Background: Accumulation of DNA damage and deficiency in DNA repair may contribute to neuronal loss in Alzheimer disease.
Results: Sublethal concentrations of aggregated β-amyloid peptides inhibit DNA-PK kinase activity in PC12 cells.
Conclusion: DNA-PK inhibition may contribute to neurodegeneration by impairing DNA repair capability, inducing DNA damage accumulation.
Significance: This represents a novel mechanism by which Aβ exerts its neurotoxic effects in Alzheimer disease.
Keywords: Alzheimer Disease, DNA Damage, DNA Repair, Neurodegeneration, Oxidative Stress, DNA-dependent Protein Kinase, beta-Amyloid Peptide
Abstract
Accumulation of DNA damage and deficiency in DNA repair potentially contribute to the progressive neuronal loss in neurodegenerative disorders, including Alzheimer disease (AD). In multicellular eukaryotes, double strand breaks (DSBs), the most lethal form of DNA damage, are mainly repaired by the nonhomologous end joining pathway, which relies on DNA-PK complex activity. Both the presence of DSBs and a decreased end joining activity have been reported in AD brains, but the molecular player causing DNA repair dysfunction is still undetermined. β-Amyloid (Aβ), a potential proximate effector of neurotoxicity in AD, might exert cytotoxic effects by reactive oxygen species generation and oxidative stress induction, which may then cause DNA damage. Here, we show that in PC12 cells sublethal concentrations of aggregated Aβ(25–35) inhibit DNA-PK kinase activity, compromising DSB repair and sensitizing cells to nonlethal oxidative injury. The inhibition of DNA-PK activity is associated with down-regulation of the catalytic subunit DNA-PK (DNA-PKcs) protein levels, caused by oxidative stress and reversed by antioxidant treatment. Moreover, we show that sublethal doses of Aβ(1–42) oligomers enter the nucleus of PC12 cells, accumulate as insoluble oligomeric species, and reduce DNA-PK kinase activity, although in the absence of oxidative stress. Overall, these findings suggest that Aβ mediates inhibition of the DNA-PK-dependent nonhomologous end joining pathway contributing to the accumulation of DSBs that, if not efficiently repaired, may lead to the neuronal loss observed in AD.
Introduction
Accumulation of DNA damage and defects in DNA repair has been documented in different neurodegenerative disorders, including Alzheimer disease (AD)2 (1–5), Huntington disease (6–9), Parkinson disease (10–13), and amyotrophic lateral sclerosis (14–17). The extent of DNA damage determines whether apoptotic cascades initiate, causing progressive neurodegeneration, or the DNA repair mechanisms become active to reverse the damage (18). Accumulation of DNA damage is thought to be particularly deleterious in post-mitotic cells, which cannot be replaced through cell division (19). For this reason, neurons are particularly susceptible to the toxic effects of reactive oxygen species (ROS), the primary mediators of oxidative stress, which may cause damages at the main macromolecular systems, including nucleic acids (20).
Oxidative stress and neuronal DNA damage are common features of neurodegenerative diseases (21–23) associated with misfolded proteins that accumulate as intracellular and/or extracellular amyloid or amyloid-like deposits (24), such as AD. Neuropathologically, AD is characterized by extensive loss of synapses and neurons, accumulation of intracellular tangles, and extracellular/intracellular β-amyloid peptide (Aβ) deposits (25). During the aggregation process, monomeric Aβ peptides form soluble oligomeric intermediates, which further associate in high molecular weight assemblies (protofibrils) and eventually generate amyloid fibrils and plaques. Soluble nonfibrillar forms of Aβ are proximate effectors of neurotoxicity and synaptotoxicity (26, 27) in AD. Studies performed on mammalian cell lines, primary cortical neurons, and AD brains suggest that Aβ peptide exerts neurotoxic effects through ROS production (28–30), generated during the early stages of protein aggregation when only protofibrils or soluble oligomers are present. DNA damage caused by ROS includes altered bases, abasic sites, and single and double strand breaks (DSBs). Interestingly, increased levels of DNA breaks and alkali-labile sites were detected in the cerebral cortex of AD patients (2), and in situ labeling methods showed the presence of single and double strand breaks in different AD brain regions (3, 31, 32). Moreover, a decreased capacity for DNA repair in fibroblasts or lymphocytes from patients with familial AD was also reported (33).
DSBs are considered the most lethal form of DNA damage that, if unrepaired, might cause cell death (34). In mammalian cells, DSB repair is achieved by two highly efficient mechanisms, homologous recombination (35, 36) and nonhomologous end joining (NHEJ). Although there is increasing evidence that these pathways compete for DSB repair (37), NHEJ is considered the predominant pathway in higher eukaryotes (38). It requires the DNA-dependent protein kinase holoenzyme (DNA-PK), which is formed by a 470-kDa catalytic subunit, DNA-PKcs, and a heterodimer of 70- and 80-kDa polypeptides, known as Ku, which binds to DNA strand breaks, recruiting and activating the DNA-PKcs (39–41). DNA-PKcs is a serine/threonine kinase belonging to the PI3K-like family of kinases, which includes ataxia telangiectasia-mutated kinase and Rad3-related kinase. End joining activity and protein levels of DNA-PKcs are significantly lower in AD brains compared with normal controls. The amount of end joining activity correlates with the expression of DNA-PKcs and is dependent on DNA-PK catalytic activity (4). In addition, immunohistochemical analysis of AD temporal cortex showed a decrease, although not significant, of DNA-PKcs expression both in neurons and astrocytes with increasing Braak stages (42). Overall, these findings suggest that repair of DNA DSBs may be deficient in AD, although the molecular candidate causing the NHEJ impairment has yet to be identified.
Because a low amount of Aβ is likely present in AD brain for extended periods prior to neuronal cell death, we investigated whether sublethal doses of Aβ might inhibit DNA-PK function, thus compromising the repair of DSBs. Addressing this issue, we found that sublethal concentrations of aggregated Aβ(25–35) inhibit DNA-PK kinase activity in PC12 cells. This inactivation was associated with down-regulation of the DNA-PKcs protein levels, caused by oxidative stress and resolved by the antioxidant NAC. We also found that exogenous Aβ(1–42) oligomers enter the nucleus of PC12 cells, accumulate as insoluble oligomeric species, and inhibit DNA-PK kinase activity. Finally, we demonstrated that Aβ-mediated DNA-PK kinase activity inhibition renders proliferating PC12 cells susceptible to nonlethal oxidative injury and attenuates DSB repair in NGF-differentiated PC12 cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
PC12 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) horse serum (EuroClone), 5% (v/v) fetal bovine serum (EuroClone), 2 mm l-glutamine (BioWest), 100 units/ml penicillin, and 100 μg/ml streptomycin (BioWest). To induce neuronal differentiation, PC12 cells (50 × 103) were plated on 12-mm glass coverslips coated with rat tail collagen type I (Sigma) in RPMI 1640 medium containing 2% serum and treated with 100 ng/ml NGF (mouse nerve growth factor 2.5S grade I, Alomone Labs) for 9 days. Medium was replaced every 2 days to ensure maximal viability.
For β-amyloid peptide treatments, Aβ(25–35) and the control reversed sequence peptide Aβ(35–25) (Bachem) were dissolved in sterile double-distilled H2O at 1 mm. To prepare aggregated Aβ peptides, the stock peptide solution was incubated at 37 °C with continuous shaking (160 rpm) for 7 days and added at its final concentrations to PC12 cells plated in complete growth medium. Oligomeric preparations of Aβ(1–42) and Aβ(42–1) peptides (Bachem) were prepared in F-12 medium containing 1% DMSO as described previously (43) and added to PC12 cells plated in F-12 medium containing 1.5% horse serum, 0.5% fetal bovine serum, and 0.5, 0.1, or 0.01% DMSO, depending on the final concentration of Aβ(1–42). The oligomerization level was verified by Western blot analysis.
