Background: Bacterial elongation factor P (EF-P) was proposed to undergo unique post-translational modification by YjeA and YjeK.
Results: We identified β-lysylation in native EF-P and show that β-lysyl-EF-P is the active form.
Conclusion: β-Lysylation occurs in EF-P in vivo and is required for activity.
Significance: β-Lysylation of EF-P may influence this factor binding to ribosome and regulate bacterial protein synthesis.
Keywords: EF-P, Post-translational Modification, Lysylation, YjeA, YjeK, eIF5A, Hypusine
Abstract
Bacterial elongation factor P (EF-P) is the ortholog of archaeal and eukaryotic initiation factor 5A (eIF5A). EF-P shares sequence homology and crystal structure with eIF5A, but unlike eIF5A, EF-P does not undergo hypusine modification. Recently, two bacterial genes, yjeA and yjeK, encoding truncated homologs of class II lysyl-tRNA synthetase and of lysine-2,3-aminomutase, respectively, have been implicated in the modification of EF-P to convert a specific lysine to a hypothetical β-lysyl-lysine. Here we present biochemical evidence for β-lysyl-lysine modification in Escherichia coli EF-P and for its role in EF-P activity by characterizing native and recombinant EF-P proteins for their modification status and activity in vitro. Mass spectrometric analyses confirmed the lysyl modification at lysine 34 in native and recombinant EF-P proteins. The β-lysyl-lysine isopeptide was identified in the exhaustive Pronase digests of native EF-P and recombinant EF-P isolated from E. coli coexpressing EF-P, YjeA, and YjeK but not in the digests of proteins derived from the vectors encoding EF-P alone or EF-P together with YjeA, indicating that both enzymes, YjeA and YjeK, are required for β-lysylation of EF-P. Endogenous EF-P as well as the recombinant EF-P preparation containing β-lysyl-EF-P stimulated N-formyl-methionyl-puromycin synthesis ∼4-fold over the preparations containing unmodified EF-P and/or α-lysyl-EF-P. The mutant lacking the modification site lysine (K34A) was inactive. This is the first report of biochemical evidence for the β-lysylation of EF-P in vivo and the requirement for this modification for the activity of EF-P.
Introduction
In all cells, protein synthesis depends on several protein factors that cyclically associate with and dissociate from the ribosome. In bacteria, the factors that promote protein synthesis include initiation factors (IF3-1, IF-2, and IF-3), elongation factors (EF-Tu, EF-Ts, EF-G, and EF-P), and termination factors (RF-1, RF-2, RF-3, and RRF) (1, 2). The eubacterial EF-P stimulates formation of the first peptide bond of N-formylmethionyl-initiated peptides (3) or N-formylmethionyl-puromycin (4) and also, to a limited extent, the translation of a natural mRNA (5) or of poly(U)-directed polyphenylalanine synthesis primed by N-acetylphenylalanyl-tRNA (6). The stimulatory effects of EF-P on the first peptide bond formation seem to vary depending on the second aminoacyl-tRNA acceptor, suggesting that it may have differential effects on different mRNAs (3). The crystal structure of a Thermus thermophilus 70 S ribosome complex with EF-P, the initiator N-formyl-methionyl-tRNAi, and a short mRNA revealed that EF-P binds between the peptidyl-tRNA and the exit tRNA binding sites, further supporting a role for EF-P in the synthesis of the first peptide bond (7).
EF-P is the bacterial ortholog of eukaryotic/archaeal translation initiation factor 5A (eIF5A/aIF5A) (8, 9), and the efp gene is present in all bacteria. Although eIF5A is a putative translation initiation factor (10–12), it has also been reported to be involved in translation elongation (13, 14). eIF5A consists of two domains, and a specific lysine residue (Lys-50 for the human protein) in the N-terminal domain undergoes post-translational modification to hypusine (Nϵ-(4-amino-2-hydroxybutyl)lysine). This modification occurs in two steps. Deoxyhypusine synthase (15) first catalyzes the transfer of the aminobutyl moiety of the polyamine spermidine to a specific lysine residue to form deoxyhypusine, which is subsequently hydroxylated by deoxyhypusine hydroxylase (16). Hypusine is essential for the activity of this factor, and deoxyhypusine/hypusine is absolutely required for archaeal and eukaryotic cell proliferation (9, 17, 18).
EF-P has a tRNA-like L shape with three domains. The first two domains show strong amino acid sequence homology to eIF5A, whereas domains II and III are repeats of similar sequence, indicative of partial gene duplication. The crystal structure of EF-P domains I and II (Protein Data Bank entry 1UEB (19)), is superimposable on the two domains of aIF5A (Protein Data Bank entry 1EIF (20)) and of eIF5A (Protein Data Bank entry 3CPF) (21). Despite conservation of the structures of EF-P, aIF5A, and eIF5A, EF-P does not undergo hypusine modification, and bacteria lack the two hypusine modification enzymes. Although the deoxyhypusine/hypusine modification site is a strictly conserved lysine in aIF5A and eIF5A, conservation is relaxed in EF-P to allow Lys, Arg, Asn, or Gln (22). Interestingly, mass spectrometric analyses of the tryptic peptides of Escherichia coli native EF-P suggested a novel post-translational modification of EF-P (different from the hypusine modification) by the addition of a mass of 144 Da at Lys-34 (23), the site corresponding to the hypusine modification site of eIF5A.
Based on a combination of comparative genomics, literature mining, and phylogenetic analyses, two genes closely linked to the efp gene, yjeA and yjeK, have been identified in the genome of a variety of eubacteria (22). It was predicted that in a subset of bacterial species that contain EF-P with the conserved target Lys, YjeA and YjeK protein families are involved in the EF-P modification. YjeA (GenX or PoxA) proteins are members of the class II lysyl-tRNA synthetase (LysRS) family that contain the catalytic domain of LysRS (24, 25) but that lack the N terminus tRNA anticodon binding domain. The sequence alignment of YjeA with E. coli LysRS shows conservation of the active site amino acids involved in the binding of lysine and ATP (24). Indeed, YjeA was shown to activate lysine to form AMP-lysine but was devoid of Lys-tRNA lysylating activity (24). YjeK is a truncated form of lysine-2,3-aminomutase lacking the C-terminal multimerization domain. E. coli YjeK catalyzes the conversion of l-lysine to d-β-lysine (26) but at a low efficiency.
The cellular functions of the YjeA and YjeK families of enzymes remained obscure until recent reports of their roles in the common metabolic pathways critical for Salmonella virulence and resistance to various stresses, including acids, detergents, and several classes of antibiotics (27). The E. coli yjeA and yjeK mutant strains displayed a similar growth defect as the efp mutant (25), providing strong evidence for the importance of the EF-P modification in vivo. Furthermore, it was demonstrated that recombinant YjeA could indeed catalyze incorporation of radiolabeled lysine into the recombinant EF-P protein in an ATP-dependent manner in vitro (25, 27). Two pathways of the EF-P modification were proposed (22). YjeA may first catalyze the ATP-dependent activation of lysine to AMP-lysine, from which the lysyl residue is transferred to the ϵ-amino group of a specific lysine (Lys-34 for E. coli EF-P) to form an α-lysyl-EF-P, which then is converted by YjeK to a β-lysyl-EF-P. Alternatively, YjeK may first act on l-lysine to convert it to d-β-lysine, which then is activated by YjeA to AMP-d-β-lysine for the transfer of the β-lysyl moiety to the specific lysine residue of EF-P. After completion of this study, it was reported that d-β-lysine is a superior substrate for YjeA in vitro compared with α-lysine and l-β-lysine and that d-β-lysine can complement a growth defect of a yjeK mutant of Salmonella, lending support for the latter pathway (28).
In E. coli, coexpression of EF-P with YjeA, or with YjeA and YjeK resulted in partial or full lysylation as judged from an increase in EF-P molecular mass by 128 Da (25). Although it was inferred that YjeA and YjeK modify EF-P to form β-lysyl-EF-P, direct biochemical evidence has not been reported for such β-lysylation in native EF-P; nor has the effect of β-lysylation on EF-P activity been demonstrated. In this report, we provide definitive evidence, by mass spectrometric analyses and by identification of β-lysyl-lysine peptide in the Pronase digests, for the β-lysylation of EF-P at lysine 34 in the native EF-P and also in the recombinant EF-P produced in E. coli by co-expression of EF-P, YjeA, and YjeK. Furthermore, we demonstrate that β-lysylation is required for the activity of EF-P, by comparing the activity of native EF-P with that of the recombinant proteins, unmodified EF-P and α- and β-lysylated EF-P.
