Background: BCL2 repression occurs in normal and leukemic B-lymphocytes in lymph nodes.
Results: miR-155 and miR-125b repress BCL2 translation and, in part, mediate the proliferative response to CD40 ligand (CD154).
Conclusion: miR-155 and miR-125b contribute to proliferative responses to CD154 and one of their targets is BCL2.
Significance: CD154 drives proliferation of mature B-cell malignancies and abrogating the effects of miRNA induced by CD154 may be therapeutically useful.
Keywords: Bcl-2, Blood, Leukemia, Lymphoma, MicroRNA, Stromal Cell, Translation Control, CD40
Abstract
Developmental stage-specific regulation of BCL2 occurs during B-cell maturation and has a role in normal immunity. CD40 signaling promotes proliferation and rescues B-cells from apoptosis, partly through induction of BCL2L1 and BCL2A1 and repression of BCL2. We previously showed that a stromal cell/CD40 ligand (CD154) culture system reproduced this switch in survival protein expression in primary human leukemic B-cells and we employed this model system to investigate BCL2 repression. BCL2 was post-transcriptionally regulated and the repressed BCL2 mRNA was associated with non-polysomal, but dense fractions on sucrose density gradients. Microarrays identified a set of miRNA that were induced by culture conditions and potentially able to bind to the BCL2 3′-UTR. Luciferase reporter assays demonstrated that miR-125b and miR-155 repressed BCL2 mRNA but while stromal cell contact alone was sufficient to induce strongly miR-125b this did not cause BCL2 repression. miR-155, which is the most abundant miRNA under basal conditions, specifically required CD154 for further induction above a threshold to exert its full repressive effects. Anti-miR-125b and anti-miR-155 prevented CD154-mediated repression of BCL2 and reduced CD154-mediated proliferation in the MEC1 B-cell line. We suggest that miR-155 and miR-125b, which are induced by CD154 and stromal cell signals, contribute to regulating proliferation and that BCL2 is one of their target mRNAs.
Introduction
BCL2 family pro-survival protein expression alters during normal B-cell maturation. Peripheral blood B-cells express BCL2 but following encounter with antigen and T-cells, on entry into the germinal center, BCL2 is repressed and BCL2L1 (also called BCL-XL), and BCL2A1 (also called A1 or Bfl1), are induced (1). As well as a role in apoptosis, BCL2 inhibits cell cycle entry (2), and its overexpression reduces proliferation (3). Constitutive expression of BCL2 in transgenic mice enhances specific production of memory B cells (4), suggesting that regulation of BCL2 in the germinal center may be required for normal immunity. T-cell induced CD40 signaling causes expression of BCL2L1 and BCL2A1, in part through increased NF-κB driven transcription (5), but the mechanisms for BCL2 repression are not known.
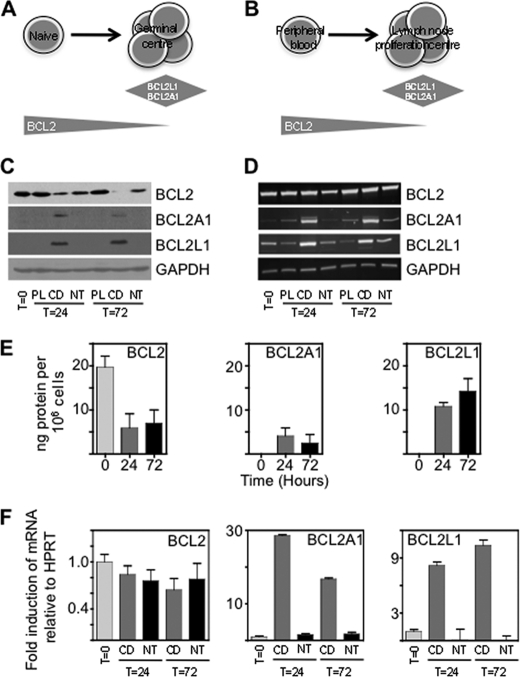
Chronic lymphocytic leukemia (CLL)2 is a lymphoproliferative disorder in which BCL2 is expressed (6) from an unrearranged locus. CLL cells are very sensitive to small molecule inhibitors of BCL2, which are currently being trialled, but toxicities such as thrombocytopenia (due to effects on normal platelets) may limit usefulness. CLL cells circulate in the peripheral blood but also exist in lymph nodes. The cellular, growth factor, and chemokine components of the lymph node microenvironment (7) are responsible for strong survival of CLL cells (1, 8) and are considered targets for therapy (9). Within CLL lymph nodes regions containing larger cells, with a relatively high growth fraction, are identified as proliferation centers. Leukemic cells in proliferation centers demonstrate down-regulation of BCL2 (10) and induction of other pro-survival proteins, BCL2L1 and BCL2A1 (1, 8) similar to the changes occurring during normal B-cell maturation (Fig. 1, A and B).
FIGURE 1.
BCL2 is translationally repressed by stromal cell/CD154 culture. Diagram showing changes in expression of BCL2 pro-survival family proteins in (A) normal B-cells on entering the germinal center, and (B) CLL cells in the lymph node proliferation center. C, Westerns showing expression of BCL2, BCL2L1, and BCL2A1 in freshly isolated CLL cells (T = 0) and at 24 and 72 h of culture on plastic (PL), stromal cells (NT), and stromal cells with CD154 (CD154); representative of six patients. D, RT-PCR to show amounts of BCL2, BCL2L1, and BCL2A1 mRNA in freshly isolated CLL cells (T = 0) and at 24 and 72 h of PL, NT, or CD154 culture (n = 6). E, quantitation of BCL2, BCL2L1, and BCL2A1 protein at 24 and 72 h of CD154 culture (n = 4). F, semi-quantitative real time PCR showing amounts of BCL2, BCL2L1, and BCL2A1 mRNA in freshly isolated cells and after 24 and 72 h of CD or NT culture (n = 6).
The regulation of BCL2 is controversial. While BCL2 mRNA has been detected in situations in which there is no BCL2 protein suggesting that BCL2 is post-transcriptionally regulated (11–13), microarray and real-time PCR gene expression studies have demonstrated an association between mRNA and protein levels (14–16). Support for post-transcriptional repression of basal BCL2 expression, through the action of miRNA, in purified peripheral blood CLL cells has come from the finding that BCL2 is a target for mir-15a and miR-16–1 (17), which are encoded in a region of chromosome 13 that is frequently deleted in CLL (18, 19). Although these miRNA have been implicated in the regulation of BCL2 under basal conditions, analysis of mice bearing homozygous deletions of the miR-15a/miR16–1 locus (20) or colon cancer cell lines (21) do not support a direct repressive effect.
We observed that culture of CLL cells on stromal cells expressing the T-cell surface protein CD40 ligand (CD154) causes BCL2 repression (10). In this report we investigated lineage specific mechanisms of BCL2 repression. We employed CLL cells in stromal cell/CD154 culture as a primary cell model system and show a novel mechanism of translational regulation through the coordinated action of miRNA including miR-155.
