COMMD1 cDNA constructs. Proteins were fractionated and immunoblotted with anti-ATP7A, anti-ATP7B, anti-clusterin, anti-COMMD1, and anti-β-actin antibodies. A (ii) and B (ii), densitometric analysis of ATP7A and ATP7B levels, expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different, *, p < 0.05. WB, Western blot.
In addition, both clusterin knockdown and overexpression affected the levels of COMMD1. Clusterin knockdown increased COMMD1 levels (Fig. 5A), whereas clusterin overexpression decreased COMMD1 levels (Fig. 5B). This result is consistent with a previous report, which demonstrated that clusterin targets COMMD1 for degradation (23). Our data showed that clusterin and COMMD1 act similarly in modulating ATP7A and ATP7B protein levels.
Role of COMMD1 in Regulating the Stability of ATP7A
The data presented above demonstrated that overexpression of clusterin and COMMD1 decreased endogenous ATP7A levels in HEK293T cells. This result was in contrast to a recent study by Vonk et al. (27), which demonstrated that COMMD1 overexpression stabilized WT and mutant ATP7A proteins that were transiently expressed in HEK293T cells. We sought to clarify the reason for this discrepancy. Full-length ATP7A variants with CXXC to SXXS mutations in MBD1–6 (ATP7Am1–6) and MBD4–6 (ATP7Am4–6) (Fig. 6A) were previously generated (43). These variants fail to traffic with elevated copper levels, and remain at the TGN (43). To confirm the data by Vonk et al. (27), HEK293T cells were transiently transfected with plasmids encoding WT-ATP7A or ATP7Am4–6, and either COMMD1 or an empty vector control plasmid (Fig. 6B). The levels of both WT-ATP7A and ATP7Am4–6 were increased with the addition of COMMD1 when compared with the empty vector control. This result was consistent with the observations made by Vonk et al. (27) that COMMD1 can stabilize ATP7A proteins that have been transiently overexpressed.
FIGURE 6.
COMMD1 plays a role in determining the stability of ATP7A. A, schematic representation of the N terminus of WT-ATP7A, ATP7Am4–6, and ATP7Am1–6. B (i), HEK293T cells were transiently transfected with the empty vector or cDNA constructs encoding WT-ATP7A, WT-ATP7A, and COMMD1-FLAG, or ATP7Am4–6 and COMMD1-FLAG for 48 h. Proteins were fractionated and immunoblotted with anti-ATP7A, anti-COMMD1, and anti-β-actin antibodies. C, CHO-K1 cells stably expressing WT-ATP7A, ATP7Am4–6, and ATP7Am1–6 were transiently transfected with the empty vector or cDNA constructs encoding COMMD1-FLAG for 48 h. Proteins were fractionated and immunoblotted with anti-ATP7A, anti-COMMD1, and anti-β-actin antibodies. D (i), co-immunoprecipitation of endogenous COMMD1 and either WT-ATP7A, ATP7Am4–6, or ATP7Am1–6 stably expressed in CHO-K1 cells. Cell lysates and co-immunoprecipitated proteins were fractionated and immunoblotted with anti-ATP7A, anti-COMMD1, and anti-β-actin antibodies. Preimmune serum was used as a negative control. B (ii) and D (ii), densitometric analysis of ATP7A levels or co-immunoprecipitated COMMD1 levels relative to ATP7A, respectively, expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different, *, p < 0.05. WB, Western blot.
To expand on this work and to further explore the role of COMMD1 in ATP7A stability, we conducted a similar experiment to investigate the effect of COMMD1 protein levels on ATP7A abundance using CHO-K1 cells that stably expressed WT-ATP7A, ATP7Am4–6, or ATP7Am1–6. A plasmid construct that encoded COMMD1 or an empty vector control plasmid was transiently transfected into these cells before proteins were analyzed by immunoblotting (Fig. 6C). The levels of stably expressed WT and mutant ATP7A protein decreased with the overexpression of COMMD1, similar to our observations with endogenous WT-ATP7A (Fig. 5B). Furthermore, the decrease in the amount of ATP7Am4–6 and ATP7Am1–6 was greater than that of WT-ATP7A, suggesting that COMMD1 has a more substantial effect on mutant ATP7A protein than on WT-ATP7A (Fig. 6C). We concluded that COMMD1 facilitates degradation of the ATP7A protein that is endogenously or stably expressed, whereas it acts to apparently stabilize transiently expressed ATP7A.