EXPERIMENTAL PROCEDURES
Materials and Strains
The polycistronic expression vector pST39 and transfer vector pET3aTr (29) were kindly provided by Dr. Song Tan (Pennsylvania State University). l-[4,5-3H]Lysine (105.4 Ci/mmol) was purchased from PerkinElmer Life Sciences. Chemically synthesized β-lysyl-lysine, d-β-lysine, and l-β-lysine were kind gifts from Dr. J. E. Folk (NIDA, National Institutes of Health). l-β-Lysine was also derived from acid hydrolysis of Viomycin (Tocris Bioscience) and showed the same ion exchange chromatic properties as l- d-β-lysine synthesized by Dr. J. E. Folk. 5-Hydroxy-d-β-lysine was kindly provided by Dr. Yoshio Hayashi (Kyoto Pharmaceutical University, Kyoto, Japan). The E. coli efp disruption strain (JW4107) and the yjeA disruption strain (JW4116) and their parent wild type (BW25113) were obtained from the Keio collection (National BioResource Project, Japan), and colonies were screened for isolation of a single colony with disruption of each gene by PCR. The Probond purification kit, precast Tris-glycine and NuPAGE (BisTris) gels, electrophoresis buffers, and Simply Blue staining solution were from Invitrogen; the ECL Plus Western blotting detection system, phenyl-Sepharose, and Q Sepharose, DEAE-Sephacel, Sephacryl S-200, and SP Sepharose were from GE Healthcare. Hydroxyapatite was from Calbiochem. Other chemicals and reagents were from Sigma-Aldrich. Detection of 3H radioactivity was with a Beckman scintillation counter LS6000LC.
Methods
Isolation and Subcloning of E. coli efp, yjeA, and yjeK
DNA for the genes efp, yjeK (kamA), and yjeA (genX, poxA) was amplified from E. coli K-16 MG1655 by PCR with gene-specific primer pairs. Amplification of efp by PCR was achieved using HotTub polymerase (GE Healthcare Life Sciences), and amplification of yjeA and yjeK was achieved by PCR with Phusion polymerase (Finnzymes) in the supplied buffers supplemented with 5% DMSO. The PCR fragments after digestion with NcoI and BamHI (efp and yjeA) or BspHI and BamHI (yjeK) were ligated into pET30a(+) (Novagen) to create pBL1, pBL4, and pBL5, respectively. The EF-P Lys-34 to Ala mutation (K34A) was created by PCR using mutated oligonucleotides. Correct clones were identified by restriction digestion and by bidirectional sequencing.
Construction of Polycistronic Vectors Encoding His-tagged or Non-tagged EF-P, Alone or Together with YjeA and YjeK
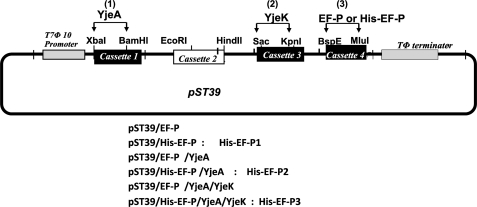
Recombinant pST39 polycistronic vectors were constructed (29, 30) for expression of EF-P or His-tagged EF-P (with insertion of GSSHHHHHHSS after the initial methionine) alone or for co-expression with YjeA and YjeK (Scheme 1). In order to subclone YjeA and YjeK into the XbaI/BamHI and SacI/KpnI sites, respectively, of the pST39 vector, the internal NdeI site of YjeA ORF and BspEI and MluI sites of YjeK ORF were removed by site-directed silent mutagenesis of PBL4 and PBL5 vectors to generate PBL4m and PBL5m. The ORFs of EF-P, YjeA, or YjeK were PCR-amplified using PBL1, PBL4m, and PBL5m as templates, respectively, and each PCR product was digested with NdeI and BamHI and inserted into the pET3aTrm plasmid (29) at NdeI and BamHI sites. Each cassette was excised with the indicated set of enzymes and subcloned into the pST39 vector at the indicated restriction sites in the order indicated. The sequence of recombinant vectors was confirmed at each step before proceeding to the next step. We generated six recombinant vectors, pST39/EF-P, pST39/EF-P/YjeA, pST39/EF-P/YjeA/YjeK, pST39/His-EF-P, pST39/His-EF-P/YjeA, and pST39/His-EF-P/YjeA/YjeK, and also a mutant plasmid, pST39/His-EF-P (K34A).
Scheme 1.
Construction of a polycistronic vector for expression of EF-P alone or its coexpression with YjeA and YjeK for production of unmodified or modified recombinant EF-P. The coding sequences of EF-P, YjeA, and YjeK were first cloned into pET3aTrm vector (29), and each cassette was excised and subcloned into pST39 vector (29), in the order of YjeA, YjeK, and EF-P, at the indicated restriction sites to generate the recombinant vectors listed. Recombinant proteins, His-EF-P1, His-EF-P2, and His-EF-P3, were purified from E. coli harboring the respective recombinant vectors (29). For production of unmodified EF-P, only the EF-P cassette was inserted into pST39. For production of α-lysyl-EF-P, the YjeA cassette was first inserted into pST39, followed by the EF-P cassette. A recombinant vector encoding all three proteins, EF-P, YjeA, and Yjek, was used for generation of β-lysyl-EF-P.
Purification of Recombinant His-tagged EF-P from E. coli
E. coli, BL21(DE3)pLys, was used for expression from the pST39 recombinant vectors, and BL21(DE3) was used for expression from the pET30 recombinant vectors. Protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 4 h or at 18 °C overnight, the cells (from 100 ml of culture) were harvested, and the cell pellet was sonicated. The His-tagged proteins were purified using the Probond purification kit following the manufacturer's protocol.
Preparation of [3H]Lys-labeled EF-P by the YjeA Reaction in Vitro
Non-tagged recombinant EF-P was overexpressed using pST39/EF-P in E. coli BL21(DE3)pLys cells. The lysate protein (containing 300 μg of total proteins) was used, after removal of low molecular weight fractions by 60% ammonium sulfate precipitation and dialysis, as a substrate for the YjeA-mediated radiolabeling with [3H]lysine. The YjeA reaction mixture contained 20 μl of the lysate protein (containing ∼50 μg of EF-P), 0.1 m Tris-HCl, pH 9.5, 5 mm ATP, 200 μCi of [3H]lysine (2.2 nmol), 1 mm DTT, 10 mm MgCl2, 0.1 m KCl, and 50 μg of YjeA in 0.5 ml. The mixture was incubated at 37 °C for 2.5 h, at which time ∼25% of the EF-P was modified. Unreacted [3H]lysine was removed by repeated ammonium sulfate precipitation (60% saturation) and dialysis, resulting in 1.5 × 108 dpm of [3H]Lys-labeled EF-P. This non-tagged, YjeA-modified EF-P was used as a radioactive tracer in the purification of native EF-P from E. coli MRE600 cells.