Antioxidant NAC (Sigma) was used at final concentrations of 0.5 mm for 28 h (4-h pretreatment followed by a 24-h co-incubation with or without Aβ). Hydrogen peroxide (Sigma) was added at final concentrations of 100 μm for 30 min to induce sublethal oxidative stress. In both conditions, RPMI 1640 medium without serum was used as incubation buffer. DNA-PK inhibitor NU7026 was dissolved in DMSO at 7.1 mm and used at a final concentration of 10 μm in complete medium for 24 h (44, 45). DSB inducer doxorubicin (DOX) was used on proliferating and differentiated PC12 cells at 1 μm for 8 h. Analysis of dose-response (0.01–10 μm) and time-response (4–24 h) curves revealed that this experimental condition had maximal DNA damage response in proliferating and NGF-differentiated PC12 cells, evaluated by histone H2AX phosphorylation (γH2AX), without significant cell death.
Cell Viability Assays
The effects of aggregated Aβ(25–35) and oligomeric Aβ(1–42) on mitochondrial metabolism were assessed by MTT assay (Chemicon). Briefly, PC12 cells were plated onto 96-well plates (10 × 103 cells/well) and after 24 h were subjected to Aβ(25–35) or Aβ(1–42) treatments, followed by incubation with 10 μl of MTT solution (5 mg/ml) for 4 h at 37 °C to ensure formazan salt formation. To dissolve salts and to acidify the red medium obtained from the reaction, cells were solubilized with isopropyl alcohol/HCl ice-cold solution, and the absorbance was read at 570 nm in a spectrophotometer microplate reader (Tecan). Values of cells treated with Aβ(25–35), Aβ(1–42), or reverse phase Aβ(35–25) and Aβ(42–1) were normalized against values for vehicle-treated controls and expressed as a percentage of control set at 100%. Data are representative of 4–6 independent experiments.
The effects of Aβ(25–35) and Aβ(1–42) on plasma membrane integrity were evaluated by a LDH assay (Promega). PC12 cells, cultured and treated in the same experimental conditions used for MTT assay, were centrifuged at 250 × g for 10 min at room temperature (RT), and then 50 μl of supernatant was added to an isovolume of Substrate Mix and incubated for 30 min at RT and light-covered. After adding the Stop Solution, absorbance readings were measured by a spectrophotometer microplate reader (wavelength, 490 nm, reference wavelength 630 nm). Results were expressed as % maximum LDH release, obtained by adding 0.9% Triton X-100 for 60 min, subtracted by values corresponding to base-line LDH release (i.e. untreated cells). Data are representative of 4–6 independent experiments.
The percentage of apoptotic cells was evaluated by the DeadEndTM fluorometric TUNEL assay (Promega) and Hoechst 33342 nuclear staining. Cells (∼3 × 105) were cultured onto poly-l-lysine (Sigma)-coated coverslips (12 mm) placed at the bottom of 35-mm culture dishes and after 24 h were exposed to Aβ(25–35) or Aβ(1–42) over a time range of 24–72 h. Cells were washed three times with PBS (137 mm NaCl, 2.7 mm KCl, 8.1 mm sodium phosphate dibasic, 1.9 mm potassium phosphate monobasic, pH 7.5) and fixed with 4% (w/v) paraformaldehyde solution in PBS for 25 min at 4 °C. After washing in PBS, cells were permeabilized with 0.2% Triton® X-100 in PBS at RT for 5 min, incubated with equilibration buffer for 20 min at RT, and labeled with TdT reaction mix for 1 h at 37 °C. After washing with PBS, cells were incubated for 15 min at RT with Stop Solution, washed thoroughly, and analyzed by fluorescence microscopy.
Nuclei were stained with 0.5 μg/ml Hoechst 33342 for 4 min at RT in distilled water, and condensed and/or fragmented nuclei were counted as apoptotic nuclei.
At least 500 cells for each coverslip were counted in both assays, and data were representative of three independent experiments.
Subcellular Fractionation
The presence of oligomeric Aβ(1–42) in the nuclear compartment of PC12 cells was verified by nuclear and cytosolic fractionation. Cells (∼17 × 105), treated for 24 h with 50 μm oligomeric Aβ(1–42), were washed twice with ice-cold PBS, harvested, and homogenized on ice by Dounce disruption in 10 mm Tris-HCl, pH 7.5, and 5 mm EDTA containing 1 mm PMSF and mixture protease inhibitors (Sigma). Cell integrity was microscopically checked using trypan blue (Sigma) staining. The solution was brought up to 320 mm sucrose and centrifuged four times at 1000 × g for 10 min at 4 °C to obtain the nuclear fraction. The supernatant was then centrifuged at 14,000 × g at 4 °C for 10 min to separate soluble (supernatant) and insoluble (pellet) cytosolic proteins. Isolated nuclei were lysed and centrifuged as described in “DNA-PK kinase activity assays” paragraph to separate soluble (supernatant) and insoluble (pellet) nuclear proteins. Insoluble proteins of nuclear and cytosolic compartments were extracted by incubation for 10 min at RT with Novex® Tricine SDS sample buffer (Invitrogen), boiled for 10 min, and then spun 2 min at 14,000 × g. Soluble and insoluble proteins were then separated by 10–20% Novex® Tricine gel (Invitrogen) and analyzed by Western blot for Aβ(1–42) oligomers.
To assess the effect of DOX and Aβ(25–35) on Ku70/Ku86 compartmentalization, treated proliferating PC12 cells (∼5 × 106) were washed twice with ice-cold PBS, harvested, spun at 4 °C for 5 min at 600 × g, and lysed on ice with NE-PER® nuclear and cytoplasmic extraction reagents (Thermo Scientific) following the manufacturer's instructions. For each fraction, 40 μg of proteins were separated by SDS-PAGE and analyzed by Western blot.
SDS-PAGE and Western Blot Analysis
Appropriate amounts of protein extracts were boiled for 5 min in SDS-PAGE Laemmli buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1% bromphenol blue, 50 mm DTT) and separated by SDS-PAGE (5% polyacrylamide for DNA-PKcs, 10% for Ku70, Ku86, tubulin, and actin, and 12% for histone 1 and protein carbonylation detection). Proteins were electrotransferred to nitrocellulose membrane (HybondTM C-extra, GE Healthcare) at 30 V overnight at 4 °C for DNA-PKcs detection and at 100 V for 1 h at 4 °C for the other proteins analyzed. Membranes were blocked for 1 h at RT with 10% (w/v) milk in TBS-T solution (Blocking buffer, 0.1% Tween 20 in 1.3 m NaCl, 200 mm KCl, 250 mm Tris, pH 7.5) and incubated with primary antibodies using the following dilutions: anti-actin 1:1000 (Sigma A3853); anti-DNA-PKcs Ab-4 mixture 1:400 (Neo Markers MS-423-P); anti-Ku70 1:400 (Santa Cruz Biotechnology sc1486); anti-ku86 1:500 (Santa Cruz Biotechnology sc1484); anti-histone 1 1:300 (Upstate 05-457); anti-α tubulin 1:500 (Santa Cruz Biotechnology sc8035) in TBS-T containing 2 or 5% milk for 2 h at RT or overnight at 4 °C with gentle shaking. After extensive washing in TBS-T, membranes were probed for 1 h at RT with HRP-conjugated antibodies (anti-rabbit IgG 1:100,000 (711-035-152) and anti-mouse IgG 1:100,000 (715-035-151) Jackson ImmunoResearch; anti-goat IgG 1:500,000 (sc-2768) Santa Cruz Biotechnology) diluted in TBS-T containing 2% milk and washed thoroughly with TBS-T. For Aβ(1–42) and Aβ(25–35) detection, proteins were separated by 10–20% Novex® Tricine gel and electrotransferred to a 0.2-μm nitrocellulose membrane (GE Healthcare) at 160 mA for 30 min, followed by 100 mA for 45 min. Membranes were blocked for 1 h at RT with Blocking buffer and incubated with 4G8 1:1000 (mouse monoclonal against Aβ residues 17–24; Signet) and anti-Aβ(25–35) 1:1000 (Abcam) in TBS-T containing 5% milk overnight at 4 °C with gentle shaking. After extensive washing in TBS-T, membranes were probed for 1 h at RT with the HRP-conjugated antibodies anti-mouse and anti-rabbit IgG. Immunoreactive bands were visualized by enhanced chemiluminescence detection system (EuroClone) on Amersham Biosciences HyperfilmTM ECL. Images were acquired using a Typhoon 9200 variable-mode imager (GE Healthcare), and densitometric analysis was performed using ImageQuant software (GE Healthcare).
