Background: S-phase checkpoint is important for maintaining genome stability upon DNA damage in S-phase.
Results: A replication essential protein Dbf4 is phosphorylated by checkpoint kinases when DNA is damaged.
Conclusion: Dbf4 is a downstream target of the S-phase checkpoint to mediate DNA damage responses.
Significance: These studies help understand how the genome is protected from DNA damage to prevent tumorigenesis.
Keywords: Cell Cycle, Checkpoint Control, DNA Damage Response, DNA Repair, DNA Replication, ATM, ATR, Cdc7, Dbf4, S-phase Checkpoint
Abstract
Dbf4/Cdc7 (Dbf4-dependent kinase (DDK)) is activated at the onset of S-phase, and its kinase activity is required for DNA replication initiation from each origin. We showed that DDK is an important target for the S-phase checkpoint in mammalian cells to suppress replication initiation and to protect replication forks. We demonstrated that ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) proteins directly phosphorylate Dbf4 in response to ionizing radiation and replication stress. We identified novel ATM/ATR phosphorylation sites on Dbf4 and showed that ATM/ATR-mediated phosphorylation of Dbf4 is critical for the intra-S-phase checkpoint to inhibit DNA replication. The kinase activity of DDK, which is not suppressed upon DNA damage, is required for fork protection under replication stress. We further demonstrated that ATM/ATR-mediated phosphorylation of Dbf4 is important for preventing DNA rereplication upon loss of replication licensing through the activation of the S-phase checkpoint. These studies indicate that DDK is a direct substrate of ATM and ATR to mediate the intra-S-phase checkpoint in mammalian cells.
Introduction
The initiation of DNA replication requires a series of coordinated events during the cell cycle (1, 2). In late mitosis and G1, MCM2-7 proteins are loaded onto chromatin to form prereplicative complexes. When cells enter S-phase, S-phase cyclin-dependent kinases and Dbf4/Cdc7 (DDK)4 are required for activating prereplicative complexes by recruiting replication initiation proteins, such as Cdc45 and GINS, to origins (3, 4).
Cdc7 is a serine/threonine kinase, conserved from yeast to humans (5). It is activated by binding to its regulatory protein Dbf4 (6, 7). Cdc7 protein levels appear constant during the cell cycle, whereas its kinase activity peaks at the G1/S transition due to the cell cycle-regulated oscillations in Dbf4 protein levels (6, 8–10). DDK is required for the initiation of DNA replication from each individual origin, including both early and late firing origins (11, 12). The MCM2-7 complex is an important target of DDK for DNA replication initiation (13).
The S-phase checkpoint is activated in response to replication stress or DNA lesions that occur in S-phase and is critical for the maintenance of genome stability (14, 15). The S-phase checkpoint inhibits late replication origin firing to slow down DNA replication in response to DNA damage in S-phase. It also stabilizes and protects stalled replication forks upon replication stress and facilitates replication restart during recovery from fork stalling or damage. Ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related protein (ATR) are two major phosphatidylinositol 3-like protein kinases that are critical for detecting aberrant DNA structures (16). ATM in collaboration with the Mre11/Rad50/Nbs1 complex participates in the detection of DNA double-stranded breaks (DSBs), leading to the activation of the Chk2 pathway. ATR with its partner ATR-interacting protein detects DNA lesions by recognizing replication protein A (RPA)-coated single-stranded DNA (ssDNA) and activates the Chk1 pathway (17, 18).
In yeast, substantial evidence suggests that DDK is a direct target of the S-phase checkpoint. Dbf4/Dfp1 (Dfp1:Dbf4 ortholog in Schizosaccharomyces pombe) undergoes Rad53/Cds1 (orthologs of Chk2 in yeast)-dependent phosphorylation upon HU treatment (19–21). The chromatin binding of Dbf4 is reduced after HU treatment in a Rad53-dependent manner (10), and Rad53-mediated phosphorylation of Dbf4 attenuates the kinase activity of DDK (21). Recent studies further demonstrated that Rad53-mediated phosphorylation of Dbf4 directly inhibits origin firing to mediate the intra-S-phase checkpoint in yeast (22, 23). Analysis of various Dbf4 and Cdc7 mutants and genetic interactions suggest that DDK plays a critical role in preserving fork integrity and recovery from replication fork arrest in yeast (20, 24, 25).
In the Xenopus cell-free system, it was described that etoposide, a DNA topoisomerase II inhibitor, activates the ATR-dependent S-phase checkpoint and inhibits the kinase activity of DDK (26). Similar results of inactivation of DDK activity were obtained in human leukemia cells treated with etoposide (27). However, recent studies in both Xenopus cell-free systems and mammalian cells demonstrated that DDK remains active during the damage-induced S-phase checkpoint response (28–33). In addition, a Dbf4-related factor, Drf1, was found to play a role more important than that of Dbf4 in mediating the S-phase checkpoint (32, 34). These studies raise a question as to whether DDK is a critical S-phase checkpoint target to inhibit replication initiation if its kinase activity is not altered after DNA damage in higher organisms.
In this study, we demonstrated that Dbf4 is a direct downstream target of ATM and ATR when the S-phase checkpoint is activated. We identified ATM/ATR phosphorylation sites on Dbf4 and showed that ATM/ATR-dependent phosphorylation of Dbf4 is important for inhibiting replication initiation to mediate the intra-S-phase checkpoint through a mechanism independent of attenuating DDK kinase activities. Importantly, we also found that the kinase activity of DDK is required for protecting replication forks upon replication stress. Therefore, Dbf4 plays dual roles to mediate the S-phase checkpoint responses in mammalian cells. Although it is a direct target of the S-phase checkpoint to inhibit DNA replication, DDK remains active to protect stalled replication forks. Consistent with its role in inhibiting replication initiation upon the activation of S-phase checkpoint, we also demonstrated that ATM/ATR-mediated phosphorylation of Dbf4 is important for the suppression of DNA rereplication when the licensing control is impaired.
EXPERIMENTAL PROCEDURES
Cell Culture, Cell Synchronization, Transfection, and Retroviral Infection
T98G, 293T, U2OS, U2OS-ATR-WT, U2OS-ATR-KD, GM847, and GM847-ATR-KD cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. The expression of ATR-WT/KD in the U2OS or GM847 fibroblasts was induced by the addition of 1 μg/ml doxycycline to the media for 24 h (35, 36). T98G cells were synchronized in G0 by culturing in DMEM supplemented with 0.1% fetal bovine serum for 48 h and then releasing to G1 or S by adding 10% fetal bovine serum and harvesting cells at the indicated time points. Stable U2OS cell lines expressing tagged Dbf4 wild type or phospho-Dbf4 mutants were generated by retroviral infection using pBabe vector, followed by selection of puromycin, G418, or hygromycin as described previously (37).
Short Hairpin RNA (shRNA) and Retroviral Infection
Silencing of endogenous ATM, ATR, Cdc7, or Dbf4 in U2OS cells was performed by retroviral infection using the vector pMKO expressing corresponding shRNAs (38). pMKO-based shRNA plasmids were constructed by inserting the annealed and phosphorylated shRNA target sequences into pMKO. The shRNA target sequences used were as follows: ATM, GCACCAGTCCAGTATTGGCTT and AACATCTACTCAAAGACATT; ATR, CGAGACTTCTGCGGATTGCAG and AACCTCCGTGATGTTGCTTGA; Cdc7, GCTCAGCAGGAAAGGTGTTCA; Dbf4, GAGCAGAATTTCCTGTATA.
Whole Cell Lysate and Chromatin Isolation
Cells were lysed in NETN (150 mm NaCl, 1 mm EDTA, 20 mm Tris-Cl, pH 8.0, 0.5% Nonidet P-40 (v/v)) containing protease and phosphatase inhibitors (50 mm sodium fluoride (NaF) and 0.1 mm sodium orthovanadate (NaVO4)). Phosphatase treatment of cell lysates was performed as described (39). For chromatin isolation, cells were washed with phosphate-buffered saline (PBS), collected, and resuspended in CSK buffer (10 mm PIPES, pH 6.8, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 1 mm EGTA, 50 mm sodium fluoride, 0.1 mm sodium orthovanadate, 0.1% Triton X-100, and protease inhibitors), and incubated on ice for 10 min. Cytoplasmic proteins were separated from nuclei by low speed centrifugation at 1,300 × g for 5 min. Isolated nuclei were washed once in CSK buffer and lysed in solution (3 mm EDTA, 0.2 mm EGTA, 1 mm dithiothreitol, and protease inhibitors). After centrifugation at 1,700 × g for 5 min, pellets were resuspended in CSK buffer. 2× SDS loading buffer was added, and samples were boiled for 10 min.
Adenovirus Construction and Infection
Adenoviruses encoding GFP, human Dbf4–5A (S226A/T265A/T449A/S502A/S539A), Cdt1, and Cdc7KD (D196N) were generated by using the AdEasy system (40, 41). Adenovirus plasmids were constructed by inserting the corresponding cDNAs into pAd-track-CMV shuttle vector. Then in vivo recombination was performed by transforming pAd-track-CMV plasmid together with pAd-Easy-1 adenoviral vector into BJ5813 competent cells by electroporation. The recombinant adenoviral plasmids were transfected into 293 cells to generate corresponding recombinant adenoviruses. Large scale purification of adenoviruses from 293 cells was obtained by CsCl density gradient centrifugation. The concentration of purified virus was measured by A260 reading using the equation 1A260 ≈ 1012 pfu (42). Recombinant adenoviruses encoding p27 were provided by S. Reed (The Scripps Research Institute).