EXPERIMENTAL PROCEDURES
Cell Culture
Chronic lymphocytic leukemia (CLL) cells were isolated from heparinized venous whole blood using density gradient centrifugation after Local Research Ethics Committee approval was obtained (supplemental Table S1). CLL cells were cultured alone on tissue culture plastic or co-cultured with 80–90% confluent and 35 Gy irradiated non-transfected (NT culture condition) mouse fibroblast cells or human CD40 ligand (CD154) expressing mouse fibroblast cells (with rh-IL4 (10 ng/ml) (R&D Systems, Minneapolis, MN) in the medium (CD154 culture condition). MEC1 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with fetal bovine serum (Invitrogen) and penicillin/streptomycin (Invitrogen).
Quantitation of BCL2 Family Protein Expression
Protein extracts were made from freshly isolated CLL cells and CLL cells cultured on CD or NT cells using RIPA buffer supplemented with commercial protease inhibitors (Sigma). Recombinant BCL2, BCL2L1 and BCL2A1 proteins were a gift from Professor David Huang (WEHI, Melbourne, Australia) and recombinant MCL1 protein was purchased from Bioclone Inc, San Diego, CA. Antibodies used were MCL1 (Calbiochem, Notts, UK), BCL2A1, BCL2, BCL2L1, BCL2L2, and GAPDH antibodies at 1:1000 dilutions (New England Biolabs, Hertfordshire, UK). Anti-rabbit HRP-conjugated secondary antibody was used at 1:2000 (New England Biolabs). Densitometry was performed using Image J software (NIH).
Determining BCL2 Family mRNA Expression
BCL2 family mRNA levels were determined using Taqman real-time PCR. Real-Time PCR was performed using an Applied Biosystems 7500 Real-Time PCR machine (Applied Biosystems, Foster City, CA), Taqman Universal PCR Master Mix and commercial primer/probe sets (Applied Biosystems: BCL2 #Hs00153350_m1, MCL1 #Hs03043898_m1, BCL2A1 #Hs00187845_m1, BCL2L2 #Hs01573809_g1, HPRT #4333768F).
Polysome Profiling of BCL2 Family mRNAs and miRNAs
Polysome profiles were generated from freshly isolated CLL cells and CLL cells cultured on CD and NT. Cycloheximide (CHX) (100 μg/ml) was added to whole blood prior to density gradient centrifugation. For time-course experiments, CHX (100 μg/ml) was added to culture media immediately prior to CLL cell harvesting. To confirm that mRNAs in the heavy polysome fractions were bound to ribosomes, EDTA (15 mm) was added to lysis buffer instead of CHX. To determine whether mRNAs in heavy polysome fractions were being actively translated or not, puromycin (100 μg/ml) was added to CLL cells for 3 or 30 min prior to the addition of CHX and subsequent harvesting.
To make polysome extracts CLL cells were harvested, centrifuged, washed in ice-cold PBS containing 100 μg/ml CHX and lysed in 500 μl of polysome extraction buffer (15 mm Tris (pH 7.5), 15 mm MgCl2, 300 mm NaCl, 1% Triton X-100, 100 μg/ml CHX, 50 μg/ml heparin, 5 mm DTT, RNase inhibitors). Lysates were centrifuged at maximum speed for 5 min at 4 °C and the resulting supernatant immediately layered onto a 10–50% linear sucrose gradient (containing 15 mm Tris, pH 7.5, 15 mm MgCl2, 300 mm NaCl, 1 mm DTT, 50 μg/ml heparin) and centrifuged at 38,000rpm for 2 h at 4 °C using a Beckman SW40 rotor. Linear sucrose gradients were made using a Biocomp Gradient Station (Wolf Laboratories Ltd., York, UK) and validated using a sucrose refractometer.
Luciferase Reporter Assays
The entire BCL2 3′-UTR was PCR amplified from cDNA (forward primer: GATCTCTAGAACATGCCTGCCCCAAACAAATATG, reverse primer: GATCTCTAGAACAGACAAGGAAAGTTTAATGGCAATGTG) and ligated into the XbaI site in the pGL3-Promoter vector (Promega, Madison, WI) in both the correct and reverse orientations. Mutation of the miR-15b (position 2521–2527), miR125b (position 2411–2417) and miR155 (position 1568–1574) miRNA binding sites was carried out using the QuikChange II Site-directed Mutagenesis kit (Agilent Technologies UK Ltd., Wokingham, UK) and confirmed by sequencing. Primers for mutagenesis were miR-15b 5′-GGAATATCCAATCCTGcatcattaTCCTGCCAAAATCATTTTAATGGAG;miR-125b 5′-AGACCTCCCCGGCGGGCtatctttaACAGAATGATCAGACC and miR-155 5′-TCTGTACATCCTGGGatgcggaAAAAAAAATCAATGGTGGGG. Mutated seed sequences are presented in lowercase.
Transfections were carried out in 96-well plates in triplicate. HEK-293 cells were seeded at 2 × 104 cells/well 24 h prior to transfection. The pGL3 Promoter vector (0.25 μg) was co-transfected with the control pRL-TK vector (0.25 μg) using polyethylenimine (PEI) (Sigma) and serum-free media. After 4 h, media was supplemented with 10% FBS, and cells were re-transfected with a scrambled pre-miR or pre-miR-15b, pre-miR-17, pre-miR-20a, pre-miR-125b and pre-miR-155 (Applied Biosystems) at 30 nm and 100 nm using NeoFX transfection reagent (Applied Biosystems). After 24 h, luciferase and Renilla levels were measured using the Dual-Glo Luciferase assay system (Promega) and a Wallac plate reader.
Taqman Low Density Array (TLDA) miRNA Array Profiling
MicroRNA expression profiles of freshly isolated CLL cells (PB) and CLL cells cultured on CD or NT feeder layers for 24 h were determined using TLDA arrays. MicroRNA extracts were made using a miRvana miRNA isolation kit (Applied Biosystems), and reverse transcribed using MegaPlex RT Primers and a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). Human MicroRNA Array (A) plates (containing assays for 377 miRNA) were used (Applied Biosystems, #4398965) and reactions carried out on a 7900HT Real Time PCR system. Results were normalized to the internal control RNU44. Data analysis was performed using Multiple Experiment Viewer (MeV v10.2). The microarray data has been deposited with GEO (GSE24694 and GPL11050).
miRNA Inhibition
CLL cells were cultured on CD in the presence or absence of a peptide nucleic acid (PNA) directed against miR-155 with terminal lysines (K-PNA-K3) in order that it can be internalized by cells without the use of transfection reagents (a gift from Drs. Martin Fabani and Mike Gait, MRC-Laboratory of Molecular Biology, Cambridge, UK). CLL cells were cultured in serum-free media containing 10 μm PNA for 4 h, followed by culture for a further 24 and 72 h in complete media. BCL2 expression was determined by Westerns. MEC1 cells were transfected with anti-miR-155 (Applied Biosystems) using siPORT NeoFX (Ambion, Austin, TX) following which they were cultured for 24 h and harvested.