Protein Carbonylation Analysis and ROS Detection
OxyBlot protein oxidation detection kit (Chemicon) was used to detect carbonyl groups introduced into proteins by oxidative reaction. Briefly, ∼5 × 106 cells were grown onto poly-l-lysine-coated 100-mm polystyrene dishes and treated with Aβ(25–35) or Aβ(1–42) as indicated. Cells were washed twice with ice-cold PBS, harvested, centrifuged at 4 °C for 5 min at 600 × g, and lysed on ice with RIPA buffer containing protease and phosphatase inhibitors. Five μg of each sample was denatured by adding an isovolume of 12% SDS (w/v) before the derivatization reaction, where carbonyl groups of the protein side chain were derivatized into 2,4-dinitrophenylhydrazone by reaction with 2,4-dinitrophenylhydrazine. Each sample was mixed with an isovolume of 2,4-dinitrophenylhydrazine and incubated at RT for 15 min. The reaction was blocked by adding the Neutralization Solution and mixing with 2-mercaptoethanol to achieve a final concentration of 0.74 m. Proteins were loaded onto 12% polyacrylamide gel, separated by SDS-PAGE, and transferred to a nitrocellulose membrane for 1 h at 100 V at 4 °C. After blocking with 10% milk in PBST (PBS containing 0.05% Tween 20), blots were probed 1 h at RT with rabbit anti-dinitrophenyl antibody (1:150). Immunoreactive bands were detected with goat anti-rabbit IgG (HRP-conjugated) (1:300) and visualized by enhanced chemiluminescence detection system. The effective correct isoloading of samples was checked by staining acrylamide gel with Coomassie Brilliant Blue R-250 solution (Bio-Rad) for 1 h with gentle shaking at RT. The solution was drained off, and gel was washed at RT with destaining solution (40% (v/v) methanol and 10% (v/v) glacial acetic acid) until the protein bands were visible without background. At the end of the washings, gel was soaked in 2% (v/v) glycerol for 15 min and dried. Blots are representative of three independent experiments.
ROS intracellular accumulation was evaluated by the conversion of dihydroethidium (DHE) to ethidium. Briefly, 10 μm DHE (Invitrogen) were added to 2 × 105 PC12 cells grown onto poly-l-lysine-coated coverslips placed at the bottom of 35-mm dishes and incubated at 37 °C for 30 min to allow for the conversion of the DHE. Cells were then washed twice with PBS, fixed with 4% paraformaldehyde, and counterstained for 4 min at RT with 0.5 μg/ml Hoechst 33342.
At least 500 cells for each coverslip were counted and analyzed by fluorescence microscopy. Data are representative of 3 independent experiments.
Immunofluorescence Analysis
The apoptotic effects of DNA-PK kinase activity inhibition under oxidative stress conditions were assessed in proliferant PC12 cells. Cells (∼7 × 105) were grown onto poly-l-lysine-coated coverslips placed at the bottom of 35-mm culture dishes for 24 h and treated as indicated. At the end of treatments, cells were rinsed twice with ice-cold PBS and incubated for 20 min at −20 °C with ice-cold 70% acetone, 30% methanol solution. Cells were then re-hydrated for 10 min in PBS at RT and washed twice with PBS, and nuclei were stained with 0.5 μg/ml Hoechst 33342 in distilled water for 4 min at RT. Condensed and/or fragmented nuclei were counted as apoptotic nuclei.
To analyze the effects of Aβ(25–35) on DNA damage and DSB repair activity, proliferating and NGF-differentiated PC12 cells were analyzed for γH2AX foci formation and repair. Treated cells were rinsed and fixed as described above and incubated with 1 μg/ml anti-phospho-H2AX (Ser-139) monoclonal antibody (Millipore) in 0.2 mg/ml BSA in PBS for 2 h at RT. After three washes in PBS, PBST (0.05% (v/v) Tween 20 in PBS), and PBS, Alexa Fluor 594 goat anti-mouse IgG antibody, 1:1000 (Invitrogen), was used as secondary antibody by incubation at RT for 25 min. Cells having more than 10 foci/nucleus were scored as positive. Nuclei counterstaining was performed as described above.
At least 500 cells for each coverslip were examined with Nikon Eclipse TE2000-U fluorescence microscope (×60 immersion oil objective) equipped with a CCD camera. Data are representative of 3–4 independent experiments.
DNA-PK Kinase Activity Assays
PC12 cells (∼5 × 106) were grown onto poly-l-lysine-coated 100-mm polystyrene dishes for 24 h and treated as indicated. Cells were then washed twice with ice-cold PBS, harvested, and centrifuged at 4 °C for 5 min at 600 × g. Resulting pellet was resuspended on ice with a high salt whole cell extract (WCE) buffer (20 mm Hepes, pH 7.6, 450 mm NaCl, 25% glycerol, 0.2 mm EDTA, 0.2 mm DTT) containing 1 mm PMSF, mixture protease inhibitors, and 50 mm sodium fluoride and subjected to five freeze/thaw cycles (ethanol and dry ice, 30 °C). After centrifugation at 15,000 × g for 10 min at 4 °C, protein concentration in the supernatants (WCE) was determined. Forty μg of proteins were used for Western blot analysis of DNA-PK complex protein levels as described under “SDS-PAGE and Western Blot Analysis.” DNA-PK from cellular lysates was isolated by two different protocols, dsDNA-cellulose (Sigma) pulldown and immunoprecipitation with anti-DNA-PKcs Ab-4 mixture.
200–400 μg of WCE were incubated with 40 μl of preswollen dsDNA-cellulose for 30 min at 4 °C. After a centrifugation at 500 × g for 1 min at 4 °C, the supernatant was collected, and the dsDNA-cellulose was washed six times with ice-cold low salt buffer (20 mm Hepes, pH 7.6, 0.2 mm EDTA, 0.2 mm DTT) containing 0.5 mm PMSF, mixture protease inhibitors, and 50 mm sodium fluoride.
200–400 μg of WCE were precleared by incubation with 40 μl of protein G bead slurry (50%) (Pierce) for 60 min at 4 °C with end over end rotation. After centrifugation at 1000 × g at 4 °C for 1 min, the protein G was separated from the supernatant and discarded. Cleared lysate was incubated for 14–16 h at 4 °C in slow rotation with 2.5 μg of DNA-PKcs Ab-4, and protein-antibody complex was precipitated by adding 40 μl of protein G beads and incubation for 2–4 h at 4 °C under slow rotary agitation. At the end of incubation time, the sample was centrifuged at 1000 × g at 4 °C for 1 min, and bound proteins were washed as described above.
Samples obtained from both protocols were then processed with SignaTECTTM DNA-dependent protein kinase assay system (Promega) following the manufacturer's instructions. Kinase reactions were conducted with 20-μl aliquots of the resuspended DNA-PK-absorbed cellulose/protein G beads for 30 min at 30 °C and were performed in both the presence and absence of a biotinylated DNA-PK p53-derived substrate. Both assays were performed in triplicate.
For cell-free DNA-PK kinase activity assay, different concentrations of aggregated Aβ(25–35), oligomeric Aβ(1–42) (1–100 μm), and H2O2 (10–1000 μm) were preincubated with 20 units of purified human DNA-PK from HeLa cells (Promega) at 30 °C for 10 min before performing kinase reactions as described above. Control reversed sequence peptide Aβ(35–25) and Aβ(42–1) were used as controls. Data presented are representative of three independent experiments.