Purification of Native EF-P from E. coli MRE600 Cells
Native EF-P was purified following a protocol modified from that published previously (31) by omission of the centrifugation step to sediment ribosomes and extraction of the EF-P from the ribosomal wash. The steps are as follows: 110 g of E. coli MRE600 cell pellet was resuspended in 500 ml of Buffer A (50 mm Tris-HCl, pH 7.5, 1 mm DTT, 1 mm PMSF) and lysed by using a French press. The lysate was centrifuged at 35,000 × g (JA20 rotor) for 40 min, and cell debris was removed. To the clarified supernatant, radiolabeled tracer EF-P (2 × 106 dpm) was added. Proteins were precipitated by ammonium sulfate (30–50%), dissolved in 200 ml of Buffer A, and dialyzed against Buffer A (4 h) at 4 °C. Dialyzed protein was applied to a column of Q Sepharose (2.6 × 14 cm, 100 ml), washed with 100 ml of Buffer A, and eluted with a stepwise gradient of 0.1, 0.2, 0.3, 0.4, and 0.5 m KCl in Buffer A (100 ml each step), and 25-ml fractions were collected. The radioactive fractions were pooled, concentrated by ammonium sulfate precipitation (60%), and dissolved in 30 ml of Buffer B (20 mm potassium phosphate, pH 7.2, 1 mm DTT, and 1 mm PMSF) and dialyzed against Buffer B for 3 h. The protein was applied to a column of hydroxyapatite (30 ml), washed with 30 ml of Buffer B, and then eluted with increasing concentrations of potassium phosphate buffer (15 ml each step of 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, and 0.5 m), and 5-ml fractions were collected. The proteins in the radioactive fractions eluting at 0.15 m potassium phosphate buffer were pooled, precipitated with 60% ammonium sulfate, dissolved in 1 m ammonium sulfate, and applied to a phenyl-Sepharose column (20 ml). An ammonium sulfate stepwise gradient was applied (15 ml for each step of 0.8, 0.7, 0.6, 0.5, 0.4, 0.3, 0.2, and 0.1 m), and 5-ml fractions were collected. The radioactive fractions were pooled and concentrated to 2 ml and applied to a column of Sephacryl S-200 HR (80 ml, 50 × 1.5 cm) in Buffer A containing 0.1 m KCl. 1.5-ml fractions were collected. The fractions containing radioactivity were pooled, concentrated, and equilibrated with Buffer C (50 mm Tris-HCl, pH 6.0, 1 mm DTT). The protein fractions were applied to an SP-Sepharose column (10 ml, 0.5 × 13 cm) in Buffer C and washed with 10 ml of Buffer C. The proteins were eluted with a KCl stepwise gradient (6 ml for each step of 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, and 0.5 m) in Buffer C, and 1.5-ml fractions were collected. The purity of the EF-P protein in the radioactive fraction was checked by SDS-PAGE at each step.
YjeA in Vitro Assay
A typical 20-μl reaction mixture contained 100 mm Tris-HCl buffer (pH 9.5), 1 mm DTT, 5 mm ATP, 10 mm MgCl2, 100 mm KCl, 3 μg of EF-P (∼7.5 μm), 5 μCi (2.5 μm) of [3H]lysine, and YjeA (1 μg) and was incubated for 1 h at 37 °C, unless otherwise indicated. At the end of the incubation, an aliquot (10 μl) was taken out for SDS-PAGE. Carrier BSA (500 μg) was added to the rest of the reaction mixture and immediately precipitated with 1 ml of ice-cold 10% TCA. The TCA-precipitated proteins were washed twice with 1 ml of 10% TCA to remove all unincorporated [3H]lysine, and the pellet was completely dissolved in 0.1 ml of 0.2 n NaOH for determining the radioactivity incorporated into EF-P.
Identification of β-Lysyl-lysine and β-Lysine in Proteolytic Digest or Acid Hydrolysate of EF-P by Ion Exchange Chromatography
Purified His-EF-P proteins (radiolabeled by culture of E. coli cells in a medium containing 10 μCi/ml [3H]lysine) were exhaustively digested with Pronase (consecutive addition of 1 mg at 0 and 24 h) in 0.1 ml of 0.1 m Tris-HCl buffer, pH 8.0 (containing 1 mm DTT and 0.1 mg/ml thimerosal) at 37 °C. After 48 h, the digest was precipitated with 10% TCA. For separation of radioactive lysine from β-lysyl-lysine, the TCA supernatant was analyzed by ion exchange chromatography on an amino acid analyzer (32) using a two-buffer system, Buffer 1 (0.2 n sodium citrate buffer, pH 4.31) for 10 min, Buffer 2 (1.5 n sodium citrate buffer, pH 5.55) for 10 min, and Buffer 1 for 20 min. In this program, lysine elutes at 20 min, and β-lysyl-lysine elutes at 32 min. Fluorometric identification of amino acids/peptides was accomplished by postcolumn derivatization with orthophthaldehyde, using a buffer program with the sequence of Buffer 1 for 10 min, Buffer 3 (0.4 n sodium citrate buffer, pH 5.0) for 20 min, Buffer 4 (0.6 n sodium citrate buffer, pH 5.8) for 10 min, Buffer 2 for 10 min, and Buffer 1 for 20 min. In this program, 5-OH-lysine elutes at 17 min, lysine at 23 min, 5- hydroxy-β-lysine at 37 min, β-lysine at 40 min, and β-lysyl-lysine at 56 min, as determined by analyses of authentic compounds.
Mass Spectrometric Analyses of EF-P
Native EF-P purified from E. coli and recombinant His-EF-P intact proteins were desalted using a C-18 ZipTip microcolumn (Millipore); eluted in 1% formic acid, 50% methanol; and subjected to off-line static nano-ESI mass spectrometric analysis using an LTQ-Orbitrap XL mass spectrometer equipped with a nanospray source (Thermo Scientific). The spray voltage was set at 1.5 kV in the positive mode. A full Fourier Transform Mass Spectrometry scan was acquired over an m/z range of 600–2000 with resolution of 60,000 at m/z 400. The resulting spectra were deconvoluted to determine the molecular weight of the intact proteins using the Xtract tool within Xcalibur version 2.1.0 software (Thermo Scientific). The modification at Lys-34 was confirmed by nano-LC-ESI-MS/MS analyses of chymotryptic digests of EF-P as follows. After SDS-PAGE or two-dimensional gel electrophoresis and Coomassie Blue staining, gel bands containing EF-P proteins were subjected to in-gel reduction with DTT, alkylation with iodoacetamide, and digestion using chymotrypsin (Promega, Madison, WI). The resulting peptides were extracted with 5% formic acid, acetonitrile (1:1), concentrated with a SpeedVac, and desalted using a C-18 ZipTip microcolumn (Millipore) prior to MS analysis. Nano-LC-ESI-MS/MS was performed on the LTQ-Orbitrap XL mass spectrometer equipped with an Eksigent nano-LC one-dimensional system. The mobile phase consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in 95% acetonitrile (solvent B). Peptides were loaded onto a C18 trap column (Zorbax 5 × 0.3 mm; Agilent) and separated by a 15-cm IntegraFrit column (ProteoPepTM, New Objective) using a solvent gradient (A/B) from 95:5 to 60:40 in 80 min at a flow rate of 300 nl/min. The LC eluant was sprayed into the mass spectrometer with an emitter tip (PicoTip, New Objective) using a spray voltage of 2.0 kV in the positive ion mode. Eluted peptides were detected in a high resolution (60,000 at m/z 400) precursor MS scan (from m/z 300 to 2000), followed by sequential data-dependent MS/MS scans for the five most intense ions. The normalized collision energy value was set at 35%. Acquired MS/MS data were searched against the E. coli database using Bioworks (Thermo Scientific) for protein identification. Search parameters include carbamidomethylation of cysteine (+57.0215 Da), lysylation or hydroxylysylation of lysine (+128.0950 or 144.0899 Da, respectively), and oxidation of methionine (+15.9949 Da), with a maximum of two missed cleavages allowed and a peptide mass tolerance of ±20 ppm.
Methionyl-puromycin Synthesis Assay
Peptidyltransferase activity was assayed by the formation of N-formyl[3H]methionyl-puromycin from 70 S·AUG·N-formyl[3H]Met-tRNA complexes and puromycin in an assay similar to that described previously (33). In a typical experiment, 30 pmol of 70 S ribosomes in 50 μl of 15 mm Tris-HCl (pH 7.5), 8 mm MgCl2, 55 mm KCl, and 3 mm 2-mercaptoethanol was incubated with 5 μl of AUG (0.17 μg) and 5 μl of N-formyl[3H]Met-tRNA (12,000 dpm) at 35 °C for 10 min. At zero time, EF-P (0.1–0.5 μg) and 5 μl of puromycin (7 μm) were preincubated at 30 °C for 30 s. A 20-μl aliquot of ribosome·AUG·N-formyl[3H]Met-tRNA complex was added, and the mixture was incubated at 35 °C for 1.5–10 min. The reaction was terminated by adding 400 μl of 1 m sodium phosphate buffer (pH 7.2), and the N-formyl[3H]methionyl-puromycin formed was extracted with 800 μl of ethyl acetate. 400 μl of the ethyl acetate extract was counted in a Beckman scintillation counter.