Antibodies
Antibodies to human Dbf4 were provided by the laboratory of Wei Jiang (28) or generated using OVA-conjugated peptide (SDFSTDNSGSQPKQKSDTV). Antibody to human Dbf4 Ser-539 phospho-specific was generated by immunizing rabbits with the OVA-conjugated peptide (ITINSpSQEHLTV, where pS represents phosphoserine). An antibody to Cdt1 was described earlier (43).
Other antibodies used in this study were purchased as indicated: antibody to Cdc7 (DCS-341) from Thermo Scientific; phospho-Chk1 (Ser-317) from R & D Systems; phospho-Chk2 (Thr-68), phospho-Nbs1 (Ser-343), and phospho-p53 (Ser-15) from Cell Signaling Technology; Chk1, Chk2, and Ku70 from Santa Cruz Biotechnology, Inc.; RPA1, RPA2, ATM, and ATR antibodies from Calbiochem Merck; Myc (9E10), actin, and γ-H2AX from Millipore; MCM2 from BD Biosciences; and FLAG (M2) from Sigma-Aldrich.
Fluorescence-activated Cell Sorting (FACS) Analysis
Cells were washed twice with PBS, collected by trypsinization, and fixed with ice-cold 70% ethanol overnight at 4 °C. After fixation, cells were stained with propidium iodide solution, which contained 38 mm sodium citrate, 10 μg/ml RNase A, and 15 μg/ml propidium iodide (Sigma-Aldrich). The labeled cells were analyzed with a FACScan flow cytometer (BD Biosciences) using CELLQUEST software (BD Biosciences).
In Vitro Kinase Assays and Radioresistant DNA Synthesis Analysis
pcDNA3 containing FLAG-tagged ATM, ATR, or Myc-Cdc7/FLAG-Dbf4 was transiently transfected into 293T cells and immunoprecipitated with anti-FLAG M2-conjugated-agarose beads (Sigma-Aldrich). Immunoprecipitates were washed with NETN buffer followed by kinase buffer and then incubated with the indicated purified GST fusion proteins as substrates in either ATM/ATR kinase buffer (25 mm HEPES, pH 7.5, 50 mm NaCl, 10 mm MgCl2, 10 mm MnCl2, 1 mm DTT, 10 μm ATP) or Cdc7 kinase buffer (25 mm HEPES, pH 7.5, 50 mm NaCl, 10 mm MgCl2, 1 mm DTT, 10 μm ATP) in the presence of 10 μCi of [γ-32P]ATP, phosphatase inhibitors (10 mm NaF, 50 mm β-glycerophosphate) at 30 °C for 30 min, followed by the addition of SDS-PAGE sample buffer. Phosphorylated radioactive proteins were separated by SDS-PAGE and detected by autoradiography of the dried gels. A Dbf4 autophosphorylation assay was performed by incubating anti-FLAG M2 immunoprecipitates from 293T cells expressing Myc-Cdc7WT/FLAG-Dbf4 or Myc-Cdc7KD/FLAG-Dbf4 using Cdc7 kinase buffer in the presence of [γ-32P]ATP.
A series of Dbf4 fragments and mutational variants of these fragments were cloned into the pGEX-4T vector (GE Healthcare). GST fusion proteins were expressed in BL21 (DE3) Escherichia coli (Invitrogen), purified according to the manufacturer's instructions, and used as kinase substrates. Radioresistant DNA synthesis analysis was performed as described (44).
Comet Assays
The neutral comet assay was performed by using the comet assay kit (4250-050-K, Trevigen, Gaithersburg, MD) according to the manufacturer's recommendations. U2OS cells were treated with or without HU (1 mm, 8 h at 37 °C). The cells were stained with SYBR Green (1:10000 dilution; Trevigen), visualized by fluorescence microscopy, and analyzed using CometScore (TriTek Corp.). Typically, about 50 cells/sample were examined.
RESULTS
Dbf4 Is Phosphorylated in Response to Various Kinds of DNA Damage in Mammalian Cells
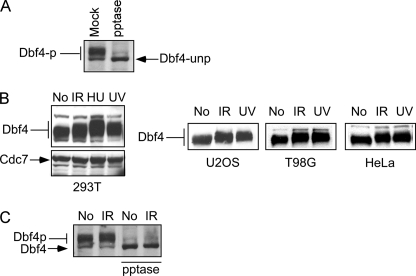
Human Dbf4 is phosphorylated by Cdc7 during the normal cell cycle (7, 45, 46). Consistently, phosphatase treatment converts Dbf4 to faster migrating unphosphorylated species (Fig. 1A). Upon DNA damage, including IR, UV, and HU, the migration of human Dbf4, but not that of Cdc7, is further retarded in multiple cell lines (Fig. 1B). Phosphatase treatment abolishes the migration difference before and after DNA damage (Fig. 1C), suggesting that various kinds of DNA damage induce Dbf4 hyperphosphorylation in addition to cell cycle-regulated phosphorylation of Dbf4 by Cdc7 in mammalian cells.
FIGURE 1.
Dbf4 is hyperphosphorylated in response to DNA damage in various cell lines. A, whole cell extracts were prepared from 293T cells and treated with or without protein phosphatase (pptase or mock) as described (39). Anti-Dbf4 Western blot analysis was performed. B, 293T, U2OS, T98G, and HeLa cells were treated without (No) or with IR (20 Gy, 30 min after), HU (1 mm, 24 h after), or UV light (30 J/m2, 1 h after). Whole cell extracts were prepared, and Western blot analysis for Dbf4 and Cdc7 was performed. C, whole cell extracts were prepared from 293T cells before (No) or after IR treatment (20 Gy, 30 min after), followed by treatment with or without protein phosphatase. Anti-Dbf4 Western blot analysis was performed.
Dbf4 Is Direct Substrate of ATM/ATR at Multiple Sites
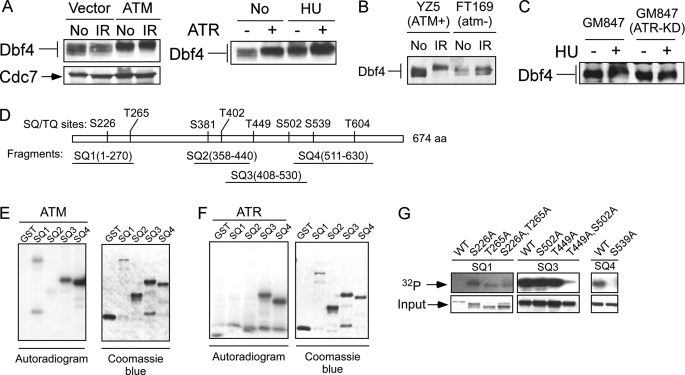
Overexpression of ATM or ATR stimulates the phosphorylation of Dbf4 before and after DNA damage as revealed by slower migration on SDS-PAGE (Fig. 2A), suggesting that ATM and ATR can phosphorylate Dbf4 in vivo. To examine whether damage-induced Dbf4 phosphorylation depends on ATM in vivo, we used ATM-deficient cell line FT169 and its isogenic derivative YZ5, which was reconstituted with ATM (47). IR-induced Dbf4 migration retardation is much reduced in FT169 in a comparison with YZ5 (Fig. 2B), suggesting that IR-induced Dbf4 hyperphosphorylation requires ATM activity. We also examined whether Dbf4 hyperphosphorylation induced by replication stress requires ATR activity. We used an SV40-transformed fibroblast cell line, GM847, and its derivative cell line expressing a doxycycline-inducible kinase-inactive allele of ATR (GM847-KD) (35). Overexpression of the kinase-inactive allele of ATR (ATR-KD) substantially decreased Dbf4 hyperphosphorylation after HU treatment (Fig. 2C). Similar results were obtained following UV and aphidicolin treatment (data not shown). These results suggest that ATM and ATR play important roles in mediating Dbf4 hyperphosphorylation upon DNA damage.
FIGURE 2.