Co-immunoprecipitation Experiments
PC12 cells (∼5 × 106) were grown on poly-l-lysine-coated 100-mm polystyrene dishes for 24 h and treated for 24 h with 50 μm oligomeric Aβ(1–42). Cells were then washed twice with ice-cold PBS, harvested, and centrifuged at 4 °C for 5 min at 600 × g. Resulting pellet was homogenized on ice by Dounce disruption as described under “Subcellular Fractionation” to isolate soluble and insoluble proteins of nuclear and cytoplasmic compartments. Insoluble proteins were extracted by incubation for 10 min with 50 mm Tris, pH 8.0, 2% SDS, 10% glycerol, followed by sonication for 20 s. After dilution (1:20 v/v) with 50 mm Tris-Cl, pH 8.0, 150 mm NaCl containing mixture protease inhibitors, samples were precleared by incubation with 25 μl of protein G beads for 30 min at 4 °C with end over end rotation. After centrifugation at 1000 × g at 4 °C for 1 min, the protein G was separated from the supernatant and stored. Cleared lysate was incubated for 14–16 h at 4 °C in slow rotation with 2.5 μg of DNA-PKcs Ab-4, and the protein-antibody complex was precipitated by adding 25 μl of protein G beads and incubated for 4 h at 4 °C under slow rotary agitation. At the end of incubation time, the sample was centrifuged at 1000 × g at 4 °C for 1 min, and bound proteins were washed five times with 50 mm Tris, pH 8.0, 150 mm NaCl, 0.5% Nonidet P-40 and twice with 5 mm Tris, pH 8.0. Immunoprecipitates were then separated by 10–20% Novex® Tricine gel (Invitrogen) and analyzed with 4G8 antibody by Western blot for Aβ(1–42) oligomer detection. The same protocol was applied for immunoprecipitation of Aβ(1–42) oligomers by using 2 μg of 4G8 antibody, and DNA-PKcs Ab-4 antibody was used to reveal DNA-PKcs protein.
Statistical Analysis
Data were expressed as means ± S.E. Results were analyzed using one-way analysis of variance followed, where appropriate, by Tukey's post hoc test and were considered significant when p < 0.05.
RESULTS
Effects of Aβ Peptides on Cell Viability
To carry out this study, we used both differentiated and proliferating PC12 cells as they represent a well known cell model for studying pathophysiological effects of Aβ (46, 47). As Aβ peptides, we employed Aβ(1–42) oligomers, which are considered the proximate effectors of neurotoxicity and synaptotoxicity observed in AD (26, 27), and aggregated Aβ(25–35). This is a synthetic peptide of 11 amino acids that corresponds to a fragment of Aβ(1–40) and Aβ(1–42) and retains both physical and biological properties of the full-length Aβ (48). Aβ(25–35) has a time-dependent propensity to aggregate into fibrils with β-structure (49), shows maximal toxicity in the aggregated state (50, 51), and represents the biologically active region of Aβ able to induce apoptosis depending on dose and time exposure (48, 51, 52).
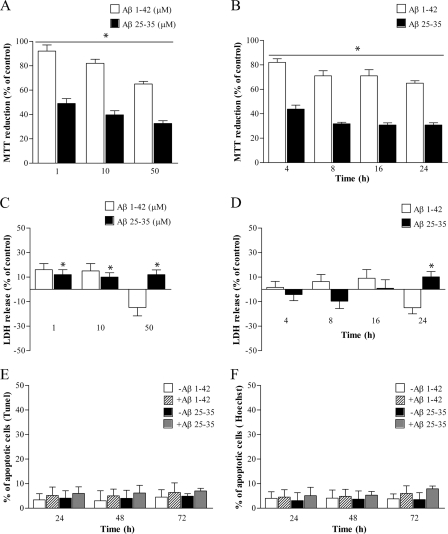
Experimental evidence demonstrates that DNA-PKcs is degraded during the apoptotic process (53–56). To analyze the effects of Aβ on the DNA-PK complex in the absence of cell death, we assessed cell viability of proliferating PC12 cells over a range of aggregated Aβ(25–35) and oligomeric Aβ(1–42) concentrations (1–50 μm). PC12 cells exposed to Aβ(25–35) or Aβ(1–42) for 24 h showed a dose-dependent inhibition of MTT reduction (Fig. 1, A and B). For both Aβ peptides, the effect was maximal after exposure for 24 h to 50 μm concentration, with a decrease in MTT reduction capacity by ∼70% for Aβ(25–35) and 35% for Aβ(1–42) versus control cells (Fig. 1A). This effect was already evident after 4 h of Aβ treatment, with a reduction by ∼60% for Aβ(25–35) and 20% for Aβ(1–42) versus nontreated cells (Fig. 1B).
FIGURE 1.
Effects of aggregated Aβ(25–35) and oligomeric Aβ(1–42) on cell viability. A and B, MTT reduction following 24 h of Aβ(25–35) (black columns) and Aβ(1–42) (white columns) treatment at different doses (1–50 μm, A) and for different times (50 μm, 4–24 h, B). Values are average ± S.E. expressed as % of control (100%). n = 4–6, asterisks identify p < 0.05 versus controls. C and D, LDH release following 24 h of Aβ(25–35) and Aβ(1–42) treatment at different doses (1–50 μm, C) and for different times (50 μm, 4–24 h, D). Values are expressed as % maximal LDH ± S.E. release. n = 4–6, asterisks identify p < 0.05 versus controls (base-line LDH release, i.e. untreated cells). E and F, percentage of apoptotic nuclei following 50 μm Aβ(25–35) and Aβ(1–42) treatment for 24–72 h detected by TUNEL assay (E) and Hoechst 33342 staining (F). At least 500 cells for each coverslip were counted in both assays, and data (expressed as average ± S.E.) are representative of three independent experiments.
Although one of the main cellular systems impaired by the presence of Aβ peptide is mitochondrial metabolism (46), this phenomenon is essentially reversible, and it is not an effective measure of cell viability. Hence, parallel experiments using LDH assays were also performed. As shown in Fig. 1, C and D, PC12 cells exposed to Aβ(25–35) showed only 10% of LDH release with respect to control cells within 24 h of treatment, whereas Aβ(1–42) had no statistically significant effects. To assess apoptotic effects following Aβ peptide treatments, we also carried out TUNEL assay and Hoechst 33342 nuclear staining over a period of 24–72 h. No significant changes were observed, even at maximal doses of both Aβ(25–35) and Aβ(1–42) peptides (50 μm) and after a prolonged time of exposure (Fig. 1, E and F). Western blot analysis of poly(ADP-ribose) polymerase cleavage, a well known hallmark of apoptotic status (57), confirmed these data (data not shown).
It has been reported that Aβ peptide is capable of inducing DNA damage (58). We evaluated the effect of Aβ(25–35) treatment on DNA damage response by immunostaining for γH2AX, an early sensitive biomarker of radiation and drug-induced DSBs (59). Cells having more than 10 γH2AX foci/nucleus were scored positive. PC12 cells treated with different doses of Aβ(25–35) (1–50 μm for 4–24 h) did not show a significant increase of γH2AX-positive nuclei as compared with untreated cells (data not shown). Exposure to reversed sequence peptides Aβ(35–25) and Aβ(42–1) had no significant effects in all experimental conditions used (data not shown).
Overall, these results show that Aβ peptides, up to 50 μm and within 24 h, inhibit mitochondrial metabolism without disrupting membrane integrity, triggering apoptotic cell death or causing DSBs. Therefore, 1–50 μm Aβ(25–35) and Aβ(1–42) concentrations were defined as sublethal and used in all subsequent experiments.