RESULTS
Purification of Native EF-P, Recombinant His-EF-P, and His-YjeA
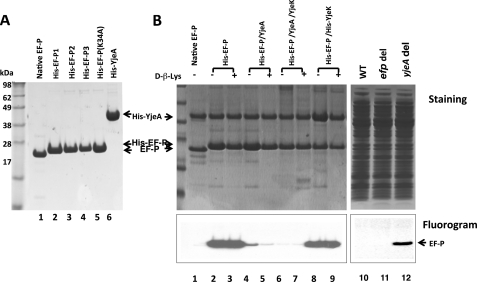
Similar growth defects for the efp, yjeA, and yjeK gene disruption mutants of E. coli (25) and the common features of the phenotypes of the yjeA and yjeK mutants in Salmonella (27) suggested that the modification of EF-P by YjeA and YjeK is required for the biological activity of EF-P in cells. In order to obtain direct biochemical evidence for the nature of the EF-P modification and for its importance for EF-P activity, we purified native EF-P from E. coli MRE600 cells (shown in Fig. 1A, lane 1) and also produced different preparations of recombinant EF-P in E. coli, using a polycistronic vector pST39 (Scheme 1), namely His-EF-P1 by overexpressing His-EF-P alone (from pST39/His-EF-P), His-EF-P2 by co-expressing His-EF-P with YjeA (from pST39/His-EF-P/YjeA), and His-EF-P3 by co-expressing His-EF-P with both YjeA and YjeK (from pST39/His-EF-P/YjeA/YjeK). We also purified a mutant His-EF-P with an alanine substitution at lysine 34, the site of the EF-P modification (Fig. 1A, lane 5). We cloned and expressed E. coli YjeA and purified recombinant His-YjeA protein (Fig. 1A, lane 6) to use in the modification of EF-P in vitro (Fig. 1B).
FIGURE 1.
Purity of native and recombinant EF-P produced in E. coli (A) and testing of these proteins and total cell lysate proteins as substrates for YjeA in vitro (B). A, purity of native and recombinant EF-P proteins used as substrates for the YjeA reaction. Recombinant EF-P preparations His-EF-P1, His-EF-P2, HisEF-P3, and His-EF-P(K34A) were purified from E. coli harboring the recombinant vectors pST39/His-EF-P, pST39/His-EF-P/YjeA, pST39/His-EF-P/YjeA/YjeK, and pST39/His-EF-P(K34A), respectively, but do not necessarily represent one homogenous EF-P with respect to modification status. B, the native EF-P (lane 1) and recombinant EF-P proteins from E. coli (cultured in LB medium without or with 0.8 mm d-β-lysine) harboring the indicated vectors (lanes 2–9) were tested as substrates for the YjeA reaction. Coexpression of His EF-P and His-YjeK was carried out by cotransformation of E. coli cells with pST39/His-EF-P and pET30a/His-YjeK and by selection and culture in medium containing ampicillin, chloramphenicol, and kanamycin. In order to determine if EF-P is the only cellular substrate for YjeA, we also tested total lysate proteins (obtained by ammonium sulfate (60%) precipitation and dialysis) from wild type BW25113 (lane 10), efp disruption strain JW4107 (lane 11), and yjeA disruption mutant and JW4116 (lane 12) as YjeA substrates. Top panels, stained pattern; bottom panels, fluorogram. The experiments were repeated twice with similar results.
Comparison of Endogenous EF-P and Recombinant EF-P Proteins as Substrates for the YjeA Reaction Reveals That Native EF-P Exists Predominantly as the Lysylated Form in Vivo
To determine the modification status of native and recombinant EF-P proteins biochemically, we first tested the purified proteins as substrates for the YjeA-catalyzed reaction in vitro (Fig. 1B, lanes 1–9). Only unmodified EF-P precursor is expected to be radiolabeled upon incubation with [3H]lysine and YjeA in vitro, and this reaction would measure the levels of unmodified EF-P substrate in different EF-P preparations or cell lysates. No radiolabeling was observed with purified native EF-P (Fig. 1B, fluorogram, lane 1), indicating that it was already fully lysylated in vivo. In contrast, robust incorporation of [3H]lysine was observed with the recombinant protein (His-EF-P1, expressed from pST39/His-EF-P) (Fig. 1B, lane 2). The lysine incorporation into His-EF-P1 was close to the stoichiometric level at higher concentrations of lysine and YjeA (data not shown), indicating that most of the His-EF-P1 is in the unmodified form. Although E. coli contains endogenous YjeA and YjeK enzymes, they seem to be limiting, thus leading to accumulation of most of the recombinant EF-P in the unmodified form, when EF-P is overexpressed alone. When recombinant EF-P (His-EF-P2) purified from E. coli co-expressing His-EF-P with YjeA, was used as a substrate, there was a dramatic decrease in radiolabeling (Fig. 1B, compare lane 4 with lane 2) by over 6-fold, suggesting that a majority (>85%) of His-EF-P2 was already lysylated in vivo by coexpressed YjeA in these cells. No or little radiolabeling was detected with the recombinant His-EF-P3 produced in cells co-expressing YjeA and YjeK (Fig. 1B, lane 6), indicating that His-EF-P3 proteins in this case were all in the lysylated form. In contrast, His-EF-P isolated from E. coli coexpressing His-EF-P and His-YjeK (Fig. 1B, lanes 8 and 9) was a fully reactive substrate, indicating that it is mostly unmodified EF-P as is His-EF-P1 (Fig. 1B, lane 8 versus lane 2). Thus, overexpression of YjeK alone does not facilitate lysylation of overexpressed EF-P, probably because endogenous YjeA is limiting. In accordance with this finding, the addition of d-β-lysine to the medium did not enhance EF-P lysylation in the absence of YjeA coexpression (no reduction in unmodified EF-P; Fig. 1B, lane 3 versus lane 2 and lane 9 versus lane 8). However, in cells overexpressing YjeA with EF-P, the addition of d-β-lysine further depleted unmodified His-EF-P (lane 5 versus lane 4) by converting it mainly to β-lysyl-EF-P (not shown), indicating that it facilitated EF-P β-lysylation as did overexpressed YjeK (lane 6).
No lysylation was observed with a mutant His-EF-P (K34A) under any reaction condition, including higher level of YjeA or lysine or longer incubation times (not shown), confirming Lys-34 as the sole lysylation target site in E. coli EF-P.
Lysylation by YjeA Occurs Only in One Bacterial Protein, EF-P
Because the hypusine modification is strictly specific for the eukaryotic ortholog, eIF5A, we wondered whether the lysylation reaction by YjeA is restricted to EF-P in bacteria. Thus, crude lysate proteins of E. coli were used as substrate sources for the YjeA reaction in vitro. Strong labeling of EF-P was observed with the lysates from cells overexpressing EF-P alone (not shown). In contrast, no labeling was observed with the control lysate or with the lysate of cells expressing all three proteins, consistent with the results obtained with the purified proteins. The lack of any labeling with the control lysate (Fig. 1B, lane 10) suggests that normally there is no accumulation of unmodified substrate protein and that all of the endogenous YjeA substrate protein(s), including EF-P, exists as the lysylated form and not as the unmodified precursor. Thus, we tested the total lysate proteins from the yjeA disruption strain as substrate(s) in vitro, because unmodified YjeA substrate protein(s) would accumulate in the absence of the YjeA enzyme. Indeed, strong labeling of EF-P was observed only in the lysate proteins of the yjeA disruption mutant (Fig. 1B, lane 12) and not in those from wild type (Fig. 1B, lane 10) or in those from the efp disruption mutant (Fig. 1B, lane 11). Moreover, EF-P was the only protein in the E. coli lysate radiolabeled in a YjeA-dependent manner. These findings demonstrate that EF-P is the only substrate for YjeA in bacteria and that the YjeA reaction exhibits as strict specificity toward its protein substrate as does deoxyhypusine synthase toward eIF5A.