To establish if mutations in ATP7A affected its interaction with COMMD1, co-immunoprecipitation of stably expressed WT or mutant ATP7A with endogenous COMMD1 was carried out (Fig. 6D). Although the levels and amount of immunoprecipitated WT-ATP7A were significantly higher than that of both ATP7Am4–6 and ATP7Am1–6, the amount of COMMD1 that co-precipitated with WT-ATP7A, ATP7Am4–6, and ATP7Am1–6 were very similar. This result demonstrated that the ratio of COMMD1:ATP7A in the immunoprecipitates was significantly higher for the mutant ATP7A than for WT-ATP7A. This data were consistent with that shown in Fig. 6C and suggests that COMMD1 binds to a greater extent to mutant ATP7A than to the WT-ATP7A, potentially facilitating degradation of the mutant proteins. A further conclusion is that the apparent mode of action of COMMD1 depends on whether its target proteins are transiently or stably expressed.
Clusterin and COMMD1 Facilitate ATP7B Degradation by Different Pathways
To date, the investigations of clusterin and COMMD1 in the context of their role in copper homeostasis and ATP7B function have focused on their interaction with the ATP7B N terminus. We have demonstrated in this study that COMMD1 also interacts with ATP7B beyond the N terminus. We therefore investigated the effect of clusterin and COMMD1 on ATP7B containing WD-causing mutations that reside within the C-terminal half of ATP7B.
The toxic milk (tx) mouse is a mouse model of WD caused by an autosomal recessive point mutation in the murine ATP7B gene, leading to a methionine to valine change in the eighth transmembrane domain of ATP7B (49). This mutation abrogated the copper-induced trafficking of ATP7B in cells and conferred defective copper transport activity, which explained the hepatic copper accumulation in the tx mice (44, 50). The ATP7B-4193del human WD patient mutation results from a base pair deletion at nucleotide position 4193, which causes a frameshift, with the addition of 5 new amino acids before a premature stop codon (51). This mutation results in the truncation of the last 61 amino acids of the protein, which includes the trileucine sequence (51) that is important for localization and recycling of ATP7B (6). Mislocalization of the mutant protein to the endoplasmic reticulum is consistent with instability due to misfolding, as has been observed with many disease causing mutations in ATP7B (6, 47, 48).
CHO-K1 cells that stably expressed either Atp7b-tx (Fig. 7, A and B) or the ATP7B-4193del mutation (Fig. 7, C and D) were transiently transfected to overexpress clusterin (lanes 1–3) or COMMD1 (lanes 4–6). To compare the role of clusterin and COMMD1 in determining which pathway was primarily responsible for degradation of these mutant ATP7B proteins, the levels of the mutant ATP7B were assessed in the presence of CHX and either the proteasomal inhibitor MG132 (Fig. 7, A and C) or the lysosomal inhibitors ammonium chloride (NH4Cl) and leupeptin (Fig. 7, B and D).
FIGURE 7.
Clusterin and COMMD1 target ATP7B for degradation via different pathways. CHO-K1 cells stably expressing (A and B) Atp7b-tx or (C and D) ATP7B-4193del were co-transfected with cDNA constructs encoding either clusterin or COMMD1-FLAG for transient expression. Cells were incubated with CXH (53 μm, 8 h) in the absence and presence of (A and C) MG132 (5 μm, 8 h) (B and D) NH4Cl (5 μm, 20 min) or leupeptin (100 μm, 8 h). Cell lysates were fractionated and immunoblotted with anti-ATP7B, anti-clusterin, anti-FLAG, and anti-β-actin antibodies. A (ii), B (ii), C (ii), and D (ii), densitometric analysis of ATP7B levels, expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different, *, p < 0.05.
Overexpression of both clusterin and COMMD1 in the presence of CHX led to a significant decrease in the levels of both ATP7B mutant proteins. ATP7B levels could be rescued by MG132 only when COMMD1 was overexpressed (Fig. 7, A and C, compare lanes 4 and 6). The combination of clusterin overexpression and MG132 had no effect in rescuing ATP7B levels (Fig. 7, A and C, compare lanes 1 and 3). In contrast, NH4Cl could only rescue ATP7B levels when clusterin was overexpressed (Fig. 7, B and D, compare lanes 1 and 3) and had no effect when COMMD1 was overexpressed (Fig. 7B, i and ii, and D, compare lanes 4 and 6). Similar observations were made using an additional lysosomal inhibitor, leupeptin (Fig. 7B, iii).
COMMD1 levels were markedly decreased with the addition of CHX when compared with clusterin levels (Fig. 7, A–D, compare lanes 2 and 5). This observation is most likely due to the ∼1 h half-life of COMMD1 (52), compared with the half-life of ∼2 h for clusterin (53). In addition, COMMD1 levels only seemed to increase following the addition of MG132 (Fig. 7, A and C, lane 6) and not with NH4Cl (Fig. 7, B and D, lane 6). This observation was consistent with a previous study that showed that COMMD1 was degraded via the proteasomal and not by the lysosomal pathway (54).
We concluded that both clusterin and COMMD1 can facilitate the degradation of the same mutant ATP7B molecules derived from WD-causing mutations that reside outside of the ATP7B N terminus. Consistent with previous studies (15, 17), clusterin appears to facilitate degradation via the lysosomal pathway and COMMD1 via the proteasomal pathway.