Cell Proliferation and Cell Cycle
MEC1 cells were transfected with anti-miR-155, anti-miR-125b or scrambled miRNA (Applied Biosystems) using siPORT NeoFX (Ambion) before incubating either in medium without serum or complete medium overnight. Cells from both culture conditions were then placed in complete medium with CD154 and IL4 and [3H]thymidine. Cells were harvested at 4 h and 24 h and incorporated radioactivity was measured by liquid scintillation counting. Experimental replicates were carried out in quadruplicate. Cells were stained with propidium iodide, and analysis of the cell cycle was with ModFit (BD Biosciences).
RESULTS
BCL2 Is Post-transcriptionally Repressed by Stromal Cell/CD154 Culture
To find out the effects of culture conditions on the amounts of BCL2 protein and mRNA we compared CLL cells on plastic (PL), stromal cells (NT), or stromal cells expressing CD154 with IL4 (CD154). While protein levels were repressed on CD154 but not NT or PL (Fig. 1C), BCL2 mRNA was expressed at similar amounts in all culture conditions (Fig. 1D). By contrast induction of BCL2A1 and BCL2L1 on CD154 was accompanied by increased amounts of mRNA confirming a major transcriptional component in the regulation of these genes (5). We measured absolute amounts of pro-survival factors (supplemental Fig. S1 and Fig. 1E). On CD154 BCL2 fell from ∼20 ng/106 cells to ∼6 ng/106 cells, and was replaced as the predominant pro-survival protein by BCL2L1 (∼12 ng/106 cells). RT-PCR findings (Fig. 1D) were confirmed by real-time semi-quantitative PCR (Fig. 1F). The differences in the level of amounts of BCL2 mRNA do not change significantly across the different culture conditions (one way ANOVA), but expression of BCL2L1 (p = 0.005) and BCL2A1 mRNAs (p = 0.0006) are very highly induced. These results suggest predominantly post-transcriptional repression of BCL2 by CD154.
BCL2 mRNA Associates with Heavy Fractions of a Sucrose Density Gradient Independent of Translation
Highly translated mRNAs associate with clusters of ribosomes (polysomes), and this association can be disrupted by the peptidyl transferase inhibitor, puromycin, which causes premature polypeptide chain termination on actively translated mRNA followed by ribosome release (22). Puromycin treatment has the consequence that mRNAs associated with polysomal fractions become associated with subpolysomal fractions. By contrast, EDTA causes complete dissociation of polysomes into their component subunits. Therefore, following EDTA treatment mRNAs that were associated with polysomes will become associated with subpolysomal fractions, but mRNAs that are located with other dense ribonucleoprotein particles will not be affected.
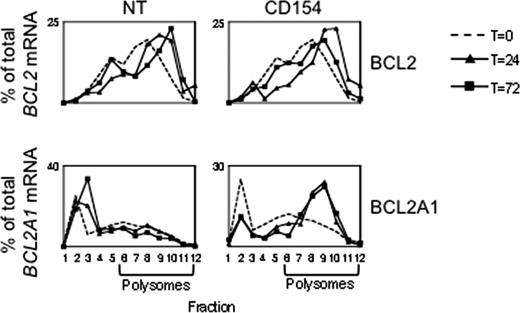
We investigated association of BCL2 mRNA with the fractions of sucrose density gradients produced from nuclear poor cell lysates and found that BCL2 mRNA associates with the dense fractions in freshly isolated CLL cells (T = 0, Fig. 2) and on NT culture (which are both conditions when BCL2 is expressed). Surprisingly BCL2 continues to associate with the dense fractions during CD154 culture, although BCL2 protein expression decreases. By contrast BCL2A1 mRNA does not bind to the dense fractions when its protein expression is absent (NT) but does become associated with these fractions when BCL2A1 protein is expressed on CD154 culture consistent with increased polysome binding and translation under this condition.
FIGURE 2.
BCL2 mRNA remains associated with the dense fractions of sucrose density gradients independent of protein expression. Relative amounts of BCL2 and BCL2A1 mRNA in fractions of a sucrose density gradient made from nuclear poor lysates from cells in either NT or CD154 culture. mRNA was measured by Taqman real time PCR at three time-points: freshly isolated cells (dotted line), 24 h (solid line with triangles and 72 h (solid line with squares). Data are representative of four independent experiments.
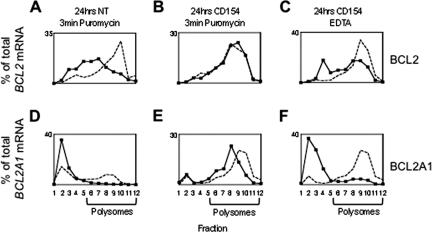
To assess peptidyl transferase activity on BCL2 mRNA cells under different culture conditions were treated with puromycin. On stromal cell culture (under which condition BCL2 is highly expressed) puromycin causes a shift of BCL2 mRNA to subpolysomal fractions (Fig. 3B) indicating ribosomal translocation along the mRNA under these conditions. However, when added after 24 h of CD154 culture (Fig. 3A) puromycin did not cause the release of BCL2, suggesting negligible peptidyl transferase activity on BCL2 mRNA (and reduced translation as compared with stromal cell culture). On CD154 culture EDTA only partially dissociated BCL2 mRNA (Fig. 3C) suggesting that it is not completely ribosome bound under these conditions.
FIGURE 3.
Association of BCL2 mRNA with the dense fractions of sucrose density gradients is puromycin insensitive. Relative amounts of BCL2 and BCL2A1 mRNA after 24 h in either (A) and (D) NT culture or (B) and (E) CD154 culture. mRNA was measured by Taqman real time PCR, either without (dotted line) or 3 min after the addition of puromycin (solid line). C and F, effects of EDTA on dissociation of mRNA from polysome fractions. The dotted line is no EDTA and solid line is following addition of EDTA. Data are representative of four independent experiments.
BCL2A1 was used as a control for puromycin and EDTA effects. On NT culture BCL2A1 is not detectable by western and BCL2A1 mRNA associates with subpolysomal fractions (Fig. 3D). On CD154 culture BCL2A1 is expressed and as expected puromycin produces a shift in BCL2A1 mRNA to subpolysomal fractions (Fig. 3E). EDTA treatment causes a complete shift to subpolysomal fractions demonstrating that BCL2A1 mRNA is polysome associated (Fig. 3F).
Overall our data suggest that BCL2 mRNA is translated on NT culture, but on CD154 culture, although BCL2 mRNA continues to associate with the dense fractions there is little ribosomal translocation along the mRNA, which is only partially polysome bound, as indicated by the incomplete effect of EDTA.