Sublethal Concentrations of Exogenous Aβ(25–35) and Aβ(1–42) Inhibit DNA-PK Activity
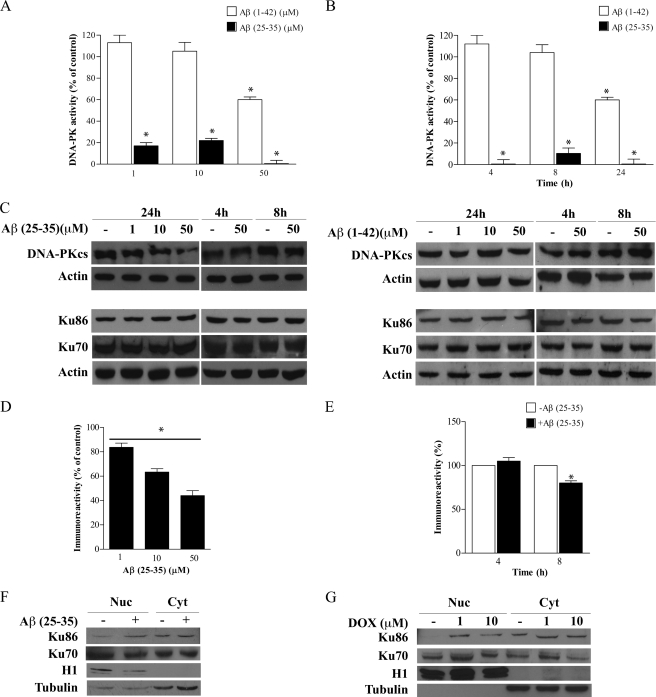
To investigate whether Aβ peptide could impair DNA-PK kinase activity, we exposed PC12 cells to the subtoxic concentration range of 1–50 μm aggregated Aβ(25–35) and oligomeric Aβ(1–42) for 4–24 h. We then analyzed kinase activity of DNA-PK derived from PC12-treated cells by assessing the ability to phosphorylate in vitro its p53 substrate. We found that Aβ(25–35) elicited a pronounced impairment of DNA-PK activity in a dose-dependent manner, with a maximal response observed at 50 μm Aβ(25–35) (Fig. 2A), after 24 h of treatment. However, a strong inhibition of DNA-PK kinase activity (99 ± 4% versus not treated and time-matched cells) was evident even after 4 h of exposure to 50 μm Aβ(25–35) (Fig. 2B). Similarly, Aβ(1–42) was able to reduce DNA-PK kinase activity (40 ± 10% versus not treated and time-matched cells), but this effect was evident only after 24 h of exposure to a concentration of 50 μm.
FIGURE 2.
Sublethal concentrations of exogenous Aβ(25–35) and Aβ(1–42) down-regulate DNA-PK activity. A and B, PC12 cells were treated with Aβ(25–35) (black columns) and Aβ(1–42) (white columns) at different doses (1–50 μm) for 24 h (A) and for different times (50 μm, 4–24 h, B). After treatments, 400 μg of WCE were subjected to dsDNA-cellulose pulldown, and a phosphorylation assay was performed. Values represent average ± S.E. of kinase activity with respect to untreated cells (100%). Asterisks identify p < 0.05 versus controls. C, 40 μg of the same protein samples were used for Western blot analysis of DNA-PK complex protein levels (DNA-PKcs, Ku70, and Ku86). Actin was used as loading control. D and E, densitometric quantitation of the immunoreactive bands corresponding to DNA-PKcs. Values represent the normalized percent changes in DNA-PKcs protein levels with respect to control (100%) after exposure to Aβ(25–35) at different doses (1–50 μm) for 24 h (D) and for different times (50 μm, 4–24 h, E) (average ± S.E.; n = 3). Asterisks identify p < 0.05 versus control values. F and G, PC12 cells were exposed (+) or not (−) to 50 μm aggregated Aβ(25–35) (F) or treated with 1 and 10 μm DOX (G) for 24 h and then subjected to biochemical fractionation to isolate nuclear (Nuc) and cytoplasmic (Cyt) proteins. Fractions were immunoblotted with antibodies for Ku70, Ku86, cytosolic marker α-tubulin, and nuclear marker Histone H1. Blots in the figure are representative of three independent experiments.
DNA-PK kinase activity is regulated by different mechanisms, including modifications in the catalytic subunit and/or the regulative subunits Ku70 and Ku86 protein levels (60, 61). To evaluate whether the Aβ-mediated decrease in DNA-PK kinase activity was dependent on protein level modifications, we performed Western blot analysis on protein extracts derived from Aβ(25–35)- and Aβ(1–42)-treated PC12 cells. We found that DNA-PKcs protein levels were down-regulated in a concentration-dependent manner (Fig. 2C, left panel) with the greatest effect of 51.4 ± 7.2% reduction after treatment for 24 h with 50 μm Aβ(25–35). Densitometric analysis of immunoreactive bands (Fig. 2, D and E) showed that the effect was detectable as early as 8 h after Aβ exposure (20 ± 2.5% decrease). On the contrary, Aβ(1–42) did not modify DNA-PKcs protein levels even at maximal dose and after 24 h treatment (Fig. 2C, right panel). Ku70 and Ku86 protein levels also remained unchanged after both Aβ peptide treatments (Fig. 2C). We then analyzed whether treatment of PC12 cells with Aβ(25–35) was able to mobilize Ku70 and Ku86 to the nucleus by Western blot analysis on nuclear and cytoplasmic PC12 cell extracts. We found that 50 μm Aβ(25–35) treatment induces a 95% increase of Ku86 in the nucleus as compared with untreated cells, whereas Ku70 compartmentalization remained unmodified (Fig. 2F). Treatment of PC12 cells with 1 and 10 μm DOX, a well established DSB inducer (62), induced a similar nuclear translocation of Ku86 (108% with 1 μm and 67% with 10 μm of increase) and a nonstatistically significant accumulation of Ku70 with only 10 μm DOX (Fig. 2G). These results indicate that sublethal doses of Aβ(25–35) induce Ku86 nuclear translocation even in the absence of DSBs. It is worth noting that DNA-PK kinase activity, DNA-PK complex protein levels, and compartmentalization were not significantly affected by exposure to the reversed sequence peptides Aβ(35–25) and Aβ(42–1) (data not shown).
Aggregated Aβ(25–35)-induced Oxidative Stress Causes the Impairment of DNA-PK Kinase Activity
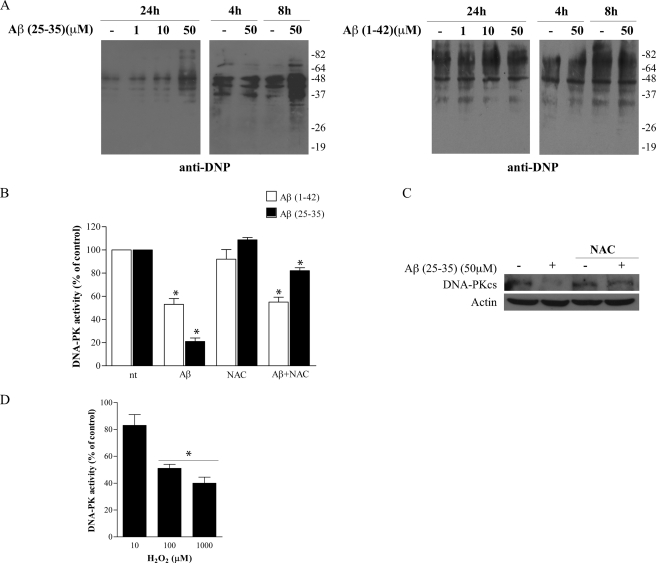
DNA-PK activity can be modulated by oxidative stress. Previous studies demonstrated that ROS levels are inversely correlated with DNA-PK kinase activity upon exposure to chemotherapeutic agents, and treatment with antioxidant reversed this inhibition (63, 64). Thus, we asked whether the induction of Aβ-mediated oxidative stress could induce the impairment of DNA-PK kinase activity. To this aim, we first verified whether aggregated Aβ(25–35) and oligomeric Aβ(1–42) were able to induce oxidative stress in PC12 cells by evaluating protein carbonylation levels and ROS intracellular accumulation by the conversion of dihydroethidium (DHE) to ethidium. As shown in Fig. 3A (left panel), maximal levels of protein carbonylation were revealed after treatment with 50 μm Aβ(25–35) for 24 h, although an increase was evident as early as 8 h. On the contrary, oligomeric Aβ(1–42) treatment did not increase protein carbonylation levels in the same experimental conditions (Fig. 3A, right panel). Interestingly, ROS production was already observed 4 h after Aβ(25–35) treatment, whereas it required 24 h of Aβ(1–42) treatment (data not shown). Exposure to reversed sequences Aβ(35–25) and Aβ(42–1) did not induce oxidative stress (data not shown).