DNA damage-induced Dbf4 hyperphosphorylation is mediated by ATM and ATR. A, 293T cells were transfected with empty vector (Vector or −), the ATM expression vector (ATM, left) or the ATR expression vector (+, right). Whole cell extracts were prepared after mock treatment (No) or exposure to IR (20 Gy, 30 min after) or HU (1 mm, 24 h after), and Western blotting for Dbf4 was performed. B, whole cell extracts were isolated from ATM-deficient cell line (FT169; atm−) or its derivative cell line (YZ5; ATM+) reconstituted with wild type ATM before (No) or after exposure to IR (20 Gy, 30 min after). Western blot analysis for Dbf4 was performed. C, fibroblast cell line GM847 and its derivative cell line carrying doxycycline-inducible ATR-KD (GM847-ATR-KD) were incubated with doxycycline (1 μg/ml) for 24 h and then treated with HU (1 mm, 24 h, +) or without (−) in the presence of doxycycline. Whole cell extracts were prepared, and Western blot analysis of Dbf4 was performed. D, schematic drawing of full-length Dbf4 containing eight ATM/ATR consensus phosphorylation sites, (S/T)Q sites, is represented. Four GST-fused Dbf4 fragments (SQ1–SQ4) containing one or two (S/T)Q sites are illustrated. E, ATM in vitro kinase assay was performed by using anti-FLAG immunoprecipitates from 293T cells that were transiently transfected with FLAG-tagged ATM. The immunoprecipitates were incubated with each of the glutathione-eluted purified GST-SQ fragments as indicated in the presence of [γ-32P]ATP. The autoradiogram shows the results of the in vitro kinase assay (left), and the Coomassie Blue-stained gel indicates the relative amount of protein added into the in vitro kinase reaction (right). F, 293T cells were transiently transfected with FLAG-tagged ATR, and the anti-FLAG immunoprecipitates were incubated with each of the glutathione-eluted purified GST-SQ fragment in the presence of [γ-32P]ATP. Phosphorylation of Dbf4 fragments and the input of the Dbf4 fragments are shown by autoradiogram (left) or Coomassie Blue-stained gel (right), respectively. G, in vitro ATM kinase assay was performed using purified GST-Dbf4 wild type fragments SQ1, SQ3, or SQ4, or the corresponding fragments with alanine substitution of serine or threonine residues at the (S/T)Q (SQ/TQ) sites as indicated. ATM-mediated phosphorylation of Dbf4 fragments was revealed by an 32P autoradiogram (top), and relative input for the GST-fused Dbf4 fragments is indicated by a Coomassie Blue-stained gel (bottom).
To examine whether Dbf4 is a direct substrate of ATM and ATR, we performed in vitro kinase assays. Dbf4 contains eight ATM/ATR consensus sites ((S/T)Q) (48, 49), as illustrated in Fig. 2D. We constructed four GST-fused Dbf4 fragments, covering different (S/T)Q sites (Fig. 2D). In vitro kinase assays showed that both ATM and ATR phosphorylate fragments SQ3 (residues 408–530) and SQ4 (residues 511–630), and ATM also weakly phosphorylates fragment SQ1 (residues 1–270) (Fig. 2, E and F). These results suggest that Dbf4 is a substrate of both ATM and ATR in vitro.
To verify that ATM and ATR phosphorylate the (S/T)Q sites in the fragments, we introduced Ser/Thr to Ala substitutions at the (S/T)Q sites in fragments SQ1, SQ3, and SQ4. Mutating the (S/T)Q sites in the SQ3 (T449A/S502A) and SQ4 (S539A) fragments dramatically reduced the phosphorylation of these fragments in the in vitro ATM kinase assay, whereas substituting Ser-226 and Thr-265 in SQ1 fragment only slightly reduced ATM-mediated phosphorylation in vitro (Fig. 2G). Similar results were obtained when an ATR kinase assay was performed using the mutated SQ3 (T449A/S502A) and SQ4 (S539A) fragments (data not shown). These data suggested that Thr-449, Ser-502, and Ser-539 are the major ATM and ATR phosphorylation sites in vitro.
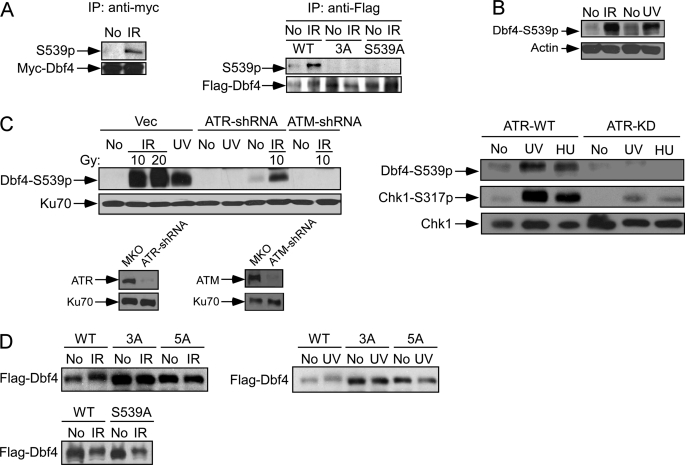
To demonstrate Dbf4 phosphorylation in vivo, we generated a phospho-specific antibody for the site Ser-539 and affinity-purified. The affinity-purified antibody specifically recognizes immunoprecipitated Myc-tagged Dbf4 wild type species after IR (Fig. 3A, left) (data not shown). The antibody specificity was also confirmed by its recognition of the anti-FLAG immunoprecipitates from U2OS cells expressing FLAG-Dbf4-WT but not FLAG-Dbf4-S449A/S502A/S539A (3A) or FLAG-Dbf4-S539A after IR (Fig. 3A, right).
FIGURE 3.
ATM and ATR phosphorylate Dbf4 at ATM/ATR consensus sites in vivo. A, U2OS cells were retrovirally infected with Myc-tagged wild type Dbf4 (left), FLAG-tagged wild type Dbf4 (WT), or the FLAG-Dbf4–3A (T449A, S502A, and S539A) and Dbf4-S539A mutants (right). Anti-Myc or anti-FLAG immunoprecipitation (IP) was performed before (No) or after IR treatment (20 Gy, 1 h after). Western blot analysis of anti-Myc immunoprecipitates (left) or anti-FLAG immunoprecipitates (right) was performed using affinity-purified polyclonal antibody recognizing phosphorylated Dbf4 at Ser-539 and monoclonal antibodies for Myc and FLAG tags. B, whole cell lysates were prepared from U2OS cells before (No) or after treatment with IR (20 Gy, 1 h after) or UV (50 J/m2, 1 h after). Western blot analysis using affinity-purified antibody recognizing phosphorylated Dbf4 at Ser-539 was performed. Western blotting against actin was used as a loading control. C, left, U2OS cells retrovirally infected with pMKO vector, ATR-shRNA, or ATM-shRNA were mock treated (No) or treated with IR (10 or 20 Gy, 1 h after) or UV light (50 J/m2, 1 h after). Whole cell lysates were analyzed by Western blot analysis using the phospho-specific antibody to Dbf4-Ser539. The expression of ATR and ATM after shRNA expression was revealed by Western blot analysis. Ku70 was used as a loading control. Right, U2OS cells expressing doxycycline-induced ATR-WT or ATR-KD were mock-treated (No) or treated with UV (50 J/m2, 1 h after) or HU (1 mm, 24 h). Whole cell extracts were prepared and immunoblotted using the indicated antibodies. D, U2OS stable cell lines were generated by retroviral infection to express FLAG-tagged wild type Dbf4 (WT) or the Dbf4 phosphomutants 3A (S449A/S502A/S539A), 5A (S226A/T265A/S449A/S502A/S539A), or S539A. Cell lysates were prepared before (No) and after treatment of IR (20 Gy, 1 h after) or UV (50 J/m2, 1 h after), and anti-FLAG Western blot analysis was performed.
By using this phospho-specific Ser-539 antibody, we showed that phosphorylation of Ser-539 occurs in response to various DNA-damaging agents (Fig. 3, B and C) (data not shown). By expressing shRNAs against ATM or ATR, we further showed that IR-induced phosphorylation of Ser-539 depends on ATM and partially on ATR, whereas UV-induced phosphorylation of Ser-539 is dependent on ATR (Fig. 3C, left). Similarly, overexpression of ATR-KD in U2OS cells substantially decreases Dbf4 phosphorylation after UV or HU treatment (Fig. 3C, right). These results suggest that Ser-539 is phosphorylated in an ATM- or ATR-dependent manner in vivo in response to various kinds of damage.
To examine the in vivo phosphorylation of the putative ATM/ATR sites other than Ser-539, which are mapped by the in vitro kinase assays as described in the legend to Fig. 2, we used the Dbf4 phosphorylation mutants with Ser/Thr to Ala substitutions at S226A/T265A/S449A/S502A/S539A (5A), S449A/S502A/S539A (3A), or S539A. Although Dbf4-5A and Dbf4-3A dramatically reduced IR- or UV-induced slower migration shift, Dbf4-S539A partially retained the shift, suggesting that besides Ser-539, other (S/T)Q sites, such as Ser-449 and Ser-502, are also phosphorylated in vivo upon DNA damage (Fig. 3D).
ATM/ATR-mediated Dbf4 Phosphorylation Is Important for Mediating Intra-S-phase Checkpoint
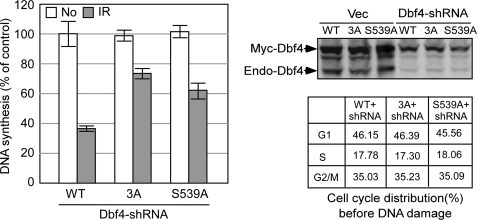
DDK is required for firing of both early and late replication origins and is an essential target of the S-phase checkpoint in yeast to suppress replication initiation (50, 51). We showed that Dbf4 is a direct target of ATM and ATR upon DNA damage. To investigate the biological function of these phosphorylation events, we expressed the Dbf4 phosphomutants Dbf4-3A (S449A/S502A/S539A) and Dbf4-S539A in U2OS cells and inactivated the endogenous Dbf4 by shRNAs. We found that the Dbf4 phosphorylation mutants exhibit radioresistant DNA synthesis with the phenotype of Dbf4-3A relatively stronger than that of Dbf4-S539A (Fig. 4). These data suggest that Dbf4 is a direct downstream target of the S-phase checkpoint in mammalian cells to suppress DNA replication in response to DNA damage, and ATM/ATR-mediated phosphorylation of Dbf4 is important for this intra-S-phase checkpoint.