Effects of pH and ATP on YjeA Reaction in Vitro
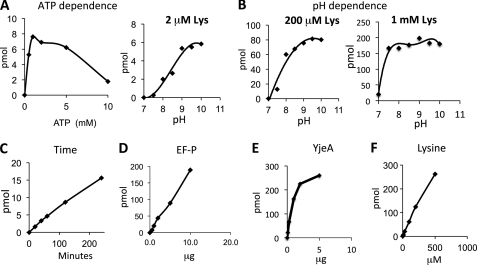
Using purified, recombinant His-EF-P1 as a substrate, we further characterized the YjeA reaction in vitro, including its dependence on ATP, pH, time of incubation, and the concentration of EF-P, YjeA, and lysine (Fig. 2). YjeA first activates lysine or its analogs to generate AMP-lysine, the carbonyl group of which is predicted to be coupled to the ϵ-amino group of a specific lysine (Lys-34 in the E. coli protein) of EF-P to form an ϵ-amino-α-lysyl amide bond. As expected, the lysylation reaction was totally dependent on ATP with an optimum between 2 and 5 mm ATP (Fig. 2A). The reaction was acutely dependent on basic pH at low lysine concentrations (2–200 μm), yielding the highest rate at pH 9–10 (Fig. 2B). However, at a higher lysine concentration (1 mm), the reaction rates were similar from pH 7.5 to 10.0. The reaction was nearly linear with time, EF-P, YjeA (up to 2 μg), and lysine (Fig. 2, C–F). YjeA displayed a low affinity for l-lysine, consistent with the high Km reported for the lysine activation reaction (Km = 6.2 mm) (24), in stark contrast to the high affinity of lysine to LysRS (Km 45 μm) (24). Nevertheless, at a high concentration of lysine (1 mm), a majority of EF-P substrate was lysylated under the reaction conditions at neutral pH.
FIGURE 2.
Dependence of the YjeA reaction on ATP (A), pH (B), time (C), EF-P (D), YjeA (E), and lysine (F). The dependence on ATP, time, and EF-P was determined in a typical reaction, as described under “Experimental Procedures” except that the levels of EF-P, ATP, Lys, and the incubation time or pH were varied as indicated. The pH dependence of the reaction was measured at three different concentrations of lysine (2, 100, and 1000 μm) in reaction mixtures containing 5 μCi (2 μm) of [3H]lysine, 5 mm ATP, and 3 μg (at 2 and 200 μm lysine) or 6 μg (at 1 mm lysine) of unmodified recombinant EF-P (His-EF-P1), 1 μg of YjeA in the 0.1 m Tris-HCl buffer with different pH values, 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, and 10. Incubation was for 1 h at 37 °C. The YjeA dependence reaction was carried out in a typical reaction mixture (pH 9.0) containing 5 mm ATP, 6 μg of EF-P, 200 μm lysine, and varying amounts of YjeA (0.1, 0.3, 1.0, 2.0, and 5.0 μg). The lysine dependence of the YjeA reaction was measured at pH 9.0 in a mixture containing 5 mm ATP, 6 μg of EF-P, 200 μm lysine, and varying amounts of YjeA. The assays were repeated twice with similar results, and representative data are shown.
Mass Spectrometric Analyses of Native and Recombinant EF-P Reveals Lysylation at Lys-34
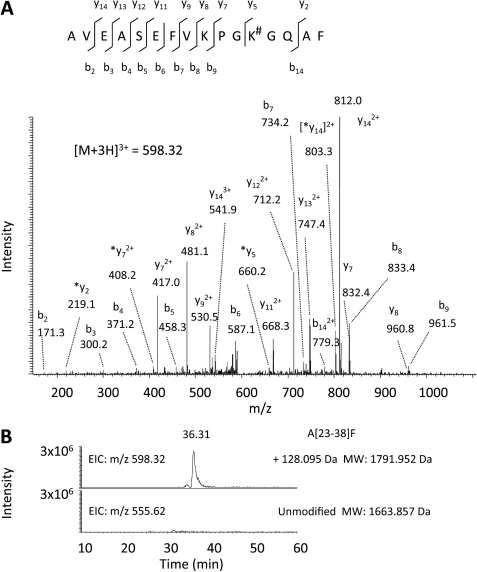
In order to determine the modification status of EF-P, we first analyzed molecular weights of native EF-P and recombinant His-EF-P1 and His-EF-P3 (expressed from pST39/EF-P/YjeA/YjeK) by static nano-ESI-mass spectrometry (data not shown). The experimental values obtained for the His-EF-P1 (21,688 Da) and His-EF-P3 (21,816 Da) closely matched the theoretical molecular weights for the unmodified His-EF-P and lysylated His-EF-P, respectively. No lysylated form (21,816 Da) was detected in His-EF-P1, and no unmodified form (21,688 Da) was detected in His-EF-P3, consistent with data in Fig. 1. Native EF-P produced signals at both 20,588.46 and 20,604.50 Da. The former corresponds to the calculated molecular mass of EF-P with the N-terminal methionine cleaved off and with addition of a lysyl moiety (+128 Da). The latter (20,604.5 with the addition of 144 Da) may represent a lysylated EF-P with an additional oxygen, possibly a methionine-oxidized or hydroxylated form. Methionine oxidation, a common artifact, may have occurred during a long process of purification of native EF-P, unlike a simple affinity purification of recombinant His-EF-P. Subsequent analysis of chymotryptic peptides from native EF-P by nano-LC-ESI-MS/MS analyses revealed the lysyl modification at Lys-34 (Fig. 3). The MS/MS data indicated that the triply charged ion at m/z 598.32 (Mr = 1791.952) originated from the chymotryptic peptide segment Ala-23 to Phe-38 with the addition of 128.095 Da at Lys-34 (Fig. 3A). There was no signal at m/z 555.62 corresponding to the unlysylated peptide (Fig. 3B) in native EF-P. The MS/MS data for His-EF-P3 also showed the lysyl modification at Lys-34 (data not shown).
FIGURE 3.
Mass spectrometric analyses of chymotryptic digest of native EF-P by nano-LC-ESI-MS/MS. A, the MS/MS spectrum of the triply charged ion at m/z 598.32 originated from the peptide segment Ala-23 to Phe-38 with the addition of 128.095 Da at Lys-34. The sequence of the peptide was assigned single-letter amino acid abbreviation based on the fragment ions observed for the peptide segments. N-terminal b ions and C-terminal y ions (including *y = y-H2O) resulting from the amide bond cleavage are labeled. #, addition of 128 Daltons. B, extracted ion chromatograms obtained from native EF-P sample digested by chymotrypsin. Mass values for the triply charged ions representing peptide segment Ala-23 to Phe-38 (A[23–38]F) with or without the addition of 128.095 Da (lysylation) were extracted.