DISCUSSION
The key findings of this study are as follows: (i) COMMD1, like clusterin, interacts with both ATP7A and ATP7B to down-regulate their levels; (ii) this down-regulation is mediated by facilitating the degradation of mutant, misfolded proteins as demonstrated for ATP7B; (iii) clusterin and COMMD1 interact independently with ATP7B (can co-exist in a complex), under different conditions (mutation versus oxidative stress), and facilitate its degradation by different pathways (lysosomal versus proteosomal, respectively).
The COMMD1 gene deletion was originally identified as the causative mutation in Bedlington terrier dogs with canine copper toxicosis (33). This, together with its interaction with the ATP7B N terminus, subcellular localization pattern, and the canine copper toxicosis phenotype, led to the suggestion of a role for COMMD1 in regulating ATP7B-mediated biliary copper excretion (16, 35). More recently, COMMD1 was shown to bind more strongly to the ATP7B N terminus, which harbored certain WD-causing mutations that rendered the protein less stable than WT-ATP7B (17). In addition, COMMD1 facilitated the degradation of ATP7B harboring an N-terminal WD-causing mutation (G85V) via the proteasomal pathway (17).
To date, most studies that have characterized COMMD1 function, in the context of ATP7B function and copper homeostasis, have focused on the interaction between COMMD1 and the ATP7B N terminus (16–18). In this study, we provide further insight and clarify several important aspects of COMMD1 function and mechanism of action. We demonstrated that COMMD1 interacts with ATP7B lacking the N terminus and therefore that this interaction is not restricted to the ATP7B N terminus as originally reported (16). In the study by Tao et al. (16), which mapped the ATP7B/COMMD1 interaction to the ATP7B N terminus, all constructs used retained the N terminus. Furthermore, we showed that both COMMD1 and clusterin could target ATP7B with C-terminal mutations for degradation. Importantly, we verified that COMMD1 interacts also with ATP7A, and therefore is not likely to be limited to regulating hepatic copper excretion, but may have a more general role in regulating copper homeostasis and protein stability.
We demonstrated previously that clusterin interacted with both Cu-ATPases and that these interactions were enhanced by misfolding induced by oxidative stress or mutation (15). In the current study, we showed that clusterin and COMMD1 differ in the conditions that promote their interaction with ATP7B. COMMD1 interaction with the same mutant ATP7B proteins was enhanced compared with the WT protein, but was not affected by oxidative stress mediated by copper or other oxidizing agents. These observations suggest that oxidative stress and mutation confer differential recognition of the Cu-ATPases by proteins such as clusterin and COMMD1. The different structural properties of clusterin and COMMD1 may influence this differential recognition.
COMMD1 and clusterin clearly have a role in down-regulating Cu-ATPase levels by facilitating their degradation. Overexpression of either protein led to decreased, whereas knockdown led to increased Cu-ATPase levels. Moreover, COMMD1 acted in a similar manner to clusterin displaying enhanced binding to unstable ATP7B N-terminal-truncated mutant proteins. Both clusterin and COMMD1 also facilitated the degradation of ATP7B harboring C-terminal WD-causing mutations (Atp7b-tx and ATP7B-4193del) via the lysosomal and proteasomal pathways, respectively, as previously shown for ATP7B with an N-terminal WD patient mutation (G85V) (15, 17). Although clusterin and COMMD1 acted similarly in the context of binding to ATP7B and its misfolded variants, they appeared to act independently of each other. Their binding to ATP7B was neither competitive nor cooperative; overexpression or knockdown of either protein had little or no effect on the interaction of the other with ATP7B. Furthermore, despite functioning independently to target ATP7B for degradation, they co-existed in a complex together. We concluded that clusterin and COMMD1 represent independent, potentially redundant systems mediating the quality control of the Cu-ATPases.
In a recent study using a mouse hepatoma cell line, COMMD1 knockdown led to decreased ATP7B levels and impaired the recruitment of ATP7B from intracellular vesicles to the TGN following the return of copper levels to normal (55). In addition, Vonk et al. (27) recently reported the interaction of COMMD1 with ATP7A and MD-causing ATP7A mutants expressed in HEK293T cells, consistent with our findings. However, the latter study also showed that increased COMMD1 levels enhanced ATP7A levels and improved the subcellular localization and copper-exporting ability of the ATP7A mutants, whereas COMMD1 depletion led to reduced ATP7A levels (27). The data from these two studies is in direct contrast to the results we report in the present study. The one main difference between the present study and that of Vonk et al. (27) is that Vonk and colleagues used a transient expression system for ATP7A, whereas we focused on endogenous and stably expressed ATP7A proteins. We reconciled the discrepancies by comparing the effect of COMMD1 on ATP7A that was expressed endogenously, stably, and transiently. We concluded that COMMD1 down-regulated the levels of both endogenous and stably expressed ATP7A (WT and mutants), but acted to stabilize these proteins when transiently overexpressed. The reason for this difference is not clear. One possibility is that the profound overexpression that is a feature of transient expression may saturate and slow progress through the COMMD1-targeted degradation pathway, so that COMMD1 binding to the mutant molecules may give the appearance of stabilization. Alternatively, saturation of the localization pathways may affect normal targeting of the overexpressed proteins to the degradation pathways; or COMMD1 may have different binding efficiencies depending on whether the transiently overexpressed proteins are mislocalized to the endoplasmic reticulum or to vesicular/endocytic pathways.