Therefore, while the observation of reduced BCL2 expression on CD154 culture (Fig. 1) could be caused by both translational or post-translational mechanisms, data obtained with puromycin and EDTA suggests a translational component to the regulation.
miRNA Potentially Capable of Binding the BCL2 3′-UTR Are Induced by the Culture System
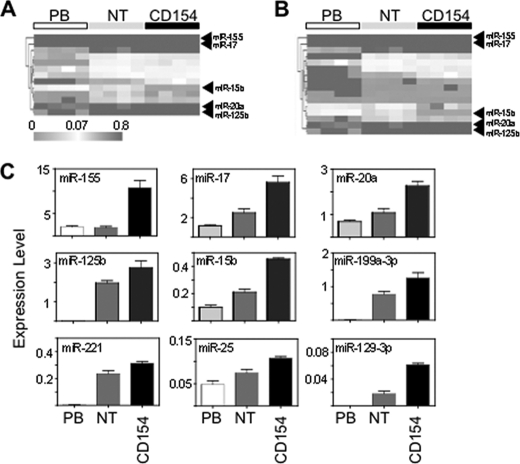
miRNA can inhibit translation of polysome-bound mRNA (23) and may also induce the formation of dense ribonucleoprotein particles that also prevent translation (24). BCL2 has a 5.2-kb 3′-UTR containing many potential miRNA binding sites. To find those sites that may be biologically relevant we used low-density microarrays and compared miRNA expression between freshly isolated peripheral blood (PB), NT and CD154 conditions. Then, using the 70 miRNA sites on the BCL2 3′-UTR that were most conserved among vertebrates (TargetScan v5.0) we found those miRNA that were significantly (ANOVA, p < 0.05)) altered across all three groups (Fig. 4A) and those significantly (Kruskal-Wallis, p < 0.05) altered between NT and CD154 conditions (Fig. 4B).
FIGURE 4.
Identification of miRNA that are induced by NT or CD154 culture and are potentially capable of binding to the BCL2 3′-UTR. Heat maps showing relative expression of miRNA expression in freshly isolated CLL cells (PB) and after 24 h of NT or CD culture (n = 4). Taqman low density microarrays for miRNA were carried out, and those miRNA potentially capable of binding the 3′-UTR of BCL2 were obtained from TargetScan. Of these 70 miRNA, those significantly changed by (A) ANOVA or (B) Wilcoxon Rank Sum test are presented. C, Taqman real time PCR of individual miRNA from freshly isolated CLL cells (PB) or cells cultured on NT or CD. The amount of miRNA relative to RNU44 is presented (n = 4).
A group of nine miRNA were significantly altered in both analyses (supplemental Fig. S2Aand Fig. 4C), and five of these were selected for further analysis. Two of these (miR-17 and miR-20a) had high context scores and two (miR-15b and miR-125b) had high conservation scores in TargetScan. miR-155 was selected because it was the most highly expressed of all miRNA on the microarry and was unique in not showing induction with the stromal cell layer alone but only in the presence of CD154. Therefore, we identify a set of miRNA induced by CD154 culture and potentially capable of regulating BCL2.
miR-155, miR-125b, and miR-15b Are Active in Luciferase Assays using the BCL2 3′-UTR
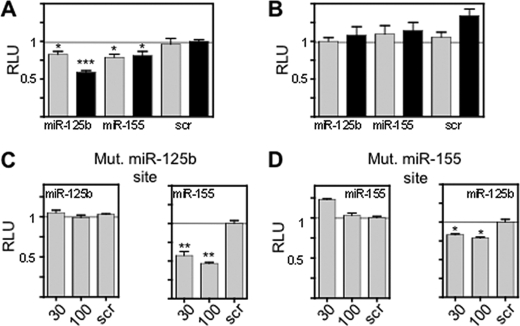
The entire BCL2 3′-UTR was cloned downstream of a luciferase reporter gene to demonstrate miRNA repressive activity. Transfected miR-155 and miR-125b mimics produced significant (Mann-Whitney U test) reductions in luciferase production as compared with a control miRNA (Fig. 5A). miR-17 and miR-20a do not show repression of luciferase although miR-15b is effective (supplemental Fig. S2C). The activity of the miR-155 and miR-125b mimics requires the 3′-UTR to be in the correct orientation (Fig. 5B). Mutation of the respective binding sites of these miRNA in the 3′-UTR cloned in the correct orientation prevents the reduction in luciferase activity produced by miR-mimics (Fig. 5C). Further evidence that the miR mimics act through their specific binding sites was obtained by transfecting the BCL2 3′-UTR bearing a mutant miR-155 site with miR-125b and the BCL2 3′-UTR bearing a mutant miR-125b site with miR-155. In both cases the expected repression was observed (Fig. 5D). Therefore, miR-155 and miR-125b are able to repress luciferase through action at specific sites on the BCL2 3′-UTR.
FIGURE 5.
miR-155 and miR-125b repress BCL2 through sites in the BCL2 3′-UTR. A, luciferase reporter constructs bearing 5. 2 kb of the BCL2 3′-UTR in the correct orientation were co-transfected into HEK293 cells with miR-125b or miR-155 mimics (30 nm light gray bars and 100 nm dark gray bars), as indicated. Scrambled miR mimic (scr) was also used at 30 and 100 nm. Luciferase activity was assayed after 24 h. Data are representative of nine independent experiments. B, luciferase reporter constructs using the indicated miR mimics and a scrambled miR (scr) utilizing the BCL2 3′-UTR cloned in the reverse orientation. Data are representative of nine independent experiments. C, HEK293 cells were transfected with the BCL2 3′-UTR, cloned into the 3′-end of a luciferase reporter gene and bearing a mutated miR-125b site with either miR-125b or miR-155 mimics. The effects of 30 or 100 nm of the respective miR mimic and of a scrambled control (scr) (30 nm) are presented. Data are representative of six independent experiments. D, HEK293 cells were transfected with the BCL2 3′-UTR, cloned into the 3′-end of a luciferase reporter gene and bearing a mutated miR-155 site together with either miR-125b or miR-155 mimics. The effects of 30 or 100 nm of the respective miR mimic and of a scrambled control (scr) (30 nm) are presented. Data are representative of six independent experiments. Significant differences were calculated using the Mann-Whitney test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
CD154 Causes Increased miR-155 Association with Polysomes
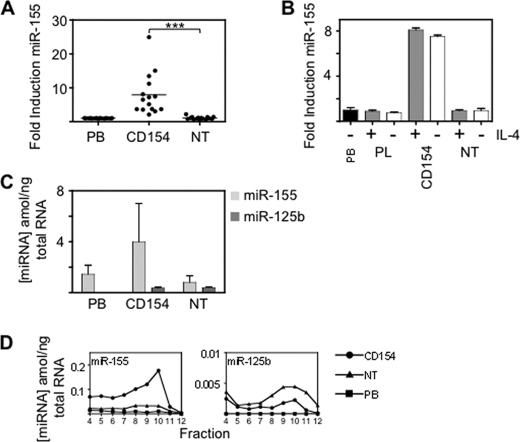
Repression of BCL2 requires CD154 stimulation. Because there was a major increase in miR-125b expression on stromal cell culture (in the absence of CD154) this miRNA alone cannot be responsible for the change in BCL2 protein expression. miR-155, in distinction to the other miRNAs, was specifically induced by CD154 culture (Fig. 6A). The induction required CD154 itself and not the other components of the culture system i.e. IL4 or the stromal cell layer, (Fig. 6B). Although miR-155 was highly expressed in PB leukemic cells (Fig. 6C) we considered the possibility that it might only associate with BCL2 mRNA when expression was increased above a threshold by CD154. To find out whether miRNA association with the polysome fractions changed with culture conditions we carried out real-time PCR of fractions from sucrose density gradients (Fig. 6D). CD154 culture caused a 3–4-fold increase in association of miR-155 with the polysome fractions, a similar induction to that observed in total cellular miR-155. NT culture produced an increase in association of miR-125b and miR-15b with the polysome fractions, as compared with PB, but there was no further increase on CD154 culture. Therefore, despite association of miR-155, miR-15b and miR-125b with the polysome fractions on NT culture, BCL2 is only reduced on CD154 culture in association with induction of miR-155 and accompanying association of this miRNA with polysome fractions.