FIGURE 4.
ATM/ATR-directed Dbf4 phosphorylation is important for mediating the intra-S-phase checkpoint. U2OS cells were retrovirally infected with Myc-tagged wild type Dbf4, Dbf4-3A (T449A/S502A/S539A), or Dbf4-S539A, followed by inactivation of endogenous Dbf4 with one round of shRNA retroviral infection. These cells were treated with IR (10 Gy, 1 h after), and the rate of DNA synthesis was determined. Left, DNA synthesis rate is expressed as the percentages by normalizing to the wild type sample without IR treatment. The expression of Myc-tagged wild type Dbf4, Dbf4-3A, or Dbf4-S539A and the inactivation of endogenous Dbf4 by shRNAs are revealed by anti-Dbf4 Western blot analysis (top right). The cell cycle distribution of the cell lines expressing Myc-tagged wild type Dbf4, Dbf4-3A, or Dbf4-S539A with endogenous Dbf4 inactivated by shRNAs before IR is shown (bottom right). Error bars, S.D.
To explore the possible mechanisms of how ATM-mediated phosphorylation of Dbf4 inhibits DNA replication initiation upon DNA damage, we performed co-immunoprecipitation of Dbf4 and Cdc7 in multiple cell lines, including U2OS, T98G, and IMR90. We found that the endogenous association of Dbf4 and Cdc7 remains at similar levels before and after various kinds of DNA damage (Fig. 5A) (data not shown), consistent with the findings from other groups (29, 33). These data suggest that the damage-induced Dbf4 phosphorylation by ATM and ATR does not regulate the Dbf4 and Cdc7 interaction in mammalian cells.
FIGURE 5.
DNA damage does not affect MCM2 phosphorylation by Dbf4/Cdc7 in vivo and in vitro. A, immunoprecipitation of Dbf4 was performed before (No) or after treatment of 293T cells with IR (20 Gy, 1 h after), UV (50 J/m2, 1 h after), or HU (1 mm, 24 h), and immunoprecipitates (IP) were subjected to Western blot analysis using antibodies to Dbf4 and Cdc7. B, DDK in vitro kinase assay. 293T cells were transfected with FLAG-Dbf4/Cdc7WT or FLAG-Dbf4/Cdc7KD. 48 h after transfection, cells were treated with IR (20 Gy, 1 h after), HU (1 mm, 24 h), or UV light (50 J/m2, 1 h after) or without (No). FLAG-Dbf4/Cdc7WT or FLAG-Dbf4/Cdc7KD were immunoprecipitated by anti-FLAG antibody and used for the in vitro kinase assays. Autophosphorylation of Dbf4 was performed by incubating the anti-FLAG immunoprecipitates with [γ-32P]ATP (left), and the amount of FLAG-Dbf4 and Cdc7 in the anti-FLAG immunoprecipitates is revealed by Western blot analysis. Phosphorylation of MCM2 by DDK was carried out by using purified GST-MCM2 (amino acids 1–169) fragment or GST as substrate in the DDK in vitro kinase assay (right). Phosphorylation was revealed by autoradiography (32P), and the relative protein input is shown by Coomassie Blue staining (Input). C, left, T98G cells were synchronized by arresting in G0 after serum starvation (0.1% FBS for 48 h) and releasing into medium containing 10% FBS. Cells were harvested at the indicated time points after releasing, and the cell cycle profile is shown. Chromatin fractions at each time point were isolated and immunoblotted with an antibody to MCM2. Ku70 was used as a loading control. Asy, asynchronized cell population; MCM2un, unphosphorylated MCM2; MCM2p, phosphorylated MCM2. Right, U2OS cells were infected with adenoviruses encoding green fluorescent protein (Ad-GFP), p27 (Ad-p27), or Cdc7KD (Ad-Cdc7KD). The chromatin fractions were prepared 48 h after infection and analyzed by immunoblotting with anti-MCM2 antibody. Western blotting of Ku70 was used as a loading control. D, T98G cells were synchronized as described in C and released to the medium containing 10% FBS with or without drug. Cells without drug treatment were harvested at 0, 12, 16, or 20 h after releasing. Cells with drug treatment were harvested at 40 h after releasing into HU (1 mm) or aphidicolin (Aph; 1 μg/ml)-containing medium. The chromatin fractions were prepared, Western blot analysis was performed using the antibody recognizing MCM2, and Ku70 was used as a loading control. The cell cycle profile is indicated. E, 16 h after releasing from serum starvation, T98G cells were mock-treated (No) or treated with IR (20 Gy). The chromatin fractions were isolated from cells without treatment (No) or 1, 3, or 6 h after IR treatment. Western blot analysis was performed using an antibody recognizing MCM2, and Ku70 was used as a loading control.
To further examine how ATM/ATR-mediated phosphorylation of Dbf4 contributes to the intra-S-phase checkpoint, we examined the kinase activity of DDK before and after DNA damage. We first assessed DDK autophosphorylation of Dbf4 by Cdc7. 293T cells were co-transfected with FLAG-Dbf4/Cdc7WT or FLAG-Dbf4/Cdc7KD (kinase dead, Cdc7-D196N (52)). FLAG-Dbf4/Cdc7 or FLAG-Dbf4/Cdc7KD was purified by anti-FLAG immunoprecipitation. In vitro kinase assays showed that phosphorylation of Dbf4 by Cdc7 remains at similar levels when FLAG-Dbf4/Cdc7 was purified from 293T cells before or after treatment of various kinds of DNA-damaging agents (Fig. 5B, left). Dbf4 phosphorylation was dramatically reduced when Cdc7KD was used, suggesting that in vitro Dbf4 phosphorylation is indeed mediated by Cdc7 and not by other kinases associated with Dbf4/Cdc7. We also examined the kinase activity of DDK to phosphorylate MCM2 before and after DNA damage. Because DDK-mediated MCM2 phosphorylation sites are mapped to the N terminus of MCM2 (52, 53), we used GST-MCM2 (amino acids 1–169) as an in vitro substrate. Dbf4/Cdc7-mediated phosphorylation of MCM2 (amino acids 1–169) was at comparable levels when FLAG-Dbf4/Cdc7 purified before or after DNA damage (Fig. 5B, right). Similar results were obtained when full-length GST-MCM2 was used as an in vitro substrate (data not shown). To exclude the possibility that overexpressed FLAG-Dbf4/Cdc7 is not regulated in the same manner as endogenous Dbf4/Cdc7, we used Cdc7 antibody to perform immunoprecipitation and showed that endogenous Dbf4/Cdc7 phosphorylates GST-MCM2 (amino acids 1–169) at similar levels before and after DNA damage (supplemental Fig. 1) (data not shown). These results support that DDK remains as an active kinase after DNA damage in mammalian cells, which is different from the observation in yeast (21).
In vivo, MCM2 is phosphorylated at the onset of S-phase, as revealed by fast migration of phosphorylated MCM2 species (Fig. 5C, left) (33, 52–54). It was also described that DDK-mediated phosphorylation of MCM2 is required for the initiation of DNA replication (52). We showed that inhibition of S-phase entry by overexpressing p27 suppressed MCM2 phosphorylation (Fig. 5C, left). Overexpression of Cdc7KD also abolished the cell cycle-regulated MCM2 phosphorylation, supporting the notion that DDK plays a significant role in mediating MCM2 phosphorylation during a normal cell cycle (Fig. 5C, right) (52). To monitor whether MCM2 phosphorylation is altered upon DNA damage, we synchronized T98G cells by serum starvation and released cells from G0 into the medium containing or not containing HU or aphidicolin. MCM2 phosphorylation remains at the similar levels when cells are synchronized to the G1/S transition 16 h after releasing from serum starvation in the absence of drug treatment or when released to HU- or aphidicolin-containing medium for 40 h (Fig. 5D, left). Cell cycle profile confirmed that most T98G cells are at the G1 to S transition after 16-h release from serum starvation or after HU or aphidicolin treatment (Fig. 5D, right). These studies suggest that replication stress does not induce obvious change of MCM2 phosphorylation. We also treated T98G cells with or without IR when they are at the G1/S transition (16 h after releasing) (Fig. 5E) and monitored MCM2 phosphorylation before and 1, 3, or 6 h after IR. The phosphorylation of MCM2 does not reduce after IR treatment. These data disfavor the idea that DNA damage suppresses DDK activity, leading to impaired phosphorylation of replication initiation proteins, such as MCM2, to mediate the intra-S-phase checkpoint. These data are consistent with the in vitro kinase assays indicating that DDK remains active to phosphorylate MCM2 after DNA damage.
Because the kinase activity of DDK is not altered after DNA damage, this result suggests that ATM-mediated phosphorylation of Dbf4 does not regulate the kinase activity of DDK, and the role of ATM-mediated phosphorylation of Dbf4 to suppress DNA replication in response to DNA damage is through a mechanism independent of suppressing DDK kinase activity in mammalian cells.