Both clusterin and COMMD1 were shown to affect intracellular copper levels in HEK293(T) cells (15, 52), a dog hepatic cell line (56) and in a mouse hepatoma cell line (55). Knockdown of either clusterin or COMMD1 would lead to increased levels of one (hepatic cell line) or both (HEK293 cells) Cu-ATPases, which in turn would lead to an increase in copper sequestration and efflux. The total cellular copper then represents the balance between copper sequestration and export. Because ATP7B is thought to be primarily involved in copper sequestration (6, 57) this would explain the more significant increase in copper retention seen with COMMD1 knockdown in the dog hepatic cell line (56). These observations are consistent with both clusterin and COMMD1 levels influencing the turnover of the Cu-ATPases, which in turn impacts on cellular copper levels. COMMD1 was also shown to bind Cu(II), but because the majority of intracellular copper is Cu(I), the significance of this Cu(II) binding by COMMD1 in intracellular copper homeostasis is unclear (58).
Since its discovery, the COMMD1 protein interaction network continues to expand (37) and includes NF-κB (52, 59), epithelial sodium channel (60, 61), and hypoxia-inducible factor (HIF-1α) (22). In all of these cases, the mechanism of action of COMMD1 in facilitating protein degradation can be unified by its involvement in the ubiquitin-proteasome pathway (35–37). The action of COMMD1 in facilitating the degradation of ATP7B via the proteasomal pathway is consistent with this. However, contrary to these studies, COMMD1 interaction with the cystic fibrosis transmembrane conductance regulator (CFTR) protein enhanced CFTR stability and cell surface expression (69). COMMD1 also was shown to exist in a complex with CCS and SOD1 and inhibits the formation of SOD1 dimers, which is the final step in SOD1 activation (62). Clearly as the number of COMMD1 binding partners continues to increase, the mode of action of COMMD1 in regulating protein stability and degradation also continues to expand.
Many studies have attempted to establish genotype/phenotype correlations in both MD and WD without success (63, 64). There is a high degree of clinical variability among patients (48) and mutation analysis of ATP7A and ATP7B has identified ∼160 and 300 patient mutations for each gene, respectively (Wilson Disease Mutation Database, University of Alberta) (64). It has been suggested that the clinical outcome depends on the type of mutation (8, 11, 48). Individuals with identical mutations, including twins (65) and also between family members, can have vastly different clinical phenotypes (66, 67), which suggests that other factors are involved in determining the clinical outcome of Menkes and Wilson diseases. Such factors may include allelic dominance, copper levels, the environment, and other interacting proteins that may serve to modify the phenotypes (68). Based on the data presented here, clusterin and COMMD1 interact with the Cu-ATPases to facilitate the degradation of these molecules, especially mutant/misfolded forms, and they have a direct impact on the maintenance of copper homeostasis. Thus, it is possible that allelic variations of the Clusterin and COMMD1 genes may play a role in modifying the clinical presentations of MD and WD.
Our data provides important insight and clarification of the roles of clusterin and COMMD1 in copper homeostasis and in regulating Cu-ATPase quality control. This is necessary for cell survival in disease states in the context of continuous synthesis of mutant Cu-ATPase molecules. This study confirms the role of clusterin as a chaperone-like molecule that targets the Cu-ATPases for degradation via the lysosomal degradation pathway. Importantly, the data presented here clarifies the role of COMMD1 in facilitating the degradation of both Cu-ATPases and suggests a role for this protein as a molecular chaperone. Based on these studies and others, the Clusterin and COMMD1 genes may represent modifying genes that modulate the phenotype of Menkes and Wilson Diseases.
Acknowledgments
We thank Saverio Bettuzzi (University of Parma, Italy) for kindly providing the full-length clusterin cDNA in pIREShygl, and Roxana Llanos, Alison Blake (Deakin University, Australia), and Willianne Vonk (University Medical Center, Utrecht, the Netherlands) for technical support.