FIGURE 6.
miR-155 is induced by CD154 culture and associates with the heavy fractions of sucrose density gradients. A, induction of miR-155 relative to RNU-44 in leukemic cells from patients (n = 15). The purified leukemic cells were cultured for 24 h on stromal cells (NT) or stromal cells with CD154 and IL-4 (CD154). PB, freshly isolated peripheral blood cells. Horizontal bar is the mean value. B, miR-155 induction does not require IL-4. Induction of miR155 relative to RNU-44 in the presence (+) or absence (−) of IL-4 in cells cultured on PL, NT or CD154. Mean ± S.E. for four patients are presented. C, absolute concentration of miR-155, miR-15b, and miR-125b (amol/ng of total RNA) in PB, NT or CD154. Mean ± S.E. for four patients are presented. Standard curves are shown in supplemental Fig. S3. D, sucrose density gradient profiles showing the amounts of miRNA (attomol/ng total RNA) in fractions 4 to 12 under different culture conditions. Squares, PB; triangles, NT; and circles, CD154. Standard curves are shown in supplemental Fig. S3. The data are representative of four patients.
Anti-miR-155 and Anti-miR-125b Prevent CD154-mediated BCL2 Repression and Proliferation in MEC1 Cells
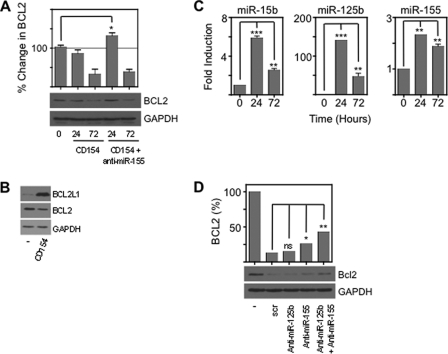
To demonstrate directly a role for miR-155 in CD154 induced BCL2 repression CLL cells were transfected with anti-miR-155 (Fig. 7A). There was a transient increase in BCL2 expression but this was not sustained either because insufficient anti-miRNA could enter the cells to antagonize the large amounts of endogenous miR-155 or because the anti-miRNA was degraded. We, therefore, employed the CLL cell line, MEC1, in further experiments, using combinations of anti-miRNAs. MEC1 expressed BCL2 without BCL2L1, similar to primary CLL cells, and on CD154 culture BCL2 was repressed and BCL2L1 induced (Fig. 7B). We measured miRNA changes in MEC1 on CD154 culture. As in primary CLL cells miR125b and miR-155 are induced (Fig. 7C). Quantification demonstrated that miR-155 was the most abundant at 5 ± 2.2 amol/ng total RNA (mean ± S.E.) as in primary cells. Therefore, MEC1 and primary CLL cells showed similar miRNA and BCL2 family protein changes in response to CD154 signaling. Next we used this cell line to investigate the combined effects of anti-miR-155 and anti-miR-125b (Fig. 7D). Anti-miR-125b had a negligible effect on BCL2 expression when used alone. Anti-miR-155 relieved repression from 14 to 26% and the combination of anti-miR-125b and anti-miR-155 relieved repression from 26 to 43%. Therefore, CD154 repression of BCL2 required miR-155 and miR-125b. We found (Fig. 1, C and E) that BCL2 was repressed to about a third of its basal level by CD154 culture and miR-155 and miR-125b may accomplish most of this effect.
FIGURE 7.
miR-125b enhances the repressive effects of miR-155, and cause reduced proliferation. Leukemic cells were cultured for 24 or 72 h in CD154 in the presence or absence of a peptide nucleic acid anti-miR-155. A, Westerns are representative of three patients. Densitometry showed that anti-miR-155 significantly (p < 0.05, Mann Whitney test) prevented CD154 repression of BCL2. Data are representative of three independent experiments. B, Westerns showing that CD154 culture causes changes to BCL2 and BCL2L1 in the CLL cell line MEC1 similar to those observed in primary leukemic cells. Data are representative of three independent experiments. C, fold induction of miR-155, miR-15b, and miR-125b in MEC1 after 24 and 72 h culture on CD154. Measurements were taken in triplicate from three independent experiments. Significant (Mann Whitney test) differences are indicated: **, p < 0.01 and ***, p < 0.001. D, miR-125b enhances the effect of miR-155. The combination of anti-miR-155 and anti-miR-125b produces a greater reduction in repression of BCL2 than either anti-miR used alone. MEC1 cells were cultured with either scrambled anti-miR (100 nm), anti-miR-125b (50 nm) with scrambled anti-miR (50 nm), ant-miR-155 (50 nm) with scrambled anti-miR (50 nm) or a combination of anti-miR-125b (50 nm) and anti-miR-155 (50 nm). Data are representative of two independent experiments. Significant (Mann Whitney test) differences are indicated: *, p < 0.05 and **, p < 0.01.
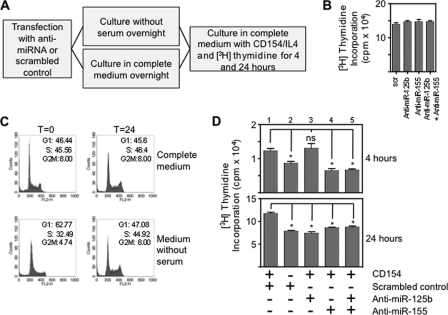
BCL2 protects against apoptosis and is required for cell cycle entry and proliferation (2, 25–29). We analyzed the effects of anti-miRNA on proliferation following either overnight culture in medium without serum (to increase the fraction of cells in G1 and slow basal proliferation) or in complete medium (Fig. 8A). Anti-miRNAs did not alter the [3H]thymidine incorporation of cells cultured continuously in complete medium (Fig. 8B) suggesting that miR-125b and miR-155 are not essential for proliferation under these conditions. We wondered whether anti-miRNAs altered the proliferative response to CD154 if cells were deprived of the stimulatory factors provided by fetal calf serum. Overnight culture without serum caused 63 ± 5% (mean ± S.E.) of cells to be in G1 whereas the corresponding proportion for cells that had been in complete medium was 46 ± 3% (Fig. 8C). After this period of culture without serum MEC1 were cultured with serum in the presence and absence of CD154. Proliferation was increased significantly by CD154 as compared with serum alone at both 4 and 24 h (p < 0.05, Mann Whitney test) (Fig. 8D, compare columns 1 and 2). At a 4 h time point anti-miR-155, but not miR-125b abolished the effect of CD154, whereas at 24 h both anti-miR-125b and anti-miR155 reduced proliferation to levels observed with serum alone.