Identification of β-Lysyl-lysine in Native EF-P and Recombinant His-EF-P3
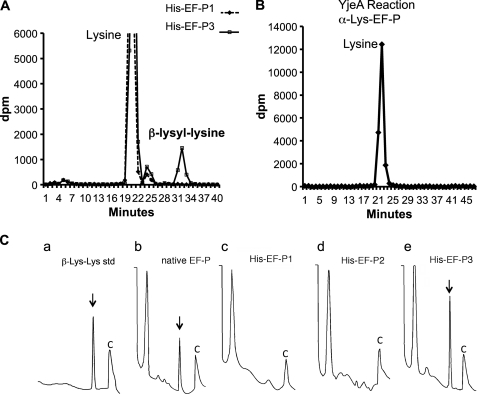
Although mass spectrometric data demonstrate complete lysylation at Lys-34 of native EF-P, the data do not distinguish α-lysyl-EF-P from β-lysyl-EF-P. Therefore, we analyzed acid hydrolysates and exhaustive proteolytic digests of native and recombinant EF-P proteins for biochemical evidence. To distinguish between the α-lysyl or β-lysyl linkage in the native and recombinant EF-P proteins, we performed ion exchange chromatographic separation of amino acids and peptides after acid hydrolysis or exhaustive proteolytic digestion. Only β-lysine (no 5-hydroxy-β-lysine) was detected in the acid hydrolysate of native EF-P and of recombinant EF-P isolated from E. coli co-expressing EF-P, YjeA, and YjeK (data not shown). We also analyzed exhaustive proteolytic digests of the EF-P proteins to identify β-lysyl-lysine isopeptide, because the β-lysyl-lysine bond is not cleaved by proteases, whereas the α-lysyl-lysine bond is. First, we purified [3H]lysine-labeled proteins, unmodified and modified EF-P (His-EF-P1, and His-EF-P3, expressed from pST39/His-EF-P, and pST39/His-EF-P/YjeA/YjeK vector, respectively), from cells cultured in a medium containing 10 μCi/ml [3H]lysine. We digested the in vivo labeled EF-P proteins exhaustively with Pronase and analyzed the digests by ion exchange chromatography. As shown in Fig. 4A, a radioactive peak was detected at the position of a β-lysyl-lysine standard (chemically synthesized and validated isopeptide) in the proteolytic digest of His-EF-P3 but not in the digest of His-EF-P1 (Fig. 4A). Acid hydrolysis of the radioactive peak at the position of β-lysyl-lysine released radioactive lysine and β-lysine in equal amounts (not shown), providing additional evidence for its identity as β-lysyl-lysine. The radioactivity in the β-lysyl-lysine peak of His EF-P3 (Fig. 4A) was 10% of the total radioactivity (dpm in lysine and β-lysyl-lysine). EF-P contains 13 lysine residues, and with lysylation of Lys-34, the percentage of radioactivity in β-lysyl-lysine would be 14.3% for the pure β-lysyl-EF-P. Thus, it is estimated that this batch of His-EF-P3 contains at least 70% (considering that the His-EF-P3 is not totally pure) as β-lysyl-EF-P and the rest as α-lysyl-EF-P. When radiolabeled α-lysyl-EF-P, produced in vitro by reaction of His-EF-P1 with YjeA and [3H]lysine, was exhaustively digested, only radioactive lysine was detected (Fig. 4B), indicating that the α-lysyl-lysyl bond is cleaved by Pronase.
FIGURE 4.
Ion exchange chromatographic identification of β-lysyl-lysine in the Pronase digest of native EF-P and recombinant His-EF-P from cells coexpressing His-EF-P, YjeA, and YjeK. A, [3H]lysine-labeled His-EF-P proteins were purified from E. coli and were exhaustively digested with Pronase. The digests were analyzed by ion exchange chromatography as described under “Experimental Procedures.” A radioactive peak corresponding to a reference β-lysyl-lysine standard was found in the digest of in vivo modified His-EF-P3 but not in the digest of unmodified His-EF-P1. B, no radioactive β-lysyl-lysine was found in the digest of α-lysyl-EF-P formed in an in vitro YjeA reaction. Only [3H]lysine was found, indicating that the α-lysyl-lysine bond is cleaved by Pronase. C, standard β-lysyl-lysine (a) and exhaustive Pronase digests of native (b) or recombinant EF-P proteins (c–e) were analyzed by ion exchange chromatography, and amino acids or peptides were measured by a fluorometric detection method (32), as described under “Experimental Procedures.” Only the latter part of the chromatogram (30–70 min) is shown with two reference peaks before and after the β-lysyl-lysine (indicated by an arrow or arrowhead). C, a reference peak (buffer change) at the end of each run.
We further analyzed the proteolytic digests of endogenous and recombinant EF-P proteins by fluorometric detection after ion exchange chromatographic separation (Fig. 4C). Again β-lysyl-lysine (standard in Fig. 4C, a) was detected in the exhaustive digest of native EF-P (Fig. 4C, b) and of the recombinant His-EF-P3 isolated from cells co-expressing EF-P, YjeA, and YjeK (Fig. 4C, e). No β-lysyl-lysine was detected in the digest of the recombinant His-EF-P proteins from E. coli expressing His-EF-P alone (Fig. 4C, c) or His-EF-P together with YjeA (Fig. 4C, d), suggesting that both YjeA and YjeK are required for β-lysylation of EF-P in cells.
β-Lysylation Is Required for Activity of EF-P in N-Formyl-methionyl-puromycin Synthesis
To assess the role of the EF-P modification in stimulating ribosomal peptidyltransferase, we compared the activities of native and recombinant EF-P proteins in the N-formyl-methionyl-puromycin synthesis assay. As shown in Figs. 5, A and B, both native EF-P and recombinant His-EF-P3 purified from cells co-expressing EF-P, YjeA, and YjeK stimulated synthesis of N-formyl-methionyl-puromycin. A mutant protein, His-EF-P(K34A), was totally inactive, whereas His-EF-P1 and His-EF-P2 displayed low levels of residual activity (20–25% of that of native EF-P). That the activity of His-EF-P3 is severalfold higher than those of His-EF-P1 and His-EF-P2 demonstrates the importance of β-lysylation in the EF-P activity. Because the residual activity of His-EF-P2 was not notably higher than that of His-EF-P1, α-lysylation alone probably is not sufficient for activation of EF-P. This is in accordance with the similar growth defects of the efp, yjeA, and yjeK disruption mutants (25), emphasizing the importance of both enzymatic steps for the EF-P activity. Although β-lysyl-EF-P was not detectable in the His-EF-P1 and His-EF-P2 by ion exchange chromatographic analyses of its Pronase digest, we cannot rule out the possibility that the two EF-P preparations contain minute amounts of β-lysyl-EF-P, formed by endogenous YjeA and YjeK, which could contribute to their residual activity. Otherwise, it may reflect limited activity inherent to the unmodified EF-P or to α-lysyl-EF-P.
FIGURE 5.
Comparison of the activities of unmodified and modified recombinant EF-P, mutant EF-P (K34A) and native EF-P in N-formyl-methionyl-puromycin synthesis. The assays were carried out as described under “Experimental Procedures.” using 0.5 μg of each purified EF-P protein shown in Fig. 1A. At the indicated time points (1.5, 3, 5, and 10 min), the reaction was stopped, and the N-formyl-methionyl-puromycin formed was measured (A) and the values at 3 min are compared as a bar graph (B). The assay was repeated three times independently, and data are shown with error bars representing S.D. From experiments illustrated in Figs. 1 and 4, His-EF-P1 is mostly unmodified EF-P, His-EF-P2 is estimated to contain α-lysyl-EF-P (>85%) and unmodified EF-P, and His-EF-P3 is estimated to contain β-lysyl-EF-P (>70%) and α-lysyl-EF-P.
DISCUSSION
EF-P is the bacterial ortholog of eIF5A, displaying conservation of both sequence and structure. The post-translational modification of EF-P is analogous to the hypusine modification of eIF5A, in that it converts a highly conserved lysine residue to an extended and more basic amino acid. YjeA and YjeK were predicted to be involved in the post-translational modification of the EF-P precursor to a β-lysyl-EF-P, and YjeA was shown to catalyze the incorporation of α-lysine (22, 24) and β-d-lysine (28) into recombinant EF-P in an ATP-dependent manner in vitro (25, 27), presumably yielding α-lysyl-EF-P and β-lysyl-EF-P, respectively. However, there has been no direct biochemical evidence for β-lysylation in native EF-P and for its role in the EF-P activity. In this report, we present definitive evidence for β-lysylation of EF-P at its conserved lysine (Lys-34) in the native E. coli protein and demonstrate that this modification catalyzed by the action of YjeA and YjeK in cells is required for the activity of EF-P. Furthermore, our data strongly suggest that β-lysylation by YjeA and YjeK is as exclusive to EF-P as is the hypusine modification to eIF5A.
