Abstract
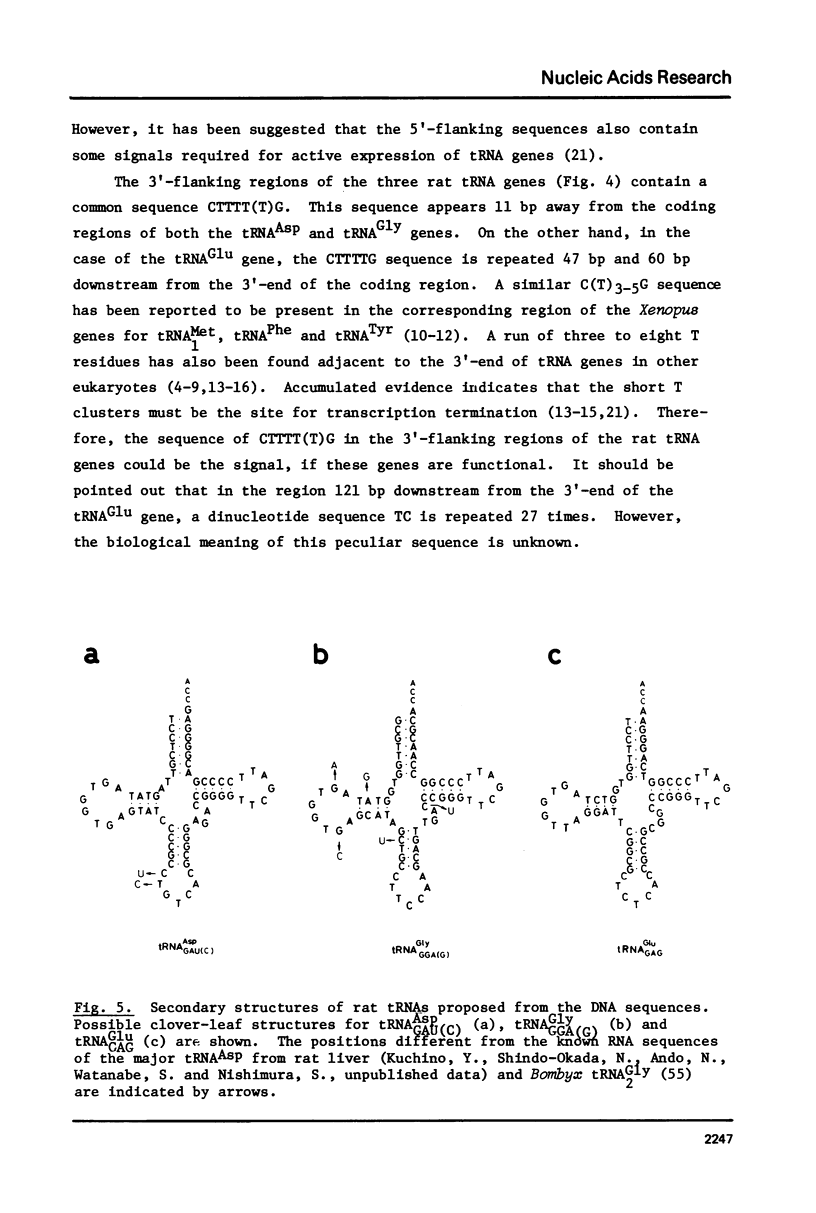
A cloned 2.1 kb fragment of rat DNA hybridized to purified tRNAAsp has been sequenced. The result revealed that in addition to the putative gene for tRNAAspGAU(C), the fragment contained the tRNAGlyGGA(G) and tRNAGluGAG genes. The genes for tRNAAsp, tRNAGly and tRNAGlu have the same polarity, are arranged in this order and are regularly separated by DNA regions of about 450 bp. These rat genes contain neither intervening sequences nor the CCA sequence expected in the 3'-end of the mature tRNA. As observed in lower eukaryotic tRNA genes, the 5'-flanking regions of the three rat genes do not have any significant sequence homology as a regulatory element. In the 3'-flanking region, the sequences CTTTTTG and CTTTTG are present 11 bp downstream from the 3'-end of the genes for tRNAAsp and tRNAGly, respectively. The same CTTTTG sequence is repeated twice in regions 47 and 60 bp away from the tRNAGlu gene. The short T cluster common to the three genes might be the transcription termination site as in lower eukaryotic tRNA genes.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckmann J. S., Johnson P. F., Abelson J. Cloning of yeast transfer RNA genes in Escherichia coli. Science. 1977 Apr 8;196(4286):205–208. doi: 10.1126/science.322282. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson S. G., Birnstiel M. L., Purdom I. F. Clustering of transfer RNA genes of Xenopus laevis. J Mol Biol. 1973 Sep 15;79(2):411–429. doi: 10.1016/0022-2836(73)90014-4. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Birnstiel M. L., Serra V. Reiterated transfer RNA genes of Xenopus laevis. J Mol Biol. 1973 Sep 15;79(2):391–410. doi: 10.1016/0022-2836(73)90013-2. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Kurer V., Smith H. O. Sequence organization of a cloned tDNA met fragment from Xenopus laevis. Cell. 1978 Jul;14(3):713–724. doi: 10.1016/0092-8674(78)90253-2. [DOI] [PubMed] [Google Scholar]
- Cranston J. W., Silber R., Malathi V. G., Hurwitz J. Studies on ribonucleic acid ligase. Characterization of an adenosine triphosphate-inorganic pyrophosphate exchange reaction and demonstration of an enzyme-adenylate complex with T4 bacteriophage-induced enzyme. J Biol Chem. 1974 Dec 10;249(23):7447–7456. [PubMed] [Google Scholar]
- De Robertis E. M., Olson M. V. Transcription and processing of cloned yeast tyrosine tRNA genes microinjected into frog oocytes. Nature. 1979 Mar 8;278(5700):137–143. doi: 10.1038/278137a0. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler R., Egg A. H., Kubli E., Artavanis-Tsakonas S., Gehring W. J., Steward R., Schedl P. Transfer RNA genes of Drosophila melanogaster. Nucleic Acids Res. 1980 Jul 11;8(13):2921–2937. doi: 10.1093/nar/8.13.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Feldman H. Arangement of transfer-RNA -genes in yeast. Nucleic Acids Res. 1976 Sep;3(9):2379–2386. doi: 10.1093/nar/3.9.