FIGURE 8.
Effects of anti-miR-125b and anti-miR155 on proliferation and the response to cell cycle arrest. A, diagram showing the experimental procedure. B, [3H]thymidine incorporation by MEC1 cells following transfection with 60 nm scrambled oligonucleotide (scr), 30 nm anti-miR-125b with 30 nm scrambled oligonucleotide, 30 nm anti-miR-155 with 30 nm scrambled oligonucleotide or 30 nm anti-miR-125b with 30 nm anti-miR-155 and culture in complete medium. There are no significant differences in proliferation. Data are representative of three independent experiments. C, cell cycle analysis by propidium iodide staining. Analysis was carried out at two time-points: immediately after overnight culture (0 h) in either complete medium or medium without serum and secondly, 24 h after culture in complete medium or medium without serum. Proportions of cells in G1/S/G2M are presented. Data are representative of two independent experiments. D, proliferation following transfection with anti-miRNAs or scrambled control (as above) and overnight culture in medium without serum (columns 3 to 5). [3H]Thymidine incorporation was measured at 4 h and 24 h after the addition of serum with or without CD154. Proliferation caused by CD154 and serum (column 1) is compared with that due to serum only (column 2). Data are representative of three independent experiments. (Significant (Mann Whitney test) differences are indicated: *, p < 0.05.
Comparing proliferation of MEC1 transfected with scrambled control at the 24 h time point (Fig. 8, B and D) showed similar rates of proliferation whether cells had been cultured in complete medium or in medium without serum. This suggested that culture without serum had not caused permanent loss of proliferative capacity. There were also no differences in cell viability (measured by annexin V binding) to account for the observed differences in [3H]thymidine incorporation. Viability of MEC1s was 85±.6% (mean ± S.E.) in complete medium, 78 ± 0.5% after culture without serum and 80 ± 0.4% after transfection with anti-miRNAs. Our data suggest that miR-125b and miR-155 reduce proliferation due to CD154, after serum starvation in which proliferation is reduced and the fraction of quiescent (G1) cells is increased.
DISCUSSION
BCL2 is a key pro-survival protein that is involved in chromosomal translocations in follicular lymphoma but is expressed from an un-rearranged locus in CLL and also non-hematological malignancies such as non-small cell lung cancer. Mice expressing a Bcl2 transgene demonstrate abnormal production of memory B-cells suggesting that the correct regulation of this survival protein is required for normal immunity (4, 30). BCL2 is a therapeutic target and small molecule inhibitors have been produced and are entering clinical trials. These agents may have clinical toxicities due to antagonism of other BCL2 family proteins or effects on other lineages (31). It would, therefore, be advantageous to exploit mechanisms of BCL2 regulation to produce therapies that are highly lineage and gene specific.
The 5.2-kb BCL2 3′-UTR contains many potential miRNA binding sites, of which a subgroup were significantly induced by the culture conditions. We focused on the most highly expressed of these, miR-125b and miR-155, but others (such as miR-15b) are likely to contribute to the observed repression of BCL2. miR-15a and miR-16–1 repress BCL2 under basal conditions (17) and the actions of other miRNA may account for the fact that anti-miR155 and anti-miR125b were not able to restore BCL2 expression fully in our experiments (Fig. 7D).
miR-155 was induced by CD154 but not by NT culture, and was also very highly expressed both under basal conditions and on CD154 culture. miR-155 is required for normal immunity and its expression is increased both by CD40 (32) and BCR signaling (33). Constitutive expression of this miRNA leads to larger than normal germinal centers whereas disruption of the locus bearing miR-155 causes reduced antibody production and small germinal centers (32). Overexpression of miR-155 leads to the development of high-grade lymphomas (34). miR-125b is also highly expressed in normal germinal center cells (35–37), and inhibits terminal B-cell differentiation through targeting PRDM1 (36). miR-125b has also recently been identified as an oncomir (38). We identify BCL2 as being a target of miR-125b and miR-155. miR-155 is expressed in lymph node proliferation centers in CLL (39), where BCL2 is expressed at low level (10) demonstrating the in vivo association between high miR-155 levels and low BCL2 expression, which is mimicked by our culture system.
The culture system we used enables dissection of the action of miRNA capable of repressing BCL2. Although miR-125b is induced by stromal cell culture alone, and associates with polysomes under these conditions, it is not sufficient to repress BCL2, which requires CD154 specific miRNA. We suggest that miR-155 is a CD154-dependent component of BCL2 regulation. We found that association of miR-155 with polysomes increases on CD154 culture, and this strongly implies that despite its high basal expression, levels must be increased above a threshold by CD154 to be effective.
Surprisingly, both normal germinal center cells and leukemic B-cells down-regulate BCL2 in lymph nodes while other pro-survival proteins are induced. Overall, these in vitro changes cause improved survival (1, 40) and BCL2 repression is not associated with apoptosis. The reasons for BCL2 repression are not clear, but we wondered whether it could have a non-apoptotic function. Several lines of evidence support a role for BCL2 family proteins in the rate of cell cycle progression and proliferation. BCL2 expressing cells that survive cytokine withdrawal are quiescent (41), thymocyte turn-over is slower in BCL2 transgenic mice and BCL2 transgenic lymphocytes enter cell cycle more slowly than their normal counterparts (3, 27–29, 42). In some experimental systems BCL2 and BCL2L1 have similar effects in preventing or delaying cell cycle entry (29, 43), but in B-cells CD40 signaling is associated with proliferation, cell cycle entry and induction of BCL2L1 expression (5). In human lymphomas BCL2 expression correlates with lower proliferative activity (44). Serum starvation can produce apoptosis or cell cycle arrest depending on the cell type. We found that serum starvation of the CLL cell line, MEC1 produced an accumulation of cells in G1 and reduced proliferation without an increase in apoptosis. Antagonism of miR-125b and miR-155 was sufficient to abrogate CD154-mediated proliferation following a period of serum deprivation (Fig. 8D). We suggest that serum deprivation blunts the proliferative response to CD154 sufficiently to allow detection of effects of anti-miR-125b and anti-miR-155. Collectively our results suggest that miR-155 and miR-125b contribute to the promotion of proliferation and we speculate that they may function in the transition from quiescence to proliferation that occurs when B-cells encounter microenvironmental signals in the lymph node.