Previous mass spectrometric analyses of tryptic peptides of E. coli native EF-P suggested the addition of 144 Daltons at Lys-34 (20). The presence of such a modification was also indicated in our analyses of native EF-P (data not shown). However, we found β-lysyl-lysine in the proteolytic digest of native EF-P (Fig. 4). In addition, β-lysine, but not 5-hydroxy-β-lysine, was detected in the acid hydrolysate (data not shown) of the native EF-P. Our data clearly indicated that both native EF-P and His-EF-P3 are lysylated (mainly β-lysylation) at Lys-34, and both showed strong activity for N-formyl-methionyl-puromycin synthesis (Fig. 5). We also identified hydroxylysyl modification (+144 Da) at Lys-34 in native EF-P (data not shown). However, quantitative comparison of these two forms by mass spectrometry is rather difficult in the absence of standard compounds, because their ionization efficiencies may differ. Further investigation is needed for the identification of the stereochemical nature of a hydroxyl-lysyl moiety at Lys-34, the position of the hydroxylated carbon, and the enzyme catalyzing the reaction and for determination of the role of hydroxylation in EF-P activity.
Two alternative pathways have been proposed for EF-P modification, with YjeA or YjeK being the first step enzyme. Obviously, YjeA can α-lysylate EF-P in vitro or in cells independently of YjeK. In contrast, YjeK alone or supplementation with d-β-lysine cannot lysylate EF-P (Fig. 1B). A majority (>85%) of His-EF-P was α-lysylated in E. coli co-overexpressing His-EF-P and YjeA without YjeK overexpression (Fig. 1). However, it has not been experimentally determined whether α-lysyl-EF-P can be converted to β-lysyl-EF-P by YjeK. YjeA exhibits a broad specificity toward lysine analogs, and its affinity for l-lysine appears to be low (24). This may be the reason why EF-P was not completely lysylated in cells harboring the pST39/EF-P/YjeA, despite the high expression of both EF-P and YjeA (Fig. 1B). Complete lysylation (including both α- and β-lysylation) of recombinant EF-P was observed only in cells co-expressing all three proteins, suggesting that YjeK facilitates lysylation. The addition of d-β-lysine to medium of E. coli coexpressing His-EF-P and YjeA also promoted lysylation (Fig. 1B, lane 5 versus lane 4) and resulted in a major β-lysylated EF-P form (not shown), indicating that d-β-lysine can substitute for the YjeK function in our E. coli overexpression system. This is consistent with a recent report that d-β-lysine can replace YjeK function in vivo and can support growth of the Salmonella yjeK mutant (28). These findings provide strong evidence that β-lysylation of EF-P can occur by sequential reactions catalyzed by YjeK followed by YjeA. However, considering the high intracellular concentration of α-lysine (versus the low concentration of d-β-lysine) and that α-lysylation of EF-P by YjeA occurs independently of YjeK, we cannot yet totally exclude the possibility that EF-P modification occurs by both pathways with either YjeA or YjeK being the first step.
The enzymatic reactions leading to the post-translational modifications of both E. coli EF-P and eIF5A (yielding β-lysyl-lysine and hypusine, respectively) are quite distinct. It is remarkable that such different mechanisms have evolved to introduce a long basic side chain into the EF-P/aIF5A/eIF5A family of proteins. YjeA bears no resemblance to deoxyhypusine synthase (34) in crystal structure or in reaction mechanism. Whereas the former uses lysine or β-lysine together with ATP as substrates, the latter uses the polyamine spermidine as a butylamine donor and NAD as a cofactor (15). As anticipated, YjeA did not incorporate lysine into eIF5A, and deoxyhypusine synthase did not incorporate the amino-butyl moiety of spermidine into the EF-P precursor (data not shown). It is interesting to note that bacteria can grow indefinitely in the absence of polyamines (35), whereas spermidine is absolutely required for the growth of the yeast Saccharomyces cerevisiae (36), and that the deoxyhypusine synthase inhibitor, monoguanyl-1,7-diaminoheptane, halts the growth of both eukaryotes and archaea, whereas the growth of bacteria seems unaffected (17). In this regard, it is conceivable that the absolute requirement for spermidine for survival and growth of eukaryotic cells has evolved primarily for hypusine synthesis in eIF5A (36).
Although the cellular functions of EF-P and eIF5A are not clearly understood, the structural and functional analogy between EF-P and eIF5A suggest substantial evolutionary conservation of the two factors (8). Both EF-P and eIF5A contain basic amino acids in an exposed loop in the N-terminal domain I (β-lysyl-lysine derived from Lys-34 in E. coli EF-P, Arg-32 in thermophilus EF-P, and hypusine from Lys-50 in human eIF5A). In the case of eIF5A, the lysine residue that is converted to hypusine is strictly conserved, and the hypusine modification is required for eIF5A activity and survival of eukaryotic cells. In contrast, the modification site lysine is not conserved in EF-P, with Arg, Asn, or Gln replacements in several bacterial genera. Although EF-P and its β-lysylation of a specific lysine are not essential, they probably are important for survival under certain conditions possibly as a translational stress alarm. It is possible that the EF-P protein is involved in the translation of a subset of mRNAs, including those related to stress resistance (27). Indeed, recently, we have observed that deletion of the E. coli efp gene by a conjugation-based approach significantly affected expression of only about 200 genes.4
The crystal structure of the T. thermophilus 70 S ribosome complex with EF-P, a short mRNA, and the initiator formyl-Met-tRNAi revealed that EF-P associates with ribosomal protein L1 and that in the presence of tRNAi, EF-P binds between the peptidyl-tRNA site and the exit tRNA site of the ribosome (7), with the basic side chain of Arg-32 directed toward the peptidyltransferase center. The interaction of EF-P with the L1 50 S ribosomal protein is consistent with its proposed action in translocation because the protein promotes the ribosomal accommodation of formyl-Met-tRNA from the aminoacyl-tRNA site to the peptidyl-tRNA site (23). It was proposed that EF-P facilitates the proper positioning of the initiator tRNA for the formation of the first peptide bond. The occurrence of β-lysine at Lys-34 in EF-P could indeed help to juxtapose the peptide chain into the catalytic core of the peptidyltransferase on the ribosome (25). The L1 protein is known to move coordinately with the translocation of the peptide chain on the ribosome (37) and is the site where the tRNA exits from the ribosome. eIF5A may bind to the eukaryotic ribosome in a manner similar to that of EF-P to enhance the formation of the first peptide bond and to affect the translocation of the relevant mRNAs. Bacterial strains in which efp has been deleted or inactivated are viable but grow at a slower rate than wild type (25, 38) and are defective in sporulation (39) and in swarming (40). Thus, EF-P may only be essential for the translation of subsets of mRNAs involved in stress resistance, sporulation, or swarming (27, 39). In view of the proposed role for EF-P as a factor involved in stress adaptation in Salmonella (27), it is interesting to note the genetic connection between several eIF5A genes that enhance tolerance to heat, osmotic, oxidative, and nutrient stresses in plants (41, 42). Under such a scenario, the exact mechanism used by EF-P and eIF5A to selectively stimulate the translocation of specific mRNAs and the nature of such mRNAs (3, 23, 43, 44) warrant future investigations.
Acknowledgments
We thank Edith C. Wolff (NIDCR, National Institutes of Health) for helpful suggestions, members of the Johansson laboratory for cloning and mutagenesis of E. coli EF-P, and Bernard Kuster for expert help with initial mass spectrometric analysis of EF-P.
This work was supported, in whole or in part, by the Intramural Research Program of NIDCR and NIAAA, National Institutes of Health. This work was also supported in part by a grant from the Carl Trygger Foundation (to H. E. J.).
This paper is dedicated to the memory of Dr. J. E. Folk (October 29, 1925 to December 27, 2010).
M. Babu and M. C. Ganoza, unpublished results.
- IF
- initiation factor
- EF
- bacterial elongation factor
- aIF5A
- archaeal initiation factor 5A
- eIF5A
- eukaryotic translation initiation factor 5A
- ESI
- electrospray ionization.