The Kinase Activity of Dbf4/Cdc7 Is Important for Protecting Fork Integrity
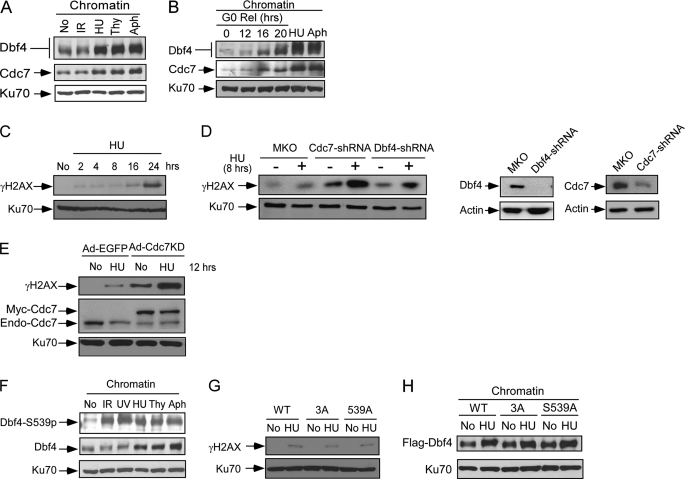
In yeast, DDK is associated with the components of the replication fork protection complex and is required for maintaining genome integrity in S-phase and G2 (24, 55). In mammalian cells, we found that treatment of HU, thymidine, and aphidicolin, but not IR, significantly increased Dbf4 and Cdc7 chromatin binding, suggesting that more Dbf4/Cdc7 is recruited to chromatin upon replication stress (Fig. 6A). To exclude the possibility that the chromatin binding enrichment of Dbf4/Cdc7 is simply due to accumulation of S-phase cells upon treatment of HU and aphidicolin, we compared chromatin binding of Dbf4 and Cdc7 at different cell cycle phases after T98G cells are released from serum starvation with that after HU and aphidicolin treatment (Fig. 6B; see Fig. 5D for cell cycle profile). Chromatin binding of Dbf4 and Cdc7 in HU- or aphidicolin-treated cells is significantly higher than that when normal cycling cells are at G1 to S transition (16 h after releasing) or in S-phase cells (20 h after releasing). These results suggest that Dbf4 and Cdc7 are enriched on chromatin upon replication stress.
FIGURE 6.
The kinase activity of Dbf4/Cdc7 is important for protecting fork integrity. A, chromatin fractions were isolated from U2OS cells after mock treatment (No), or treatment with IR (20 Gy, 1 h after), HU (1 mm, 24 h after), thymidine (Thy; 2 mm, 24 h after), or aphidicolin (Aph; 1 μg/ml, 24 h after) and were subjected to Western blot analysis with antibodies to Dbf4 and Cdc7. Western blotting of Ku70 was used as a loading control. B, T98G cells were arrested in G0 by serum starvation (0.1% FBS, 48 h) and released into medium containing 10% FBS without drug or with HU (1 mm) or Aph (1 μg/ml) for 40 h as described in the legend to Fig. 4D. Chromatin fractions were isolated and immunoblotted with antibodies to Dbf4 and Cdc7. Ku70 was used as a loading control. Cell cycle profiles are shown in Fig. 5D. C, U2OS cells were mock-treated (No) or treated with HU (1 mm), and chromatin fractions were prepared at different time points (hours) after HU treatment. The samples were immunoblotted with anti-γH2AX and Ku70 antibodies. D, U2OS cells were retrovirally infected with pMKO vector or pMKO expressing shRNAs for Dbf4 or Cdc7. Subsequently, these cells were mock-treated (No) or treated with HU (1 mm, 8 h). Chromatin fractions were prepared and immunoblotted with anti-γH2AX and Ku70 antibodies (left). The expression of Dbf4 and Cdc7 with or without infection of shRNAs was shown by Western blot analysis of cell lysates, and actin was used as a loading control (right). E, U2OS cells were infected with adenovirus encoding green fluorescent protein (Ad-GFP) or Cdc7KD (Ad-Cdc7KD). 48 h after infection, cells were mock-treated (No) or treated with HU (1 mm, 12 h). Chromatin fractions were prepared and immunoblotted with antibodies to γH2AX, Cdc7, or Ku70. F, chromatin fractions were isolated from U2OS cells after mock treatment (No) or treatment with IR (20 Gy, 1 h after), UV light (50 J/m2, 1 h after), HU (1 mm, 24 h), thymidine (Thy; 2 mm, 24 h), or aphidicolin (Aph; 1 μg/ml, 24 h). Chromatin fractions were analyzed by Western blotting using antibodies recognizing Dbf4 and phosphorylated Dbf4 at Ser-539. Western blotting of Ku70 was used as a loading control. G, U2OS cells were retrovirally infected with FLAG-tagged wild type Dbf4 (WT), the Dbf4-3A mutant (3A), or the Dbf4-S539A mutant, followed by retroviral infection with shRNAs to endogenous Dbf4. Cells were mock-treated (No) or treated with HU (1 mm, 8 h), and chromatin fractions were isolated. Western blot analysis was performed using anti-γH2AX antibodies, and Ku70 was used as a loading control. H, U2OS cells expressing FLAG-Dbf4-WT, -3A, or -S539A alleles were mock-treated (No) or treated with HU (1 mm, 24 h). Chromatin fractions were isolated, and Western blot analysis was performed using anti-FLAG antibody. Ku70 was used as a loading control.
To examine whether damage-induced Dbf4/Cdc7 chromatin binding may contribute to the maintenance of fork stability, we examined phosphorylation of H2AX (γH2AX) on chromatin by Western blot analysis. It was described that short exposure of HU or aphidicolin to normal cells does not induce significant fork collapse, and replication reassumes upon the removal of replication stress, whereas long exposure to HU (such as 24-h HU treatment) causes fork collapse and DSB formation (56). This suggests that replication forks are protected from collapsing when replication stress is below the threshold, which is important for recovery and viability upon removal of replication stress. Consistently, our studies showed that phosphorylation of H2AX (γH2AX) on chromatin is weakly detected after short exposure to HU or aphidicolin (2, 4, and 8 h) but is dramatically increased after 24 h of treatment (Fig. 6C). Importantly, we found that inactivation of Dbf4 or Cdc7 by expressing shRNAs causes significant increase of γH2AX upon short exposure to HU (1 mm, 8 h) compared with the vector-infected control cells (Fig. 6D), whereas HU-induced cell cycle arrest of these cells shows similar profile under the knockdown conditions that we used (supplemental Fig. 2). We also performed comet assays, which mainly detect DSBs under neutral conditions (57). We confirmed that DSB formation indeed increases substantially upon inactivation of Dbf4 and Cdc7 after relatively short exposure to HU by the comet assay (supplemental Fig. 3), which is in agreement with H2AX phosphorylation analysis. In addition, we consistently observed increased H2AX phosphorylation and DSB formation when Cdc7 or Dbf4 function is impaired by expressing shRNAs in normal cycling cells without HU treatment (Fig. 6D and supplemental Fig. 3, no HU). These studies suggest that Dbf4 and Cdc7 are needed for maintaining fork integrity to prevent fork collapse in a normal cell cycle and upon replication stress.
To test whether Cdc7 kinase activity is needed for fork protection, we overexpressed Cdc7KD and showed that inactivation of Cdc7 kinase activity leads to DSB formation upon replication stress, as revealed by H2AX phosphorylation and comet assays (Fig. 6E) (data not shown). These studies suggest that the kinase activity of DDK is needed for the maintenance of fork stability upon replication stress, which is consistent with the observation that DDK remains as an active kinase upon replication stress.
By using the phospho-specific antibody, we showed that chromatin-bound Dbf4 species are phosphorylated at the ATM/ATR consensus site Ser-539 upon various kinds of DNA damage (Fig. 6F). Because ATR plays a major role in damage responses to replication stress, we examined whether phosphorylation of the (S/T)Q sites Thr-449, Ser-502, and Ser-539 of Dbf4 is involved in fork protection. We used U2OS cell lines expressing Dbf4-WT, Dbf4-3A, or Dbf4-S539A with endogenous Dbf4 inactivated by expressing shRNAs. Like that of Dbf4-WT, the Dbf4-3A and Dbf4-S539A mutant cell lines do not exhibit significant H2AX phosphorylation after short exposure to HU (Fig. 6G), which is different from the results when the expression of Dbf4 or Cdc7 is silenced by shRNAs (Fig. 6D and supplemental Fig. 4). These mutants are also accumulated to chromatin to a similar extent as wild type Dbf4 upon treatment with HU and aphidicolin (Fig. 6H) (data not shown). These results suggest that ATR-mediated phosphorylation of Dbf4 at our mapped (S/T)Q sites is not necessarily needed for the recruitment of DDK to the stalled replication forks to maintain fork stability upon replication stress.
ATM/ATR-mediated Dbf4 Phosphorylation Is Important for Suppressing DNA Rereplication
DNA replication is tightly controlled by the licensing mechanism, allowing one round of DNA replication per cell cycle (58–61). Overexpression of the licensing factor Cdt1 induces massive DNA rereplication in tumor cells with an impaired S-phase checkpoint (41, 42). We demonstrated that the S-phase checkpoint is activated upon the loss of the licensing control, acting to suppress DNA rereplication (41). It is important to identify the downstream targets of the S-phase checkpoints that are used to suppress DNA rereplication.