This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRC) (to S. L. and J. F. B. M.), NHMRC Biomedical Research Fellowship 520345 (to M. A. C.), Dutch Digestive Disease Foundation Grant WS02-34, NOW Program Grant 912.04.106, and The Netherlands Organisation for Scientific Research-Earth and Life Sciences Grant 817.02.022 (to L. W. J. K.).
- Cu-ATPase
- copper-transporting P1B-type ATPase
- CHX
- cycloheximide
- MBD
- metal binding domain
- MD
- Menkes disease
- TGN
- trans-Golgi network
- WD
- Wilson disease.
REFERENCES
- 1. Lutsenko S., Petrukhin K., Cooper M. J., Gilliam C. T., Kaplan J. H. (1997) N-terminal domains of human copper-transporting adenosine triphosphatases (the Wilson and Menkes disease proteins) bind copper selectively in vivo and in vitro with stoichiometry of one copper per metal-binding repeat. J. Biol. Chem. 272, 18939–18944 [DOI] [PubMed] [Google Scholar]
- 2. Lutsenko S., Barnes N. L., Bartee M. Y., Dmitriev O. Y. (2007) Function and regulation of human copper-transporting ATPases. Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 3. Monty J. F., Llanos R. M., Mercer J. F., Kramer D. R. (2005) Copper exposure induces trafficking of the Menkes protein in intestinal epithelium of ATP7A transgenic mice. J. Nutr. 135, 2762–2766 [DOI] [PubMed] [Google Scholar]
- 4. Nyasae L., Bustos R., Braiterman L., Eipper B., Hubbard A. (2007) Dynamics of endogenous ATP7A (Menkes protein) in intestinal epithelial cells. Copper-dependent redistribution between two intracellular sites. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1181–1194 [DOI] [PubMed] [Google Scholar]
- 5. Roelofsen H., Wolters H., Van Luyn M. J., Miura N., Kuipers F., Vonk R. J. (2000) Copper-induced apical trafficking of ATP7B in polarized hepatoma cells provides a mechanism for biliary copper excretion. Gastroenterology 119, 782–793 [DOI] [PubMed] [Google Scholar]
- 6. Cater M. A., La Fontaine S., Shield K., Deal Y., Mercer J. F. (2006) ATP7B mediates vesicular sequestration of copper. Insight into biliary copper excretion. Gastroenterology 130, 493–506 [DOI] [PubMed] [Google Scholar]
- 7. Guo Y., Nyasae L., Braiterman L. T., Hubbard A. L. (2005) NH2-terminal signals in ATP7B Cu-ATPase mediate its Cu-dependent anterograde traffic in polarized hepatic cells. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G904–916 [DOI] [PubMed] [Google Scholar]
- 8. La Fontaine S., Mercer J. F. (2007) Trafficking of the copper-ATPases, ATP7A and ATP7B. Role in copper homeostasis. Arch. Biochem. Biophys. 463, 149–167 [DOI] [PubMed] [Google Scholar]
- 9. Danks D. M. (1995) in The Metabolic and Molecular Basis of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. M., Valle D., eds) pp. 2211–2235, McGraw-Hill, New York [Google Scholar]
- 10. Kim B. E., Petris M. J. (2007) Phenotypic diversity of Menkes disease in mottled mice is associated with defects in localization and trafficking of the ATP7A protein. J. Med. Genet. 44, 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaler S. G. (2011) ATP7A-related copper transport disease-emerging concepts and future trends. Nat. Rev. Neurol. 7, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Terada K., Aiba N., Yang X. L., Iida M., Nakai M., Miura N., Sugiyama T. (1999) Biliary excretion of copper in LEC rat after introduction of copper transporting P-type ATPase, ATP7B. FEBS Lett. 448, 53–56 [DOI] [PubMed] [Google Scholar]
- 13. Das S. K., Ray K. (2006) Wilsons disease. An update. Nat. Clin. Pract. Neurol. 2, 482–493 [DOI] [PubMed] [Google Scholar]
- 14. Gouider-Khouja N. (2009) Wilsons disease. Parkinsonism Relat. Disord. 15, S126–129 [DOI] [PubMed] [Google Scholar]
- 15. Materia S., Cater M. A., Klomp L. W., Mercer J. F., La Fontaine S. (2011) Clusterin (apolipoprotein J), a molecular chaperone that facilitates degradation of the copper-ATPases ATP7A and ATP7B. J. Biol. Chem. 286, 10073–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tao T. Y., Liu F., Klomp L., Wijmenga C., Gitlin J. D. (2003) The copper toxicosis gene product Murr1 directly interacts with the Wilson disease protein. J. Biol. Chem. 278, 41593–41596 [DOI] [PubMed] [Google Scholar]