REFERENCES
- 1. Ganoza M. C., Kiel M. C., Aoki H. (2002) Evolutionary conservation of reactions in translation. Microbiol. Mol. Biol. Rev. 66, 460–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell 108, 557–572 [DOI] [PubMed] [Google Scholar]
- 3. Glick B. R., Chládek S., Ganoza M. C. (1979) Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. Eur. J. Biochem. 97, 23–28 [DOI] [PubMed] [Google Scholar]
- 4. Glick B. R., Ganoza M. C. (1975) Identification of a soluble protein that stimulates peptide bond synthesis. Proc. Natl. Acad. Sci. U.S.A. 72, 4257–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Green R. H., Glick B. R., Ganoza M. C. (1985) Requirements for in vitro reconstruction of protein synthesis. Biochem. Biophys. Res. Commun. 126, 792–798 [DOI] [PubMed] [Google Scholar]
- 6. Ganoza M. C., Aoki H. (2000) Peptide bond synthesis. Function of the efp gene product. Biol. Chem. 381, 553–559 [DOI] [PubMed] [Google Scholar]
- 7. Blaha G., Stanley R. E., Steitz T. A. (2009) Formation of the first peptide bond. The structure of EF-P bound to the 70S ribosome. Science 325, 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kyrpides N. C., Woese C. R. (1998) Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. U.S.A. 95, 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park M. H., Nishimura K., Zanelli C. F., Valentini S. R. (2010) Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benne R., Hershey J. W. (1978) The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253, 3078–3087 [PubMed] [Google Scholar]
- 11. Kemper W. M., Berry K. W., Merrick W. C. (1976) Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 251, 5551–5557 [PubMed] [Google Scholar]
- 12. Henderson A., Hershey J. W. (2011) Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 6415–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gregio A. P., Cano V. P., Avaca J. S., Valentini S. R., Zanelli C. F. (2009) eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 380, 785–790 [DOI] [PubMed] [Google Scholar]
- 14. Saini P., Eyler D. E., Green R., Dever T. E. (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joe Y. A., Wolff E. C., Park M. H. (1995) Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins. J. Biol. Chem. 270, 22386–22392 [DOI] [PubMed] [Google Scholar]
- 16. Park J. H., Aravind L., Wolff E. C., Kaevel J., Kim Y. S., Park M. H. (2006) Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase. A HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. U.S.A. 103, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jansson B. P., Malandrin L., Johansson H. E. (2000) Cell cycle arrest in archaea by the hypusination inhibitor N1-guanyl-1,7-diaminoheptane. J. Bacteriol. 182, 1158–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Byers T. L., Ganem B., Pegg A. E. (1992) Cytostasis induced in L1210 murine leukaemia cells by the S-adenosyl-l-methionine decarboxylase inhibitor 5′-([(Z)-4-amino-2-butenyl]methylamino)-5′-deoxyadenosine may be due to hypusine depletion. Biochem. J. 287, 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanawa-Suetsugu K., Sekine S., Sakai H., Hori-Takemoto C., Terada T., Unzai S., Tame J. R., Kuramitsu S., Shirouzu M., Yokoyama S. (2004) Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. U.S.A. 101, 9595–9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim K. K., Hung L. W., Yokota H., Kim R., Kim S. H. (1998) Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 95, 10419–10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tong Y., Park I., Hong B. S., Nedyalkova L., Tempel W., Park H. W. (2009) Crystal structure of human eIF5A1: insight into functional similarity of human eIF5A1 and eIF5A2. Proteins 75, 1040–1045 [DOI] [PubMed] [Google Scholar]
- 22. Bailly M., de Crécy-Lagard V. (2010) Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol. Direct. 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aoki H., Xu J., Emili A., Chosay J. G., Golshani A., Ganoza M. C. (2008) Interactions of elongation factor EF-P with the Escherichia coli ribosome. FEBS J. 275, 671–681 [DOI] [PubMed] [Google Scholar]
- 24. Ambrogelly A., O'Donoghue P., Söll D., Moses S. (2010) A bacterial ortholog of class II lysyl-tRNA synthetase activates lysine. FEBS Lett. 584, 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yanagisawa T., Sumida T., Ishii R., Takemoto C., Yokoyama S. (2010) A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 17, 1136–1143 [DOI] [PubMed] [Google Scholar]
- 26. Behshad E., Ruzicka F. J., Mansoorabadi S. O., Chen D., Reed G. H., Frey P. A. (2006) Enantiomeric free radicals and enzymatic control of stereochemistry in a radical mechanism. The case of lysine 2,3-aminomutases. Biochemistry 45, 12639–12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Navarre W. W., Zou S. B., Roy H., Xie J. L., Savchenko A., Singer A., Edvokimova E., Prost L. R., Kumar R., Ibba M., Fang F. C. (2010) PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell 39, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roy H., Zou S. B., Bullwinkle T. J., Wolfe B. S., Gilreath M. S., Forsyth C. J., Navarre W. W., Ibba M. (2011) The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat. Chem. Biol. 7, 667–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan S. (2001) A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21, 224–234 [DOI] [PubMed] [Google Scholar]
- 30. Park J. H., Dias C. A., Lee S. B., Valentini S. R., Sokabe M., Fraser C. S., Park M. H. (2011) Production of active recombinant eIF5A. Reconstitution in E. coli of eukaryotic hypusine modification of eIF5A by its coexpression with modifying enzymes. Protein Eng. Des. Sel. 24, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aoki H., Adams S. L., Turner M. A., Ganoza M. C. (1997) Molecular characterization of the prokaryotic efp gene product involved in a peptidyltransferase reaction. Biochimie 79, 7–11 [DOI] [PubMed] [Google Scholar]
- 32. Folk J. E., Park M. H., Chung S. I., Schrode J., Lester E. P., Cooper H. L. (1980) Polyamines as physiological substrates for transglutaminases. J. Biol. Chem. 255, 3695–3700 [PubMed] [Google Scholar]
- 33. Aoki H., Adams S. L., Chung D. G., Yaguchi M., Chuang S. E., Ganoza M. C. (1991) Cloning, sequencing, and overexpression of the gene for prokaryotic factor EF-P involved in peptide bond synthesis. Nucleic Acids Res. 19, 6215–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Umland T. C., Wolff E. C., Park M. H., Davies D. R. (2004) A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme-NAD-inhibitor ternary complex. J. Biol. Chem. 279, 28697–28705 [DOI] [PubMed] [Google Scholar]
- 35. Chattopadhyay M. K., Tabor C. W., Tabor H. (2009) Polyamines are not required for aerobic growth of Escherichia coli. Preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J. Bacteriol. 191, 5549–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chattopadhyay M. K., Park M. H., Tabor H. (2008) Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc. Natl. Acad. Sci. U.S.A. 105, 6554–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cornish P. V., Ermolenko D. N., Staple D. W., Hoang L., Hickerson R. P., Noller H. F., Ha T. (2009) Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl. Acad. Sci. U.S.A. 106, 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aoki H., Dekany K., Adams S. L., Ganoza M. C. (1997) The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J. Biol. Chem. 272, 32254–32259 [DOI] [PubMed] [Google Scholar]
- 39. Ohashi Y., Inaoka T., Kasai K., Ito Y., Okamoto S., Satsu H., Tozawa Y., Kawamura F., Ochi K. (2003) Expression profiling of translation-associated genes in sporulating Bacillus subtilis and consequence of sporulation by gene inactivation. Biosci. Biotechnol. Biochem. 67, 2245–2253 [DOI] [PubMed] [Google Scholar]
- 40. Kearns D. B., Chu F., Rudner R., Losick R. (2004) Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 52, 357–369 [DOI] [PubMed] [Google Scholar]
- 41. Ma F., Liu Z., Wang T. W., Hopkins M. T., Peterson C. A., Thompson J. E. (2010) Arabidopsis eIF5A3 influences growth and the response to osmotic and nutrient stress. Plant Cell Environ. 33, 1682–1696 [DOI] [PubMed] [Google Scholar]
- 42. Xu J., Zhang B., Jiang C., Ming F. (2011) RceIF5A, encoding an eukaryotic translation initiation factor 5A in Rosa chinensis, can enhance thermotolerance, oxidative and osmotic stress resistance of Arabidopsis thaliana. Plant Mol. Biol. 75, 167–178 [DOI] [PubMed] [Google Scholar]
- 43. Zanelli C. F., Valentini S. R. (2007) Is there a role for eIF5A in translation? Amino Acids 33, 351–358 [DOI] [PubMed] [Google Scholar]
- 44. Zou S. B., Roy H., Ibba M., Navarre W. W. (2011) Elongation factor P mediates a novel post-transcriptional regulatory pathway critical for bacterial virulence. Virulence 2, 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]