FIGURE 3.
Effect of Aβ(25–35)-mediated oxidative stress on DNA-PK kinase activity. A, analysis of carbonylation levels of protein extracts derived from PC12 cells treated with Aβ(25–35) (left panel) and Aβ(1–42) (right panel) at different doses (1–50 μm) for 24 h and for different times (50 μm, 4–8 h). Blots were probed with anti-dinitrophenyl antibody. Molecular weights are indicated on the right of the blots. Isoloading of samples was verified by Coomassie staining of corresponding acrylamide gels (data not shown). B, DNA-PK kinase activity following 50 μm Aβ(25–35) (black columns) and Aβ(1–42) (white columns) treatment for 24 h in the presence or not of 0.5 mm NAC (4-h pretreatment). After treatments, 400 μg of WCE were subjected to dsDNA-cellulose pulldown, and a phosphorylation assay was performed. Values represent average ± S.E. of kinase activity with respect to untreated cells (100%). n = 3, *, p < 0.05 versus control values (nt). C, representative Western blot analysis (n = 3) of PC12 cells treated (+) or not (−) with 50 μm Aβ(25–35) for 24 h in presence or not of 0.5 mm NAC. Blots were probed with anti-DNA-PKcs antibody. Actin was used as loading control. D, purified human DNA-PK was incubated in the presence of increasing concentrations (10–1000 μm) of H2O2 for 10 min before performing kinase assay. Bars in the plot represent average ± S.E. of kinase activity with respect to reaction performed without H2O2 (control, 100%). n = 3, *, p < 0.05 versus corresponding control values.
To assess the contribution of oxidative stress on the alteration of DNA-PK kinase activity, we performed a DNA-PK kinase assay with protein extracts derived from PC12 cells treated with 50 μm Aβ(25–35) or Aβ(1–42) for 24 h, in the presence or not of 0.5 mm NAC, a glutathione precursor with established antioxidant properties (65). A preliminary dose-response experiment with different NAC concentrations established that this concentration did not interfere with basal DNA-PK kinase activity (data not shown).
Pretreatment of PC12 cells with 0.5 mm NAC (4 h), followed by a 24-h co-incubation with 50 μm Aβ(25–35), attenuated the impairment of DNA-PK kinase activity (82% of kinase activity with respect to 21% without NAC incubation, Fig. 3B) and restored DNA-PKcs protein levels (84% of recovery versus Aβ(25–35) treatment without NAC incubation, see Fig. 3C). Conversely, NAC did not restore DNA-PK kinase activity of PC12 cells exposed to Aβ(1–42) (53% of kinase activity with respect to 55% without NAC incubation, see Fig. 3B).
To establish whether ROS were able to directly inhibit DNA-PK activity, we performed a DNA-PK kinase activity assay using commercially available purified human DNA-PK incubated with increasing concentrations of H2O2. DNA-PK was preincubated with H2O2 for 10 min at 30 °C before the assay of kinase activity. H2O2 impaired DNA-PK kinase activity in a dose-dependent manner, reaching a maximal effect at 100 and 1000 μm (51 and 40% of kinase activity, respectively, versus control reaction without H2O2, see Fig. 3D).
These results demonstrate that Aβ(25–35) can exert its inhibitory effect via the production of ROS, whereas the impairment of DNA-PK kinase activity by Aβ(1–42) is not dependent on oxidative stress induction, at least at sublethal concentrations. Since 4 h of Aβ(25–35) treatment on PC12 cells strongly impaired DNA-PK kinase activity while the correspondent protein levels were unmodified (compare Fig. 2, B and E), it is reasonable to conclude that the early inhibition of DNA-PK kinase activity by Aβ(25–35) caused ROS production.
Exogenous Aβ(1–42) Oligomers Enter the Nucleus of PC12 Cells
It has been previously demonstrated that under both normal and oxidative DNA damage conditions, β-amyloid peptides may localize in the nuclear compartment (66–69). Importantly, Aβ(1–42) immunoreactivity has been found in both the cytosol and nuclei of some degenerating neurons in transgenic mice and AD brains (68, 69). Therefore, we asked whether in PC12 cells exogenous Aβ(1–42) oligomers, the most neurotoxic Aβ species observed in AD, can enter the nucleus where DNA-PK exerts its DNA repair activity. Because it is well known that different kinds of Aβ species have distinct biological actions, we first analyzed by Western blot our Aβ(1–42) preparation (43). As illustrated in Fig. 4B, Aβ(1–42) preparation was enriched in large and low molecular weight oligomers. In particular, we found a larger amount of Aβ(1–42) monomer and dimer as compared with trimer, tetramer, and larger oligomeric assemblies. Fig. 4B also shows that the aggregated Aβ(25–35) preparation was enriched in low molecular weight oligomers (tetramer and decamer), although a minor amount of high molecular weight species was also present.
FIGURE 4.
Exogenous Aβ(1–42) oligomers accumulate in the nucleus of PC12 cells as insoluble species. A, PC12 cells were exposed (+) or not (−) to 50 μm Aβ(1–42) oligomers for 24 h and subjected to biochemical fractionation to isolate soluble and insoluble proteins of nuclear (Nuc) and cytoplasmic (Cyt) compartments. Western blot analysis of protein extracts was carried out by using anti-Aβ 4G8 antibody. Molecular weights are indicated on the left. B, Western blot analysis of oligomeric Aβ(1–42) and aggregated Aβ(25–35) preparations. Oligomeric species (monomer, dimer, trimer, tetramer, decamer, and larger oligomeric assemblies) are indicated by arrows. Blots are representative of three independent experiments.
To assess the presence of Aβ(1–42) in the nucleus, we then carried out a biochemical fractionation on proliferating PC12 cells exposed to 50 μm Aβ(1–42) oligomers for 24 h. Soluble and insoluble nuclear and cytoplasmic proteins were extracted and analyzed for the presence of Aβ(1–42) oligomers by Western blot. We found that Aβ(1–42) oligomers (mainly monomer, dimer, and trimer) accumulate in the insoluble fractions of both nuclear and cytoplasmic compartments, although a slight amount of Aβ(1–42) dimer was present in the cytoplasmic soluble pool (Fig. 4A). These results demonstrate that exogenous Aβ(1–42) oligomers enter the nucleus of PC12 cells and accumulate as insoluble species.