- 17. de Bie P., van de Sluis B., Burstein E., van de Berghe P. V., Muller P., Berger R., Gitlin J. D., Wijmenga C., Klomp L. W. (2007) Distinct Wilsons disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology 133, 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van den Berghe P. V., Stapelbroek J. M., Krieger E., de Bie P., van de Graaf S. F., de Groot R. E., van Beurden E., Spijker E., Houwen R. H., Berger R., Klomp L. W. (2009) Reduced expression of ATP7B affected by Wilson disease-causing mutations is rescued by pharmacological folding chaperones 4-phenylbutyrate and curcumin. Hepatology 50, 1783–1795 [DOI] [PubMed] [Google Scholar]
- 19. van de Sluis B., Groot A. J., Vermeulen J., van der Wall E., van Diest P. J., Wijmenga C., Klomp L. W., Vooijs M. (2009) COMMD1 Promotes pVHL and O2-independent Proteolysis of HIF-1α via HSP90/70. PLoS One 4, e7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van de Sluis B., Mao X., Zhai Y., Groot A. J., Vermeulen J. F., van der Wall E., van Diest P. J., Hofker M. H., Wijmenga C., Klomp L. W., Cho K. R., Fearon E. R., Vooijs M., Burstein E. (2010) COMMD1 disrupts HIF-1α/β dimerization and inhibits human tumor cell invasion. J. Clin. Invest. 120, 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maine G. N., Mao X., Komarck C. M., Burstein E. (2007) COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J. 26, 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van de Sluis B., Muller P., Duran K., Chen A., Groot A. J., Klomp L. W., Liu P. P., Wijmenga C. (2007) Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol. Cell Biol. 27, 4142–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zoubeidi A., Ettinger S., Beraldi E., Hadaschik B., Zardan A., Klomp L. W., Nelson C. C., Rennie P. S., Gleave M. E. (2010) Clusterin facilitates COMMD1 and IκB degradation to enhance NF-κB activity in prostate cancer cells. Mol. Cancer Res. 8, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Humphreys D. T., Carver J. A., Easterbrook-Smith S. B., Wilson M. R. (1999) Clusterin has chaperone-like activity similar to that of small heat shock proteins. J. Biol. Chem. 274, 6875–6881 [DOI] [PubMed] [Google Scholar]
- 25. Balantinou E., Trougakos I. P., Chondrogianni N., Margaritis L. H., Gonos E. S. (2009) Transcriptional and post-translational regulation of clusterin by the two main cellular proteolytic pathways. Free Radic. Biol. Med. 46, 1267–1274 [DOI] [PubMed] [Google Scholar]
- 26. Wyatt A. R., Yerbury J. J., Berghofer P., Greguric I., Katsifis A., Dobson C. M., Wilson M. R. (2011) Clusterin facilitates in vivo clearance of extracellular misfolded proteins. Cell. Mol. Life Sci. 68, 3919–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vonk W. I., de Bie P., Wichers C. G., van den Berghe P. V., van der Plaats R., Berger R., Wijmenga C., Klomp L. W., van de Sluis B. (2011) The copper-transporting capacity of ATP7A mutants associated with Menkes disease is ameliorated by COMMD1 as a result of improved protein expression. Cell. Mol. Life Sci. 69, 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aronow B. J., Lund S. D., Brown T. L., Harmony J. A., Witte D. P. (1993) Apolipoprotein J expression at fluid-tissue interfaces. Potential role in barrier cytoprotection. Proc. Natl. Acad. Sci. U.S.A. 90, 725–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson M. R., Yerbury J. J., Poon S. (2008) Potential roles of abundant extracellular chaperones in the control of amyloid formation and toxicity. Mol. Biosyst. 4, 42–52 [DOI] [PubMed] [Google Scholar]
- 30. Leskov K. S., Klokov D. Y., Li J., Kinsella T. J., Boothman D. A. (2003) Synthesis and functional analyses of nuclear clusterin, a cell death protein. J. Biol. Chem. 278, 11590–11600 [DOI] [PubMed] [Google Scholar]
- 31. Trougakos I. P., Gonos E. S. (2006) Regulation of clusterin/apolipoprotein J, a functional homologue to the small heat shock proteins, by oxidative stress in ageing and age-related diseases. Free Radic. Res. 40, 1324–1334 [DOI] [PubMed] [Google Scholar]
- 32. Pucci S., Mazzarelli P., Nucci C., Ricci F., Spagnoli L. G. (2009) CLU “in and out.” Looking for a link. Adv. Cancer Res. 105, 93–113 [DOI] [PubMed] [Google Scholar]
- 33. van De Sluis B., Rothuizen J., Pearson P. L., van Oost B. A., Wijmenga C. (2002) Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum. Mol. Genet. 11, 165–173 [DOI] [PubMed] [Google Scholar]