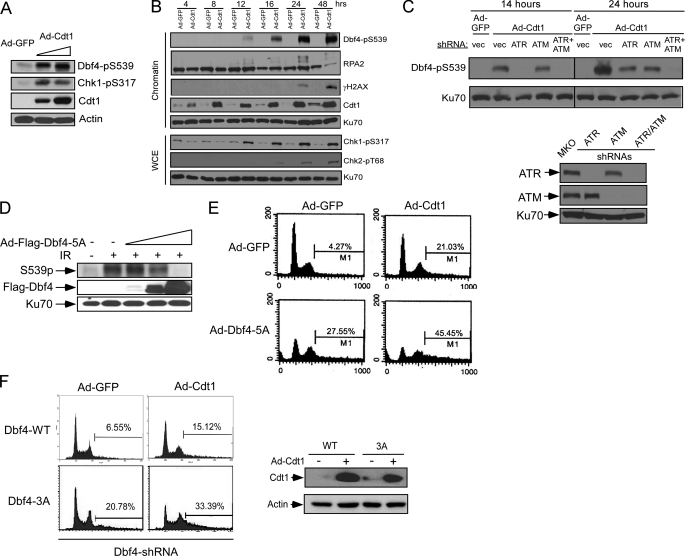
Upon Cdt1 overexpression, MCM-mediated unwinding is uncoupled from DNA synthesis, causing ssDNA accumulation and ATR/Chk1 activation (41). Subsequently, DSBs are accumulated, which activates ATM/Chk2. We demonstrated that activation of ATR at the early stage upon loss of replication licensing is critical for inhibiting DNA rereplication (41), and thus it is important to identify the downstream targets of ATR to mediate the suppression of rereplication. Dbf4 is phosphorylated at the ATM/ATR site Ser-539 upon Cdt1 overexpression (Fig. 7A). We further showed that Ser-539 is phosphorylated at the early time points after Cdt1 overexpression when Chk1 is phosphorylated by ATR prior to the phosphorylation of Chk2 by ATM and the phosphorylation of H2AX when DSBs are formed (Fig. 7B) (41). These data suggest that upon loss of replication licensing control, Dbf4 is phosphorylated when ATR is activated due to the accumulation of ssDNA before DSBs are generated. We further demonstrated that Cdt1 overexpression-induced Dbf4 phosphorylation is dependent on ATR at the early points when Cdt1 is overexpressed but depends on both ATM and ATR at the later time points when DSBs are formed (Fig. 7C). These results suggest that upon loss of the licensing control, S-phase checkpoint-mediated Dbf4 phosphorylation is an early response that is initially induced at the stage when DNA unwinding is uncoupled from DNA synthesis prior to DSB formation. Dbf4 is further phosphorylated by ATM when DSBs are formed at a relatively late stage of rereplication.
FIGURE 7.
ATM/ATR-dependent Dbf4 phosphorylation is important for suppressing DNA rereplication. A, whole cell lysates were prepared from U2OS cells infected with Ad-GFP or Ad-Cdt1 using two different viral titers for 48 h (5 × 107 or 1 × 108 pfu/ml). The lysates were analyzed by Western blotting using the indicated antibodies with actin as a loading control. B, U2OS cells were infected with Ad-GFP or Ad-Cdt1 (5 × 107 pfu/ml), and whole cell lysates and chromatin fractions were prepared at different time points after infection. The prepared lysates and chromatin fractions were immunoblotted with the indicated antibodies, and Ku70 was used as a loading control. C, U2OS cells were retrovirally infected with vector pMKO (vec) or pMKO encoding shRNAs against ATR or ATM. These cells were subsequently infected with Ad-GFP or Ad-Cdt1 (5 × 107 pfu/ml), and whole cell lysates were prepared at 14 or 24 h after infection. Western blot analysis was performed using the antibodies recognizing phosphorylated Ser-539 of Dbf4 or Ku70 (top). The expression of ATR or ATM was examined by Western blot analysis (bottom). D, U2OS cells were infected with Ad-GFP or Ad-Dbf4–5A (S226A/T265A/T449A/S502A/S539A) using increased viral titers (5 × 107, 5 × 108, or 1 × 109 pfu/ml) for 48 h and subsequently treated with IR (20 Gy, 1 h after, +) or mock treatment (−). Western blot analysis was performed using antibodies specific for Dbf4-pS539 or FLAG tag. Ku70 was used as a loading control. E, U2OS cells were infected with Ad-GFP or Ad-Dbf4-5A (5 × 108 pfu/ml) for 24 h, followed by infection using Ad-GFP or Ad-Cdt1 (5 × 107 pfu/ml) for another 48 h. Infected cells were collected for FACS analysis. F, the expression of endogenous Dbf4 in U2OS cells expressing wild type Dbf4 or Dbf4-3A was suppressed by two rounds of shRNA retroviral infection, followed by adenoviral infection with Ad-GFP or Ad-Cdt1 (5 × 107 pfu/ml). FACS analysis was performed 48 h after adenoviral infection (left). Cdt1 expression was shown by anti-Cdt1 Western blot analysis with actin as a loading control (right).
The observation that Dbf4 is phosphorylated by ATR at the early stage upon loss of the licensing control prompted us to examine whether S-phase checkpoint-induced Dbf4 phosphorylation acts to suppress DNA rereplication. We overexpressed the Dbf4 phosphomutant (Dbf4-5A) by adenoviral infection prior to IR in U2OS cells. IR-induced phosphorylation of Dbf4 at Ser-539 was impaired, suggesting that overexpressed Dbf4-5A exhibits a dominant negative effect to suppress damage-induced phosphorylation of endogenous Dbf4 at Ser-539 (Fig. 7D). We further demonstrated that overexpression of Dbf4-5A enhances Cdt1-induced rereplication (Fig. 7E, compare Ad-GFP/Ad-Cdt1 and Ad-Dbf4–5A/Ad-Cdt1). This suggests that ATM/ATR-mediated phosphorylation of Dbf4 is important for suppression of DNA rereplication upon loss of the licensing control. We also observed that rereplication is induced when Dbf4-5A was overexpressed alone in the absence of Cdt1 (Fig. 7E, compare Ad-GFP/Ad-GFP and Ad-Dbf4–5A/Ad-GFP), suggesting that checkpoint-mediated phosphorylation of Dbf4 also acts to prevent rereplication during a normal cell cycle. In addition, we used the stable cell lines expressing Dbf4 wild type and Dbf4-3A with endogenous Dbf4 inactivated by retroviral shRNA infection for analyzing DNA rereplication. Adenovirus-mediated Cdt1 overexpression induces higher rereplication in the cell lines expressing Dbf4-3A than in those expressing wild type Dbf4 (Fig. 7F). Rereplication in the Dbf4-3A mutant cell line in the absence of Cdt1 overexpression was also observed. These data suggest that Dbf4 is an important target of the S-phase checkpoint to suppress DNA rereplication through ATM/ATR-mediated phosphorylation.
DISCUSSION
DDK plays essential roles in initiating DNA replication at origins during each cell cycle (3, 62). Studies in yeast provide substantial evidence that DDK also participates in the S-phase checkpoint and fork protection upon DNA damage and replication stress (13, 50). The S-phase checkpoint function is conserved in higher eukaryotes (14), but it is controversial whether DDK is an important target for the intra-S-phase checkpoint to suppress replication initiation in mammalian cells. The exact function of DDK in fork protection in mammalian cells also remains unclear.
Recently, it was described that in Xenopus cell-free systems and in mammalian cells, DDK plays an important role in triggering replication reinitiation during the S-phase checkpoint recovery (28). In this study, we demonstrated that DDK also acts downstream of the checkpoints as a direct target of ATM and ATR to mediate the intra-S-phase checkpoint response. We observed that Dbf4 is hyperphosphorylated upon DNA damage or replication stress, and these damage-induced hyperphosphorylation events depend on ATM and/or ATR activities. ATM and ATR directly phosphorylate Dbf4 in vitro, and mutating the ATM/ATR conserved phosphorylation (S/T)Q sites dramatically reduces damage-induced hyperphosphorylation of Dbf4 in vivo, supporting the notion that Dbf4 is directly phosphorylated by ATM and/or ATR upon the activation of checkpoints. This is analogous to yeast, where Dfp1/Dbf4 is directly targeted by the S-phase checkpoint through Cds1/Rad53 (Chk2 ortholog)-mediated phosphorylation when cells are treated with HU (19–21). These studies suggest that, as in the lower organisms, Dbf4 in mammalian cells is also a downstream target of the intra-S-phase checkpoint to suppress replication initiation of late origins in an S-phase checkpoint response.
It was shown that the direct phosphorylation of Dbf4 by Rad53 in yeast inhibits the kinase activity of DDK, thereby preventing late origins from firing to mediate the S-phase checkpoint (63). Using Xenopus cell-free extracts, different results were obtained as to whether DDK activity is inhibited upon DNA damage (26, 28). We demonstrated by both in vivo and in vitro assays that in mammalian cells, checkpoint-mediated phosphorylation of Dbf4 neither induces disassociation of Dbf4 and Cdc7 nor inhibits the kinase activity of the DDK complex for Dbf4 autophosphorylation and MCM2 phosphorylation. These results suggest that inhibition of the intra-S-phase checkpoint by ATM/ATR-directed phosphorylation of Dbf4 is not through modulating the kinase activity of DDK. Currently, the mechanisms involved in this ATM/ATR-DDK-mediated S-phase checkpoint pathway remain unclear. One possibility is that phosphorylation of the ATM/ATR sites on Dbf4 provides docking sites to recruit one or more replication-inhibitory regulators to the proximity of replication origins to prevent replication initiation. Alternatively, the phosphorylation events may prevent the interaction of certain replication initiation proteins with DDK and thus prevent DNA replication initiation. It is also possible that although DDK remains active upon DNA damage, ATM/ATR-mediated phosphorylation may selectively alter its interaction with some of its substrates that are specifically involved in replication initiation. Consequently, the DDK-mediated phosphorylation of these substrates is reduced due to the disruption of the interaction of DDK with these substrates, although DDK retains full kinase activity upon DNA damage. It will be interesting if such substrates can be identified. Understanding the mechanisms underlying the role of DDK to mediate the S-phase checkpoint in mammalian cells is of great importance.