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Gage L. P. Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell. 1979 Nov;18(3):817–828. doi: 10.1016/0092-8674(79)90134-x. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigliatti T. A., White B. N., Tener G. M., Kaufman T. C., Suzuki D. T. The localization of transfer RNA5Lys genes in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3527–3531. doi: 10.1073/pnas.71.9.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbüchle O., Larson D., Hall G. I., Sprague K. U. The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell. 1979 Dec;18(4):1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- Hatlen L., Attardi G. Proportion of HeLa cell genome complementary to transfer RNA and 5 s RNA. J Mol Biol. 1971 Mar 28;56(3):535–553. doi: 10.1016/0022-2836(71)90400-1. [DOI] [PubMed] [Google Scholar]
- Hovemann B., Sharp S., Yamada H., Söll D. Analysis of a drosophila tRNA gene cluster. Cell. 1980 Apr;19(4):889–895. doi: 10.1016/0092-8674(80)90080-x. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kressmann A., Hofstetter H., Di Capua E., Grosschedl R., Birnstiel M. L. A tRNA gene of Xenopus laevis contains at least two sites promoting transcription. Nucleic Acids Res. 1979 Dec 11;7(7):1749–1763. doi: 10.1093/nar/7.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli E., Schmidt T. The localization of tRNA4Glu genes from Drosophila melanogaster by "in situ" hybridization. Nucleic Acids Res. 1978 May;5(5):1465–1478. doi: 10.1093/nar/5.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Schmidt O., Söll D. Dimeric transfer RNA precursors in S. pombe. Cell. 1980 Sep;21(2):509–516. doi: 10.1016/0092-8674(80)90488-2. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F., Jr, White E. L., Benjamin R., Huang R. C. Low molecular weight RNA species from chromatin. Biochemistry. 1975 Aug 12;14(16):3715–3724. doi: 10.1021/bi00687a031. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morrow J. F. Recombinant DNA techniques. Methods Enzymol. 1979;68:3–24. doi: 10.1016/0076-6879(79)68003-5. [DOI] [PubMed] [Google Scholar]
- Müller F., Clarkson S. G. Nucleotide sequence of genes coding for tRNAPhe and tRNATyr from a repeating unit of X. laevis DNA. Cell. 1980 Feb;19(2):345–353. doi: 10.1016/0092-8674(80)90509-7. [DOI] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensky W. The radioiodination of RNA and DNA to high specific activities. Methods Cell Biol. 1976;13:121–152. doi: 10.1016/s0091-679x(08)61800-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O., Mao J., Ogden R., Beckmann J., Sakano H., Abelson J., Söll D. Dimeric tRNA precursors in yeast. Nature. 1980 Oct 23;287(5784):750–752. doi: 10.1038/287750a0. [DOI] [PubMed] [Google Scholar]