Individual miRNAs have many target mRNAs and this has prompted the RNA operon concept whereby expression or translation of functionally related mRNAs are altered by sets of miRNA (45). We showed that BCL2 mRNA is a target of miR-125b and miR-155 and prior literature demonstrated that BCL2 has a subtle effect in delaying the rate of cell cycle progression. We have shown that BCL2 is a target of miR-125b and miR-155 and speculate that these elements form part of an RNA operon that mediates the B-cell proliferative response to CD154.
An advantage of our primary cell model system is that rather than transfecting expression constructs we could manipulate cell culture conditions to produce substantial changes in endogenous proteins, which mimic some of the changes in BCL2-family pro-survival proteins observed in lymph nodes. Our data showed that BCL2 mRNA associated with the dense fractions of sucrose density gradients both in culture conditions in which there was active translation and under conditions of repression, and this association was puromycin insensitive. The mechanisms by which miRNAs repress translation in metazoan cells continue to be investigated. Although miRNA can cause degradation of Caenorhabditis elegans mRNA (46) we did not find changes in amounts of BCL2 mRNA in different culture conditions. In other experimental systems miRNA inhibit cap-complex assembly (47, 48) or post-initiation events (23, 49). BCL2 has a weak IRES site (50) and its translation may not be absolutely dependent on cap-dependent mechanisms in all conditions. However, repressed BCL2 mRNA was resistant to the effects of puromycin suggesting that miRNA interference with post-initiation events is also not relevant to this system. A further mechanism of translational suppression by miRNA has been suggested: in a Drosophila cell free system miRNA caused sequestration of mRNA in dense ribonucleoprotein complexes (pseudo-polysomes) (24). We found that the repressed mRNA was EDTA insensitive showing that it was unlikely to be polysome associated. Overall our results are compatible with repressed BCL2 mRNA being in a non-polysome dense ribonucleoprotein fraction.
The lymph node microenvironment, including T-cells, follicular dendritic cells, chemokines, and growth factors supports the survival and proliferation of normal and leukemic B-cells and is a target for therapy (9). We have shown that BCL2 is a target of a novel mechanism of repression by miR-155 and miR-125b, potentially induced by signals from the microenvironment, and these miRNA promote the proliferative response to CD154.
Supplementary Material
Acknowledgments
We thank Drs. Martin Fabani and Mike Gait, MRC Laboratory of Molecular Biology, Cambridge, UK for the gift of anti-miR-155 and Professor David Huang and Dr. Peter Czabotar, WEHI, Melbourne, Australia for the gift of recombinant proteins.
This work was supported by grants from the Kay Kendall Leukemia Fund.

This article contains supplemental Table S1 and Figs. S1–S3.
- CLL
- chronic lymphocytic leukemia
- CHX
- cycloheximide
- PEI
- polyethylenimine
- PNA
- peptide nucleic acid.
REFERENCES
- 1. Willimott S., Baou M., Naresh K., Wagner S. D. (2007) CD154 induces a switch in pro-survival Bcl-2 family members in chronic lymphocytic leukaemia. Br. J. Haematol. 138, 721–732 [DOI] [PubMed] [Google Scholar]
- 2. Huang D. C., O'Reilly L. A., Strasser A., Cory S. (1997) The anti-apoptosis function of Bcl-2 can be genetically separated from its inhibitory effect on cell cycle entry. EMBO J. 16, 4628–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Reilly L. A., Harris A., Tarlinton D., Corcoran L., Strasser A. (1997) J. Immunol. [PubMed] [Google Scholar]
- 4. Smith K. G., Light A., O'Reilly L. A., Ang S. M., Strasser A., Tarlinton D. (2000) bcl-2 transgene expression inhibits apoptosis in the germinal center and reveals differences in the selection of memory B cells and bone marrow antibody-forming cells. J. Exp. Med. 191, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee H. H., Dadgostar H., Cheng Q., Shu J., Cheng G. (1999) NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 96, 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schena M., Larsson L. G., Gottardi D., Gaidano G., Carlsson M., Nilsson K., Caligaris-Cappio F. (1992) Growth- and differentiation-associated expression of bcl-2 in B-chronic lymphocytic leukemia cells. Blood 79, 2981–2989 [PubMed] [Google Scholar]
- 7. Caligaris-Cappio F. (2003) Role of the microenvironment in chronic lymphocytic leukaemia. Br. J. Haematol. 123, 380–388 [DOI] [PubMed] [Google Scholar]
- 8. Vogler M., Butterworth M., Majid A., Walewska R. J., Sun X. M., Dyer M. J., Cohen G. M. (2009) Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood 113, 4403–4413 [DOI] [PubMed] [Google Scholar]
- 9. Burger J. A., Ghia P., Rosenwald A., Caligaris-Cappio F. (2009) The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood 114, 3367–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmid C., Isaacson P. (1994) Proliferation centres in B-cell malignant lymphoma, lymphocytic (B-CLL): an immunophenotypic study. Histopathology 24, 445–451 [DOI] [PubMed] [Google Scholar]
- 11. Akagi T., Kondo E., Yoshino T. (1994) Leuk. Lymphoma 13, 81–87 [DOI] [PubMed] [Google Scholar]
- 12. Chleq-Deschamps C. M., LeBrun D. P., Huie P., Besnier D. P., Warnke R. A., Sibley R. K., Cleary R. K. (1993) Topographical dissociation of BCL-2 messenger RNA and protein expression in human lymphoid tissues. Blood 81, 293–298 [PubMed] [Google Scholar]
- 13. Kondo E., Nakamura S., Onoue H., Matsuo Y., Yoshino T., Aoki H., Hayashi K., Takahashi K., Minowada J., Nomura S. (1992) Detection of bcl-2 protein and bcl-2 messenger RNA in normal and neoplastic lymphoid tissues by immunohistochemistry and in situ hybridization. Blood 80, 2044–2051 [PubMed] [Google Scholar]
- 14. Husson H., Carideo E. G., Neuberg D., Schultze J., Munoz O., Marks P. W., Donovan J. W., Chillemi A. C., O'Connell P., Freedman A. S. (2002) Gene expression profiling of follicular lymphoma and normal germinal center B cells using cDNA arrays. Blood 99, 282–289 [DOI] [PubMed] [Google Scholar]
- 15. Klein U., Tu Y., Stolovitzky G. A., Keller J. L., Haddad J., Jr., Miljkovic V., Cattoretti G., Califano A., Dalla-Favera R. (2003) Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. U.S.A. 100, 2639–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen Y., Iqbal J., Huang J. Z., Zhou G., Chan W. C. (2004) BCL2 protein expression parallels its mRNA level in normal and malignant B cells. Blood 104, 2936–2939 [DOI] [PubMed] [Google Scholar]
- 17. Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., Rassenti L., Alder H., Volinia S., Liu C. G., Kipps T. J., Negrini M., Croce C. M. (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C. M. (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sellmann L., Scholtysik R., Kreuz M., Cyrull S., Tiacci E., Stanelle J., Carpinteiro A., Nückel H., Boes T., Gesk S., Siebert R., Klein-Hitpass L., Dührsen U., Dürig J., Küppers R. (2010) Gene dosage effects in chronic lymphocytic leukemia. Cancer Genet. Cytogenet. 203, 149–160 [DOI] [PubMed] [Google Scholar]