Aggregated Aβ(25–35) and Oligomeric Aβ(1–42) Directly Inhibit DNA-PK Kinase Activity in a Cell-free Assay but Do Not Interact with DNA-PKcs in PC12 Cells
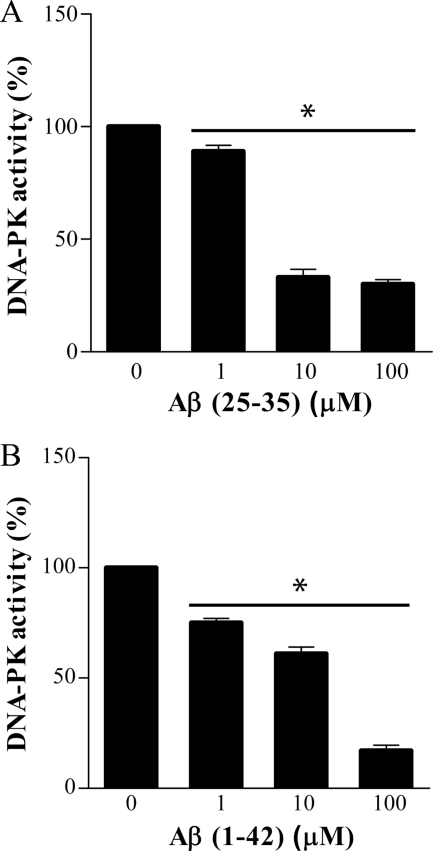
As Aβ peptide is able to enter the nucleus of PC12 cells, it is reasonable to question whether it may directly interact with DNA-PKcs and eventually inhibit its kinase activity. To address this issue, we first carried out a DNA-PK kinase activity assay in vitro using purified human DNA-PK incubated with increasing concentrations of aggregated Aβ(25–35). Aggregated Aβ(25–35), preincubated with DNA-PK for 10 min at 30 °C, inhibited DNA-PK kinase activity in a dose-dependent manner (30–33% of kinase activity at 10 and 100 μm versus control reaction without Aβ, see Fig. 5A). Analogous results were obtained with oligomeric preparations of Aβ(1–42). Incubation with 1 μm oligomeric Aβ(1–42) induced 25 ± 2% decrease of DNA-PK kinase activity that reached 83 ± 3.6% of inhibition using 100 μm (Fig. 5B). In the same experimental conditions, parallel experiments with reverse peptides Aβ(35–25) and Aβ(42–1) did not reveal any relevant changes (data not shown).
FIGURE 5.
Aggregated Aβ(25–35) and oligomeric Aβ(1–42) impair DNA-PK kinase activity in a cell-free assay. Purified human DNA-PK was incubated in the presence of increasing concentrations (1–100 μm) of synthetic aggregated Aβ(25–35) (A) and oligomeric Aβ(1–42) (B) for 10 min before performing kinase assay. Bars in the plot represent average ± S.E. of kinase activity with respect to reaction performed without Aβ (control, 100%). n = 3, *, p < 0.05 versus corresponding control values.
We next assessed the direct interaction between DNA-PKcs and oligomeric Aβ(1–42) in PC12 cells by co-immunoprecipitation experiments. To this aim, we immunoprecipitated DNA-PKcs from both soluble and insoluble fractions (nuclear and cytoplasmic compartments), followed by Western blot with anti-Aβ antibody. Surprisingly, we did not find the presence of Aβ oligomers bound to DNA-PKcs either in soluble or insoluble fractions. Immunoprecipitation of Aβ followed by Western blot analysis with anti-DNA-PKcs antibody confirmed that Aβ and DNA-PKcs do not co-immunoprecipitate in our experimental conditions (data not shown). The same results were obtained carrying out immunoprecipitation experiments with aggregated Aβ (data not shown) (25–35).
Inhibition of DNA-PK Kinase Activity Mediated by Aβ(25–35) Sensitizes PC12 Cells to Oxidative Stress Insult
The inhibition of DNA-PK activity by selective inhibitors or functional mutations increases vulnerability of mammalian cells to DNA-damaging conditions (ionizing radiations and chemotherapeutics), excitotoxicity, and oxidative stress (44, 45, 70–73). These results suggest a protective role of DNA-PK against different cell death stimuli, although a pro-apoptotic role has been also proposed (74, 75).
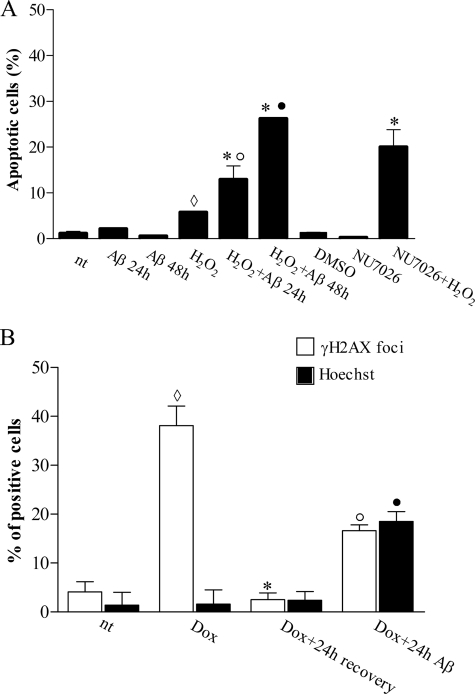
To analyze whether Aβ(25–35)-mediated DNA-PK kinase activity inhibition could increase cell vulnerability to oxidative stress conditions, we first treated PC12 cells with aggregated Aβ(25–35) and then with sublethal concentrations of H2O2. As shown in Fig. 6A, application of 100 μm H2O2 for 30 min did not induce relevant levels of apoptosis (5% apoptotic nuclei versus 1% of untreated control cells), as well as exposure to 50 μm Aβ(25–35) for 24–48 h per se (about 0.5–2% of apoptotic nuclei). In contrast, significant apoptotic response (13% of apoptotic nuclei) was observed by preincubating cells with aggregated Aβ(25–35) for 24 h before exposure to 30 min of H2O2. This effect markedly increased when the incubation with Aβ(25–35) was prolonged for another 24 h (26% apoptotic nuclei). These observations indicate that the inhibition of DNA-PK kinase activity mediated by Aβ renders PC12 cells sensible to nonlethal oxidative stress conditions, leading to apoptotic cell death, and support the hypothesis for an anti-apoptotic role of DNA-PK. To test this further, we pretreated cells with NU7026, a competitive and highly selective inhibitor of DNA-PK with no effect on ataxia telangiectasia-mutated kinase and Rad3-related kinase at concentrations <100 μm (45), before exposure to 100 μm H2O2. Treatment for 24 h with 10 μm NU7026, a noncytotoxic concentration that inhibits purified human DNA-PK and DSB repair and chemo/radiosensitizes different tumor cell lines (44, 45), resulted in 20% apoptotic cells with respect to 6% (only DMSO) or 0.3% (only NU7026) of control treated cells.
FIGURE 6.
Aβ(25–35) sensitizes PC12 cells to oxidative stress and attenuates DNA repair activity by DNA-PK activity inhibition. A, proliferating PC12 cells were treated as indicated. Hoechst 33342 nuclear staining was performed and condensed and/or fragmented nuclei were counted as apoptotic nuclei. At least 500 cells for each coverslip were counted, and data are representative of 3–4 independent experiments. ♢, p < 0.05 versus not treated (nt); *, p < 0.05 versus H2O2; ○, p < 0.05 versus Aβ 24 h; ●, p < 0.05 versus Aβ 48 h. B, NGF-differentiated PC12 cells were treated for 8 h with DOX with or without 50 μm Aβ(25–35) pretreatment (24 h). Histone H2AX phosphorylated foci were counted, and cells having more than 10 foci/nucleus were considered positive. Apoptotic nuclei were revealed by Hoechst 33342 nuclear staining. Bars in the plot represent average ± S.E. of positive cells expressed as percentage. At least 500 cells for each coverslip were counted, and data are representative of 3–4 independent experiments. ♢, p < 0.05 versus nt; *, p < 0.05 versus Dox; ○, p < 0.05 versus Dox + 24 h recovery; ●, p < 0.05 versus nt; Dox, Dox + 24 h recovery.
DNA-PK Kinase Activity Inhibition Mediated by Aβ(25–35) Attenuates DNA Repair Activity in NGF-differentiated PC12 Cells
Post-mitotic cells adopt mainly NHEJ to repair DSBs (76, 77). Hence, to evaluate the effect of Aβ-mediated DNA-PK kinase activity inhibition on DSB repair, we used NGF-differentiated PC12 cells treated with 1 μm DOX (62). The ability of cells to repair DSBs was assessed by counting γH2AX foci. Cells having more than 10 foci/nucleus were scored positive.