- 34. Klomp A. E., van de Sluis B., Klomp L. W., Wijmenga C. (2003) The ubiquitously expressed MURR1 protein is absent in canine copper toxicosis. J. Hepatol. 39, 703–709 [DOI] [PubMed] [Google Scholar]
- 35. Maine G. N., Burstein E. (2007) COMMD proteins. COMMing to the scene. Cell. Mol. Life Sci. 64, 1997–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Bie P., van de Sluis B., Klomp L., Wijmenga C. (2005) The many faces of the copper metabolism protein MURR1/COMMD1. J. Hered. 96, 803–811 [DOI] [PubMed] [Google Scholar]
- 37. van de Sluis B., Groot A. J., Wijmenga C., Vooijs M., Klomp L. W. (2007) COMMD1. A novel protein involved in the proteolysis of proteins. Cell Cycle 6, 2091–2098 [DOI] [PubMed] [Google Scholar]
- 38. Bailey R. W., Dunker A. K., Brown C. J., Garner E. C., Griswold M. D. (2001) Clusterin, a binding protein with a molten globule-like region. Biochemistry 40, 11828–11840 [DOI] [PubMed] [Google Scholar]
- 39. Nuutinen T., Suuronen T., Kauppinen A., Salminen A. (2009) Clusterin, a forgotten player in Alzheimers disease. Brain Res. Rev. 61, 89–104 [DOI] [PubMed] [Google Scholar]
- 40. Burkhead J. L., Morgan C. T., Shinde U., Haddock G., Lutsenko S. (2009) COMMD1 forms oligomeric complexes targeted to the endocytic membranes via specific interactions with phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 284, 696–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sommerhalter M., Zhang Y., Rosenzweig A. C. (2007) Solution structure of the COMMD1 N-terminal domain. J. Mol. Biol. 365, 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cater M. A., Forbes J., La Fontaine S., Cox D., Mercer J. F. (2004) Intracellular trafficking of the human Wilson protein. The role of the six N-terminal metal-binding sites. Biochem. J. 380, 805–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strausak D., La Fontaine S., Hill J., Firth S. D., Lockhart P. J., Mercer J. F. (1999) The role of GMXCXXC metal binding sites in the copper-induced redistribution of the Menkes protein. J. Biol. Chem. 274, 11170–11177 [DOI] [PubMed] [Google Scholar]
- 44. La Fontaine S., Theophilos M. B., Firth S. D., Gould R., Parton R. G., Mercer J. F. (2001) Effect of the toxic milk mutation (tx) on the function and intracellular localization of Wnd, the murine homologue of the Wilson copper ATPase. Hum. Mol. Genet. 10, 361–370 [DOI] [PubMed] [Google Scholar]
- 45. van den Berghe P. V., Folmer D. E., Malingré H. E., van Beurden E., Klomp A. E., van de Sluis B., Merkx M., Berger R., Klomp L. W. (2007) Human copper transporter 2 is localized in late endosomes and lysosomes and facilitates cellular copper uptake. Biochem. J. 407, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ke B. X., Llanos R. M., Wright M., Deal Y., Mercer J. F. (2006) Alteration of copper physiology in mice overexpressing the human Menkes protein ATP7A. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1460–1467 [DOI] [PubMed] [Google Scholar]
- 47. Payne A. S., Kelly E. J., Gitlin J. D. (1998) Functional expression of the Wilson disease protein reveals mislocalization and impaired copper-dependent trafficking of the common H1069Q mutation. Proc. Natl. Acad. Sci. U.S.A. 95, 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Bie P., Muller P., Wijmenga C., Klomp L. W. (2007) Molecular pathogenesis of Wilson and Menkes disease. Correlation of mutations with molecular defects and disease phenotypes. J. Med. Genet. 44, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Theophilos M. B., Cox D. W., Mercer J. F. (1996) The toxic milk mouse is a murine model of Wilson disease. Hum. Mol. Genet. 5, 1619–1624 [DOI] [PubMed] [Google Scholar]
- 50. Voskoboinik I., Greenough M., La Fontaine S., Mercer J. F., Camakaris J. (2001) Functional studies on the Wilson copper P-type ATPase and toxic milk mouse mutant. Biochem. Biophys. Res. Commun. 281, 966–970 [DOI] [PubMed] [Google Scholar]
- 51. Majumdar R., Al Jumah M., Al Rajeh S., Fraser M., Al Zaben A., Awada A., Al Traif I., Paterson M. (2000) A novel deletion mutation within the carboxyl terminus of the copper-transporting ATPase gene causes Wilson disease. J. Neurol. Sci. 179, 140–143 [DOI] [PubMed] [Google Scholar]