- 20. Klein U., Lia M., Crespo M., Siegel R., Shen Q., Mo T., Ambesi-Impiombato A., Califano A., Migliazza A., Bhagat G., Dalla-Favera R. (2010) The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17, 28–40 [DOI] [PubMed] [Google Scholar]
- 21. Linsley P. S., Schelter J., Burchard J., Kibukawa M., Martin M. M., Bartz S. R., Johnson J. M., Cummins J. M., Raymond C. K., Dai H., Chau N., Cleary M., Jackson A. L., Carleton M., Lim L. (2007) Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell Biol. 27, 2240–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Azzam M. E., Algranati I. D. (1973) Mechanism of puromycin action: fate of ribosomes after release of nascent protein chains from polysomes. Proc. Natl. Acad. Sci. U.S.A. 70, 3866–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nottrott S., Simard M. J., Richter J. D. (2006) Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 13, 1108–1114 [DOI] [PubMed] [Google Scholar]
- 24. Thermann R., Hentze M. W. (2007) Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 447, 875–878 [DOI] [PubMed] [Google Scholar]
- 25. Deng X., Gao F., Flagg T., May W. S., Jr. (2004) Mono- and multisite phosphorylation enhances Bcl2's antiapoptotic function and inhibition of cell cycle entry functions. Proc. Natl. Acad. Sci. U.S.A. 101, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deng X., Gao F., May W. S., Jr. (2003) Bcl2 retards G1/S cell cycle transition by regulating intracellular ROS. Blood 102, 3179–3185 [DOI] [PubMed] [Google Scholar]
- 27. Linette G. P., Li Y., Roth K., Korsmeyer S. J. (1996) Cross talk between cell death and cell cycle progression: BCL-2 regulates NFAT-mediated activation. Proc. Natl. Acad. Sci. U.S.A. 93, 9545–9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazel S., Burtrum D., Petrie H. (1996) Regulation of cell division cycle progression by bcl-2 expression: a potential mechanism for inhibition of programmed cell death. J. Exp. Med. 183, 2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Reilly L. A., Huang D. C., Strasser A. (1996) The cell death inhibitor Bcl-2 and its homologues influence control of cell cycle entry. EMBO J. 15, 6979–6990 [PMC free article] [PubMed] [Google Scholar]
- 30. Tarlinton D. M., Smith K. G. (2000) Dissecting affinity maturation: a model explaining selection of antibody-forming cells and memory B cells in the germinal centre. Immunol. Today 21, 436–441 [DOI] [PubMed] [Google Scholar]
- 31. Mason K. D., Carpinelli M. R., Fletcher J. I., Collinge J. E., Hilton A. A., Ellis S., Kelly P. N., Ekert P. G., Metcalf D., Roberts A. W., Huang D. C., Kile B. T. (2007) Programmed anuclear cell death delimits platelet life span. Cell 128, 1173–1186 [DOI] [PubMed] [Google Scholar]
- 32. Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Regulation of the germinal center response by microRNA-155. Science 316, 604–608 [DOI] [PubMed] [Google Scholar]
- 33. Yin Q., Wang X., McBride J., Fewell C., Flemington E. (2008) B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 283, 2654–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C. M. (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 103, 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Basso K., Sumazin P., Morozov P., Schneider C., Maute R. L., Kitagawa Y., Mandelbaum J., Haddad J., Jr., Chen C. Z., Califano A., Dalla-Favera R. (2009) Identification of the human mature B cell miRNome. Immunity 30, 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gururajan M., Haga C. L., Das S., Leu C. M., Hodson D., Josson S., Turner M., Cooper M. D. (2010) MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int. Immunol. 22, 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malumbres R., Sarosiek K. A., Cubedo E., Ruiz J. W., Jiang X., Gascoyne R. D., Tibshirani R., Lossos I. S. (2009) Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 113, 3754–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bousquet M., Harris M. H., Zhou B., Lodish H. F. (2010) MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. U.S.A. 107, 21558–21563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang M., Tan L. P., Dijkstra M. K., van Lom K., Robertus J. L., Harms G., Blokzijl T., Kooistra K., van T'veer M. B., Rosati S., Visser L., Jongen-Lavrencic M., Kluin P. M., van den Berg A. (2008) miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J. Pathol. 215, 13–20 [DOI] [PubMed] [Google Scholar]
- 40. Cuní S., Pérez-Aciego P., Pérez-Chacón G., Vargas J. A., Sánchez A., Martín-Saavedra F. M., Ballester S., García-Marco J., Jordá J., Durántez A. (2004) A sustained activation of PI3K/NF-κB pathway is critical for the survival of chronic lymphocytic leukemia B cells. Leukemia 18, 1391–1400 [DOI] [PubMed] [Google Scholar]
- 41. Vaux D. L., Cory S., Adams J. M. (1988) Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335, 440–442 [DOI] [PubMed] [Google Scholar]
- 42. O'Reilly L. A., Harris A. W., Strasser A. (1997) bcl-2 transgene expression promotes survival and reduces proliferation of CD3-CD4-CD8- T cell progenitors. Int. Immunol. 9, 1291–1301 [DOI] [PubMed] [Google Scholar]
- 43. Janumyan Y. M., Sansam C. G., Chattopadhyay A., Cheng N., Soucie E. L., Penn L. Z., Andrews D., Knudson C. M., Yang E. (2003) Bcl-xL/Bcl-2 coordinately regulates apoptosis, cell cycle arrest and cell cycle entry. EMBO J. 22, 5459–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winter J. N., Andersen J., Reed J. C., Krajewski S., Variakojis D., Bauer K. D., Fisher R. I., Gordon L. I., Oken M. M., Jiang S., Jeffries D., Domer P. (1998) BCL-2 expression correlates with lower proliferative activity in the intermediate- and high-grade non-Hodgkin's lymphomas: an Eastern Cooperative Oncology Group and Southwest Oncology Group cooperative laboratory study. Blood 91, 1391–1398 [PubMed] [Google Scholar]
- 45. Keene J. D. (2007) RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 46. Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A. E. (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 [DOI] [PubMed] [Google Scholar]
- 47. Mathonnet G., Fabian M. R., Svitkin Y. V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W. C., Darzynkiewicz E., Pillai R. S., Filipowicz W., Duchaine T. F., Sonenberg N. (2007) MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317, 1764–1767 [DOI] [PubMed] [Google Scholar]
- 48. Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. (2005) Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309, 1573–1576 [DOI] [PubMed] [Google Scholar]
- 49. Petersen C. P., Bordeleau M. E., Pelletier J., Sharp P. A. (2006) Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21, 533–542 [DOI] [PubMed] [Google Scholar]
- 50. Sherrill K. W., Byrd M. P., Van Eden M. E., Lloyd R. E. (2004) BCL-2 translation is mediated via internal ribosome entry during cell stress. J. Biol. Chem. 279, 29066–29074 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.