- Sekiya T., Mori M., Takahashi N., Nishimura S. Sequence of the distal tRNA1Asp gene and the transcription termination signal in the Escherichia coli ribosomal RNA operon rrnF(or G). Nucleic Acids Res. 1980 Sep 11;8(17):3809–3827. doi: 10.1093/nar/8.17.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E., Yanofsky C. A phenylalanine tRNA gene from Neurospora crassa: conservation of secondary structure involving an intervening sequence. Nucleic Acids Res. 1980 Mar 11;8(5):1033–1042. doi: 10.1093/nar/8.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S., Schmidt O., Söll D., Hovemann B. The nucleotide sequence of a cloned Drosophila arginine tRNA gene and its in vitro transcription in Xenopus germinal vesicle extracts. J Biol Chem. 1979 Oct 25;254(20):10290–10294. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Telford J. L., Kressmann A., Koski R. A., Grosschedl R., Müller F., Clarkson S. G., Birnstiel M. L. Delimitation of a promoter for RNA polymerase III by means of a functional test. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2590–2594. doi: 10.1073/pnas.76.6.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman S. M., Tiemeier D. C., Polsky F., Edgell M. H., Seidman J. G., Leder A., Enquist L. W., Norman B., Leder P. Cloning specific segments of the mammalian genome: bacteriophage lambda containing mouse globin and surrounding gene sequences. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4406–4410. doi: 10.1073/pnas.74.10.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Venegas A., Weinberg F., Bishop R., Rutter W. J. Structure of yeast phenylalanine-tRNA genes: an intervening DNA segment within the region coding for the tRNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):190–194. doi: 10.1073/pnas.75.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas A., Quiroga M., Zaldivar J., Rutter W. J., Valenzuela P. Isolation of yeast tRNALeu genes. DNA sequence of a cloned tRNALeu3 gene. J Biol Chem. 1979 Dec 25;254(24):12306–12309. [PubMed] [Google Scholar]
- Weber L., Berger E. Base sequence complexity of the stable RNA species of Drosophila melanogaster. Biochemistry. 1976 Dec 14;15(25):5511–5519. doi: 10.1021/bi00670a015. [DOI] [PubMed] [Google Scholar]
- Yen P. H., Davidson N. The gross anatomy of a tRNA gene cluster at region 42A of the D. melanogaster chromosome. Cell. 1980 Nov;22(1 Pt 1):137–148. doi: 10.1016/0092-8674(80)90162-2. [DOI] [PubMed] [Google Scholar]
- Yen P. H., Sodja A., Cohen M., Jr, Conrad S. E., Wu M., Davidson N. Sequence arrangement of tRNA genes on a fragment of Drosophila melanogaster DNA cloned in E. coli. Cell. 1977 Aug;11(4):763–777. doi: 10.1016/0092-8674(77)90290-2. [DOI] [PubMed] [Google Scholar]
- Zasloff M., Santos T. Reiteration frequency mapping: analysis of repetitive sequence organization within cloned DNA fragments containing the human initiator methionine tRNA gene. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5668–5672. doi: 10.1073/pnas.77.10.5668. [DOI] [PMC free article] [PubMed] [Google Scholar]