Utilizing a mechanism to promote the intra-S-phase checkpoint through the ATM/ATR-DDK pathway without inhibiting DDK kinase activity may be important in several aspects of DNA damage responses for the maintenance of genome integrity. First, we showed that DDK as well as its kinase activity is needed for fork protection upon replication stress. The chromatin loading of DDK is increased when replication forks are stalled by HU and aphidicolin treatment. Inhibition of the expression of Dbf4 or Cdc7 by shRNAs leads to DSB accumulation under replication stress, and the kinase activity of Cdc7 appears important for protecting fork stability. This is consistent with the findings that certain yeast Cdc7 and Dbf4 mutants, although proficient in cell growth and intra-S-phase checkpoint, are hypersensitive to HU and other damaging agents and exhibit chromosomal instability and nuclear fragmentation following replication stress (20, 21, 64). It was also observed that DDK binds to Swi1/Tof1 and Swi3/Csm3, the components of the fork protection complex, and its kinase activity is required for induction and maintenance of the hyperphosphorylation of another fork protection protein, Mrc1, upon replication fork block (24, 55, 65). Similarly, in mammalian cells, recent studies showed that Cdc7 interacts with the fork-stabilizing factor Claspin and that Cdc7-mediated phosphorylation of Claspin is needed for ATR-mediated phosphorylation upon replication stress (66, 67). Second, the kinase activity of DDK may be needed to attenuate checkpoint signaling and promote DNA repair. Recent studies showed that in fission yeast, DDK phosphorylates Rad9, a checkpoint protein, in response to replication-induced DNA damage, and this phosphorylation promotes the release of Rad9 from damage sites (68). The Rad9 mutants defective in DDK-mediated phosphorylation are impaired in damage-induced foci formation and exhibit decreased viability after replication stress, suggesting a defect in DNA repair. Third, the kinase activity of DDK is expected to be important for activating replication initiation during S-phase recovery (28). Upon removal of replication stress and/or completion of DNA repair in S-phase, reactivation of DNA replication requires DDK-mediated phosphorylation of critical replication initiation proteins; thus, DDK and its kinase activity will be important for replication restart.
DNA replication occurs once and only once per cell cycle, which is ensured by the replication licensing control mechanism. Upon loss of the replication licensing control, DNA rereplication is induced, which would inevitably lead to genome instability. We showed that the S-phase checkpoint is activated to suppress DNA rereplication caused by overexpression of the licensing protein Cdt1 (41). In this study, we found that Dbf4 is phosphorylated by ATR at the early time points when Cdt1 is overexpressed, and this phosphorylation is important for suppressing DNA rereplication. We propose that DDK is an important target of the S-phase checkpoint to inhibit DNA rereplication, and the mechanism used in this response is similar to that of the intra-S-phase checkpoint acting to suppress DNA replication initiation after Dbf4 is phosphorylated by ATM/ATR. However, although ATM plays a predominant role to phosphorylate Dbf4 in response to IR to mediate the intra-S-phase checkpoint, ATR is more important for phosphorylating Dbf4 at the early stage upon loss of the licensing control, promoting inhibition of DNA rereplication (41). ATM is activated relatively late to phosphorylate Dbf4 when DSBs are generated during rereplication .
Our studies suggest that DDK is an important target of the intra-S-phase checkpoint through direct phosphorylation of Dbf4 by ATM/ATR. However, we also need to note that DDK is not the only critical target for the intra-S-phase checkpoint because other replication proteins, such as RPA and MCM proteins, are also found involved in the intra-S-phase checkpoint (41, 69, 70). It is thus important to understand how these replication initiation proteins are modified upon checkpoint activation and function together to effectively promote the intra-S-phase checkpoint. Our studies also suggest that the mechanisms underlying the function of DDK in mediating the intra-S-phase checkpoint and in the fork protection are different. Whereas ATM/ATR-directed phosphorylation of Dbf4 regulates the intra-S-phase checkpoint response through a mechanism without affecting its kinase activity, the fork protection mechanism of DDK requires its kinase activity. By using these different mechanisms, DDK maintains its kinase activity for mediating fork protection, facilitating DNA repair, and promoting checkpoint recovery in response to replication stress while still being a critical target for inhibiting replication initiation upon the activation of S-phase checkpoint. Our studies provide a mechanistic basis for further understanding the coordination of the regulation of different DDK function to mediate the S-phase checkpoint and to maintain genome stability in S-phase.
Supplementary Material
Acknowledgments
We thank S. Reed (The Scripps Research Institute) for kindly providing p27 adenovirus and W. Jiang (Sanford-Burnham Medical Research Institute) for kindly providing Dbf4 antibody. We also thank T. Tsuji (Sanford-Burnham Medical Research Institute) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA102361, GM080677, CA140972, and CA102361–07S1 (to X. W.) and Training Grant DK007022-30 (to L. T.). This work was also supported by National Science Council Grant NSC 98-2311-B-037-001-MY3, National Health Research Institutes Grant CA-100-PP-08, and Department of Health Grant DOH99-TD-C-111-004), Taiwan, (to A. Y.-L. L.). This work was also supported by a grant from the Takeda Science Foundation and Grant-in-Aid for Young Scientists (B) 17790269 from the Japan Society for the Promotion of Science (to T. C.).

This article contains supplemental Figs. 1–4.
- DDK
- Dbf4-dependent kinase
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3-related
- DSB
- double-stranded break
- RPA
- replication protein A
- HU
- hydroxyurea
- Gy
- grays
- Ad
- adenovirus.
REFERENCES
- 1. Kelly T. J., Brown G. W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem. 69, 829–880 [DOI] [PubMed] [Google Scholar]
- 2. Bell S. P., Dutta A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 3. Lei M., Tye B. K. (2001) Initiating DNA synthesis. From recruiting to activating the MCM complex. J. Cell Sci. 114, 1447–1454 [DOI] [PubMed] [Google Scholar]
- 4. Labib K. (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston L. H., Masai H., Sugino A. (2000) A Cdc7p-Dbf4p protein kinase activity is conserved from yeast to humans. Prog. Cell Cycle Res. 4, 61–69 [DOI] [PubMed] [Google Scholar]
- 6. Jackson A. L., Pahl P. M., Harrison K., Rosamond J., Sclafani R. A. (1993) Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell Biol. 13, 2899–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang W., McDonald D., Hope T. J., Hunter T. (1999) Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 18, 5703–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoon H. J., Loo S., Campbell J. L. (1993) Regulation of Saccharomyces cerevisiae CDC7 function during the cell cycle. Mol. Biol. Cell 4, 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng L., Collyer T., Hardy C. F. (1999) Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell Biol. 19, 4270–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pasero P., Duncker B. P., Schwob E., Gasser S. M. (1999) A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 13, 2159–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donaldson A. D., Fangman W. L., Brewer B. J. (1998) Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 12, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bousset K., Diffley J. F. (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 12, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jares P., Donaldson A., Blow J. J. (2000) The Cdc7/Dbf4 protein kinase. Target of the S phase checkpoint? EMBO Rep. 1, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartek J., Lukas C., Lukas J. (2004) Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5, 792–804 [DOI] [PubMed] [Google Scholar]
- 15. Aguilera A., Gómez-González B. (2008) Genome instability. A mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217 [DOI] [PubMed] [Google Scholar]
- 16. Abraham R. T. (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177–2196 [DOI] [PubMed] [Google Scholar]
- 17. Cimprich K. A., Cortez D. (2008) ATR. An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J. H., Paull T. T. (2007) Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26, 7741–7748 [DOI] [PubMed] [Google Scholar]
- 19. Brown G. W., Kelly T. J. (1999) Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc. Natl. Acad. Sci. U.S.A. 96, 8443–8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snaith H. A., Brown G. W., Forsburg S. L. (2000) Schizosaccharomyces pombe Hsk1p is a potential cds1p target required for genome integrity. Mol. Cell Biol. 20, 7922–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weinreich M., Stillman B. (1999) Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18, 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez-Mosqueda J., Maas N. L., Jonsson Z. O., Defazio-Eli L. G., Wohlschlegel J., Toczyski D. P. (2010) Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zegerman P., Diffley J. F. (2010) Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467, 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumoto S., Ogino K., Noguchi E., Russell P., Masai H. (2005) Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J. Biol. Chem. 280, 42536–42542 [DOI] [PubMed] [Google Scholar]