- 52. Burstein E., Ganesh L., Dick R. D., van De Sluis B., Wilkinson J. C., Klomp L. W., Wijmenga C., Brewer G. J., Nabel G. J., Duckett C. S. (2004) A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO J. 23, 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rizzi F., Caccamo A. E., Belloni L., Bettuzzi S. (2009) Clusterin is a short half-life, polyubiquitinated protein, which controls the fate of prostate cancer cells. J. Cell. Physiol. 219, 314–323 [DOI] [PubMed] [Google Scholar]
- 54. Huang Y., Wu M., Li H. Y. (2008) Tumor suppressor ARF promotes non-classic proteasome-independent polyubiquitination of COMMD1. J. Biol. Chem. 283, 11453–11460 [DOI] [PubMed] [Google Scholar]
- 55. Miyayama T., Hiraoka D., Kawaji F., Nakamura E., Suzuki N., Ogra Y. (2010) Roles of COMM-domain-containing 1 in stability and recruitment of the copper-transporting ATPase in a mouse hepatoma cell line. Biochem. J. 429, 53–61 [DOI] [PubMed] [Google Scholar]
- 56. Spee B., Arends B., van Wees A. M., Bode P., Penning L. C., Rothuizen J. (2007) Functional consequences of RNA interference targeting COMMD1 in a canine hepatic cell line in relationship to copper toxicosis. Anim. Genet. 38, 168–170 [DOI] [PubMed] [Google Scholar]
- 57. Barnes N., Bartee M. Y., Braiterman L., Gupta A., Ustiyan V., Zuzel V., Kaplan J. H., Hubbard A. L., Lutsenko S. (2009) Cell-specific trafficking suggests a new role for renal ATP7B in the intracellular copper storage. Traffic 10, 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sarkar B., Roberts E. A. (2011) The puzzle posed by COMMD1. A newly discovered protein binding Cu(II). Metallomics 3, 20–27 [DOI] [PubMed] [Google Scholar]
- 59. Thoms H. C., Loveridge C. J., Simpson J., Clipson A., Reinhardt K., Dunlop M. G., Stark L. A. (2010) Nucleolar targeting of RelA(p65) is regulated by COMMD1-dependent ubiquitination. Cancer Res. 70, 139–149 [DOI] [PubMed] [Google Scholar]
- 60. Biasio W., Chang T., McIntosh C. J., McDonald F. J. (2004) Identification of Murr1 as a regulator of the human δ-epithelial sodium channel. J. Biol. Chem. 279, 5429–5434 [DOI] [PubMed] [Google Scholar]
- 61. Ke Y., Butt A. G., Swart M., Liu Y. F., McDonald F. J. (2010) COMMD1 down-regulates the epithelial sodium channel through Nedd4–2. Am. J. Physiol. Renal Physiol. 298, F1445–1456 [DOI] [PubMed] [Google Scholar]
- 62. Vonk W. I., Wijmenga C., Berger R., van de Sluis B., Klomp L. W. (2010) Cu,Zn superoxide dismutase maturation and activity are regulated by COMMD1. J. Biol. Chem. 285, 28991–29000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Horslen S., Hahn S. (2010) Genotype-phenotype correlation in Wilson disease. J. Clin. Gastroenterol. 44, 387–388 [DOI] [PubMed] [Google Scholar]
- 64. Møller L. B., Mogensen M., Horn N. (2009) Molecular diagnosis of Menkes disease. Genotype-phenotype correlation. Biochimie 91, 1273–1277 [DOI] [PubMed] [Google Scholar]
- 65. Senzolo M., Loreno M., Fagiuoli S., Zanus G., Canova D., Masier A., Russo F. P., Sturniolo G. C., Burra P. (2007) Different neurological outcome of liver transplantation for Wilsons disease in two homozygotic twins. Clin. Neurol. Neurosurg. 109, 71–75 [DOI] [PubMed] [Google Scholar]
- 66. Borm B., Møller L. B., Hausser I., Emeis M., Baerlocher K., Horn N., Rossi R. (2004) Variable clinical expression of an identical mutation in the ATP7A gene for Menkes disease/occipital horn syndrome in three affected males in a single family. J. Pediatr. 145, 119–121 [DOI] [PubMed] [Google Scholar]
- 67. Takeshita Y., Shimizu N., Yamaguchi Y., Nakazono H., Saitou M., Fujikawa Y., Aoki T. (2002) Two families with Wilson disease in which siblings showed different phenotypes. J. Hum. Genet. 47, 543–547 [DOI] [PubMed] [Google Scholar]
- 68. Gupta A., Bhattacharjee A., Dmitriev O. Y., Nokhrin S., Braiterman L., Hubbard A. L., Lutsenko S. (2011) Cellular copper levels determine the phenotype of the Arg-875 variant of ATP7B/Wilson disease protein. Proc. Natl. Acad. Sci. U.S.A. 108, 5390–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Drévillon L., Tanguy G., Hinzpeter A., Arous N., de Becdelièvre A., Aissat A., Tarze A., Goossens M., Fanen P. (2011) COMMD1-mediated ubiquitination regulates CFTR trafficking. PLoS One 6, e18334. [DOI] [PMC free article] [PubMed] [Google Scholar]