- 25. Takeda T., Ogino K., Matsui E., Cho M. K., Kumagai H., Miyake T., Arai K., Masai H. (1999) A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell Biol. 19, 5535–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Costanzo V., Shechter D., Lupardus P. J., Cimprich K. A., Gottesman M., Gautier J. (2003) An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11, 203–213 [DOI] [PubMed] [Google Scholar]
- 27. Dierov J., Dierova R., Carroll M. (2004) BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell 5, 275–285 [DOI] [PubMed] [Google Scholar]
- 28. Tsuji T., Lau E., Chiang G. G., Jiang W. (2008) The role of Dbf4/Drf1-dependent kinase Cdc7 in DNA damage checkpoint control. Mol. Cell 32, 862–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heffernan T. P., Unsal-Kaçmaz K., Heinloth A. N., Simpson D. A., Paules R. S., Sancar A., Cordeiro-Stone M., Kaufmann W. K. (2007) Cdc7-Dbf4 and the human S checkpoint response to UVC. J. Biol. Chem. 282, 9458–9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu P., Barkley L. R., Day T., Bi X., Slater D. M., Alexandrow M. G., Nasheuer H. P., Vaziri C. (2006) The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J. Biol. Chem. 281, 30631–30644 [DOI] [PubMed] [Google Scholar]
- 31. Petersen P., Chou D. M., You Z., Hunter T., Walter J. C., Walter G. (2006) Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2- and Cdc7-independent DNA damage checkpoint. Mol. Cell Biol. 26, 1997–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silva T., Bradley R. H., Gao Y., Coue M. (2006) Xenopus CDC7/DRF1 complex is required for the initiation of DNA replication. J. Biol. Chem. 281, 11569–11576 [DOI] [PubMed] [Google Scholar]
- 33. Tenca P., Brotherton D., Montagnoli A., Rainoldi S., Albanese C., Santocanale C. (2007) Cdc7 is an active kinase in human cancer cells undergoing replication stress. J. Biol. Chem. 282, 208–215 [DOI] [PubMed] [Google Scholar]
- 34. Takahashi T. S., Walter J. C. (2005) Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 19, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cliby W. A., Roberts C. J., Cimprich K. A., Stringer C. M., Lamb J. R., Schreiber S. L., Friend S. H. (1998) Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17, 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nghiem P., Park P. K., Kim Ys, Y. S., Desai B. N., Schreiber S. L. (2002) ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J. Biol. Chem. 277, 4428–4434 [DOI] [PubMed] [Google Scholar]
- 37. Lee A. Y., Liu E., Wu X. (2007) The Mre11/Rad50/Nbs1 complex plays an important role in the prevention of DNA rereplication in mammalian cells. J. Biol. Chem. 282, 32243–32255 [DOI] [PubMed] [Google Scholar]
- 38. Masutomi K., Yu E. Y., Khurts S., Ben-Porath I., Currier J. L., Metz G. B., Brooks M. W., Kaneko S., Murakami S., DeCaprio J. A., Weinberg R. A., Stewart S. A., Hahn W. C. (2003) Telomerase maintains telomere structure in normal human cells. Cell 114, 241–253 [DOI] [PubMed] [Google Scholar]
- 39. Wu X., Ranganathan V., Weisman D. S., Heine W. F., Ciccone D. N., O'Neill T. B., Crick K. E., Pierce K. A., Lane W. S., Rathbun G., Livingston D. M., Weaver D. T. (2000) ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405, 477–482 [DOI] [PubMed] [Google Scholar]
- 40. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu E., Lee A. Y., Chiba T., Olson E., Sun P., Wu X. (2007) The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J. Cell Biol. 179, 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D. S., Dutta A. (2003) A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11, 997–1008 [DOI] [PubMed] [Google Scholar]
- 43. Li X., Zhao Q., Liao R., Sun P., Wu X. (2003) The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278, 30854–30858 [DOI] [PubMed] [Google Scholar]
- 44. Olson E., Nievera C. J., Liu E., Lee A. Y., Chen L., Wu X. (2007) The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol. Cell Biol. 27, 6053–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumagai H., Sato N., Yamada M., Mahony D., Seghezzi W., Lees E., Arai K., Masai H. (1999) A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell Biol. 19, 5083–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sato N., Sato M., Nakayama M., Saitoh R., Arai K., Masai H. (2003) Cell cycle regulation of chromatin binding and nuclear localization of human Cdc7-ASK kinase complex. Genes Cells 8, 451–463 [DOI] [PubMed] [Google Scholar]
- 47. Ziv Y., Bar-Shira A., Pecker I., Russell P., Jorgensen T. J., Tsarfati I., Shiloh Y. (1997) Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 15, 159–167 [DOI] [PubMed] [Google Scholar]
- 48. O'Neill T., Dwyer A. J., Ziv Y., Chan D. W., Lees-Miller S. P., Abraham R. H., Lai J. H., Hill D., Shiloh Y., Cantley L. C., Rathbun G. A. (2000) Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J. Biol. Chem. 275, 22719–22727 [DOI] [PubMed] [Google Scholar]
- 49. Kim S. T., Lim D. S., Canman C. E., Kastan M. B. (1999) Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274, 37538–37543 [DOI] [PubMed] [Google Scholar]
- 50. Duncker B. P., Brown G. W. (2003) Cdc7 kinases (DDKs) and checkpoint responses. Lessons from two yeasts. Mutat. Res. 532, 21–27 [DOI] [PubMed] [Google Scholar]
- 51. Sclafani R. A. (2000) Cdc7p-Dbf4p becomes famous in the cell cycle. J. Cell Sci. 113, 2111–2117 [DOI] [PubMed] [Google Scholar]
- 52. Tsuji T., Ficarro S. B., Jiang W. (2006) Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol. Biol. Cell 17, 4459–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. (2006) Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 281, 10281–10290 [DOI] [PubMed] [Google Scholar]
- 54. Masai H., Matsui E., You Z., Ishimi Y., Tamai K., Arai K. (2000) Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 by Cdks. J. Biol. Chem. 275, 29042–29052 [DOI] [PubMed] [Google Scholar]
- 55. Sommariva E., Pellny T. K., Karahan N., Kumar S., Huberman J. A., Dalgaard J. Z. (2005) Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol. Cell. Biol. 25, 2770–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petermann E., Orta M. L., Issaeva N., Schultz N., Helleday T. (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guo W. Y., Pan H. C., Wu H. M., Hsieh W. A., Tsai M. H., Chow Y. M., Chung W. Y., Shiau C. Y., Chen S. K., Chang W. P. (2004) Individuals' leukocyte DNA double-strand break repair as an indicator of radiosurgery responses for cerebral arteriovenous malformations. J. Radiat. Res. 45, 269–274 [DOI] [PubMed] [Google Scholar]
- 58. Diffley J. F. (1996) Once and only once upon a time. Specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 10, 2819–2830 [DOI] [PubMed] [Google Scholar]
- 59. Arias E. E., Walter J. C. (2007) Strength in numbers. Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21, 497–518 [DOI] [PubMed] [Google Scholar]
- 60. Nishitani H., Lygerou Z. (2004) DNA replication licensing. Front. Biosci. 9, 2115–2132 [DOI] [PubMed] [Google Scholar]
- 61. Truong L. N., Wu X. (2011) Prevention of DNA re-replication in eukaryotic cells. J Mol. Cell Biol. 3, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Masai H., Arai K. (2002) Cdc7 kinase complex. A key regulator in the initiation of DNA replication. J. Cell Physiol. 190, 287–296 [DOI] [PubMed] [Google Scholar]
- 63. Kihara M., Nakai W., Asano S., Suzuki A., Kitada K., Kawasaki Y., Johnston L. H., Sugino A. (2000) Characterization of the yeast Cdc7p/Dbf4p complex purified from insect cells. Its protein kinase activity is regulated by Rad53p. J. Biol. Chem. 275, 35051–35062 [DOI] [PubMed] [Google Scholar]
- 64. Fung A. D., Ou J., Bueler S., Brown G. W. (2002) A conserved domain of Schizosaccharomyces pombe dfp1(+) is uniquely required for chromosome stability following alkylation damage during S phase. Mol. Cell Biol. 22, 4477–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Matsumoto S., Shimmoto M., Kakusho N., Yokoyama M., Kanoh Y., Hayano M., Russell P., Masai H. (2010) Hsk1 kinase and Cdc45 regulate replication stress-induced checkpoint responses in fission yeast. Cell Cycle 9, 4627–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Uno S., Masai H. (2011) Efficient expression and purification of human replication fork-stabilizing factor, Claspin, from mammalian cells. DNA binding activity and novel protein interactions. Genes Cells 16, 842–856 [DOI] [PubMed] [Google Scholar]
- 67. Kim J. M., Kakusho N., Yamada M., Kanoh Y., Takemoto N., Masai H. (2008) Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene 27, 3475–3482 [DOI] [PubMed] [Google Scholar]
- 68. Furuya K., Miyabe I., Tsutsui Y., Paderi F., Kakusho N., Masai H., Niki H., Carr A. M. (2010) DDK phosphorylates checkpoint clamp component Rad9 and promotes its release from damaged chromatin. Mol. Cell 40, 606–618 [DOI] [PubMed] [Google Scholar]
- 69. Olson E., Nievera C. J., Klimovich V., Fanning E., Wu X. (2006) RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 281, 39517–39533 [DOI] [PubMed] [Google Scholar]
- 70. Cortez D., Glick G., Elledge S. J. (2004) Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 101, 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.