As shown in Fig. 6B, untreated control cells showed ∼4% of nuclei positive to γH2AX as well as cells treated with aggregated 50 μm Aβ(25–35) for 24 h (data not shown). Eight hours of exposure to DOX induced foci accumulation in 38% of nuclei without any further effect after 24 h of treatment (data not shown). Following 24 h of recovery, only 3% of nuclei showed a positive immunostaining for γH2AX, indicating that DSB repair was completed. Preincubation of differentiated PC12 cells with aggregated 50 μm Aβ(25–35) for 24 h significantly decreased the ability to repair DNA damage such that, after 24 h of recovery, 17% of nuclei remained positive to γH2AX. Parallel analysis of apoptotic cells, assessed by Hoechst staining, revealed that 8 h of DOX application alone did not induce apoptosis (2.4% DOX-treated cells versus 1.4% control cells, evaluated after 24 h), whereas pretreatment with aggregated Aβ(25–35) induced 19% of apoptotic cells (Fig. 6B). It is worth mentioning that, in this condition, most of γH2AX-positive cells showed also apoptotic nuclear morphology supporting a functional association between not repaired DNA damage and apoptotic phenotype.
DISCUSSION
Unrepaired DNA lesions may trigger apoptosis, and the consequent accumulation of DNA damage potentially contributes to the progressive neuronal loss observed in neurodegenerative diseases, including AD (4, 5, 78).
Here, we found that aggregated Aβ(25–35) inhibits DNA-PK kinase activity, the key complex for DSB repair in mammalian cells in a dose-dependent manner with effects already evident after 4 h of treatment. This inactivation was associated with the generation of ROS and down-regulation of DNA-PKcs protein levels, and it is resolved by treatment with antioxidant NAC. Sublethal doses of oligomeric Aβ(1–42) were also able to inhibit DNA-PK kinase activity, although to a minor extent and only after 24 h of treatment, without affecting DNA-PK complex protein levels. This down-regulation was not mediated by oxidative stress and could not be reversed by NAC.
Moreover, by using a cell-free assay, we showed that both aggregated Aβ(25–35) and oligomeric preparations of Aβ(1–42) directly impaired DNA-PK kinase activity. Although the presence of DSBs was reported in AD (2, 3, 31, 32), this is the first study showing that Aβ is able to interfere with DNA-PK activity and to inhibit DNA-PK-mediated DSB repair.
The induction of oxidative stress mediated by aggregated Aβ(25–35) may cause the impairment of DNA-PK activity through two mechanisms. The first mechanism is that ROS production induces the degradation of DNA-PKcs protein and consequently down-regulates its kinase activity. Accordingly, we showed that down-regulation of DNA-PKcs protein is paralleled by Aβ(25–35)-induced protein carbonylation, an irreversible and not repairable oxidative protein modification, which may render proteins more prone to proteolytic degradation by proteasomes (79, 80). Although it is well known that Aβ oligomers and fibrils can inhibit the proteasome activity (81, 82), it has been demonstrated that soluble Aβ(1–40) is able to induce the degradation of post-synaptic density-95, specifically by proteasomes (83). In addition, naturally secreted Aβ oligomers cause the reduction of neuronal EphB2 protein, and co-incubation with lactacystin, a specific proteasome inhibitor, rescues this effect with a concomitant increase of ubiquitinated EphB2 (84). Hence, Aβ may have opposite effects on proteasome-mediated degradation, and also in the case of DNA-PKcs, it is plausible to hypothesize an involvement of proteasomes in the reduction of DNA-PKcs protein levels.
The second mechanism is that ROS directly inhibit DNA-PK kinase activity. Indeed, H2O2 impairs DNA-PK kinase activity in a cell-free assay, and this may explain the early inhibition of kinase activity in PC12 cells (4 h after Aβ(25–35) treatment) regardless of down-regulation of DNA-PKcs protein levels.
We also found that Aβ(1–42) oligomers accumulate in the nucleus of PC12 cells as insoluble species. Because the nucleus is the subcellular compartment where DNA-PK exerts its DNA repair function, this finding raises the question whether Aβ may directly interact with DNA-PKcs in the nucleus. Although we observed that Aβ peptides down-regulate DNA-PK activity in a cell-free assay, co-immunoprecipitation experiments did not support a direct interaction between Aβ and DNA-PKcs in PC12 cells.
Overall, these results indicate that in PC12 cells aggregated Aβ(25–35) impairs DNA-PK activity mainly through ROS production. In contrast, in PC12 cells exposed to oligomeric Aβ(1–42), DNA-PK kinase activity down-regulation is not dependent on oxidative stress induction. These results are consistent with previous studies showing that oxidative properties of Aβ(25–35) are more pronounced and manifest earlier (few hours) with respect to those of Aβ(1–42), which usually requires 24–48 h (85, 86).
A possible explanation of DNA-PK kinase activity inhibition by oligomeric Aβ(1–42) is that specific modulators of DNA-PK mediate the inhibition of its kinase activity. Indeed, it has been reported that Aβ, in addition to specific kinases, can also modulate the function of protein phosphatases. For example, full-length Aβ(1–40) and Aβ(1–42) peptides, as well as Aβ(25–35), specifically inhibit protein phosphatase 1 (PP1) activity both in a cell-free assay and in cells (87, 88). PP1 is an abundant neuronal serine/threonine-specific phosphatase that positively modulates the kinase activity of DNA-PK (89, 90). Hence, it is possible to speculate that in our experimental conditions, PP1 activity might be inhibited by Aβ(1–42), thus leading to the down-regulation of DNA-PK kinase function. Another modulator of DNA-PK, casein kinase II (CKII) (91), might be involved in the inhibition of DNA-PK kinase activity. Accordingly, it has been reported that in AD brains CKII activity and protein levels are altered and that Aβ affects CKII activity (92, 93). Further experiments to identify the mechanism(s) by which Aβ(1–42) impairs DNA-PK kinase activity would be required.
We showed that Aβ-mediated DNA-PK kinase activity inhibition renders PC12 cells susceptible to nonlethal oxidative injury leading to cell death. A similar result was obtained following exposure to the specific DNA-PK inhibitor NU7026, indicating that this effect was specifically mediated by the abrogation of DNA-PK activity. Because of the high rate of oxidative metabolism in the brain, neurons are continuously exposed to oxidative stimuli. Moreover, neurons have low levels of antioxidant enzymes, thus particularly susceptible to the damaging and highly toxic effects of ROS (94). DNA-PK exerts a protective function under different cell death conditions, including oxidative stress and excitotoxicity (71, 73). Hence, it is possible that exposure to nonlethal oxidative injuries in the presence of Aβ, a hallmark of AD pathology, elicits neuronal cell death by suppression of DNA-PK anti-apoptotic function, contributing to the neurodegenerative process.
Finally, we showed that aggregated Aβ attenuates DNA repair activity in NGF-differentiated PC12 cells exposed to the DSB inducer DOX, suggesting an impairment of DNA-PK-mediated NHEJ. End joining activity and protein levels of DNA-PKcs are reduced in the midfrontal cortex of patients with AD (4). Similarly, a decrease of DNA-PKcs expression, although not significant, was observed in neurons and astrocytes of temporal cortices of AD cases with increasing Braak stages (42). Taken together, these results enable us to hypothesize that during AD pathology continuous accumulation of Aβ and ROS production would impair DNA-PK activity contributing to neurodegeneration through the inhibition of DNA-PK-mediated DSB repair pathway. Inhibition of NHEJ may indeed increase DSBs, potentially lethal lesions if unrepaired or not correctly restored.
Several human diseases, including xeroderma pigmentosum, Cockayne syndrome, Nijmegen breakage syndrome, ataxia telangiectasia, ataxia with occulomotor apraxia 1 (AOA1), and spinocerebellar ataxia with axonal neuropathy 1 (SCAN1) are caused by mutations in DNA repair genes and present important neurological implications (77). Moreover, it has been recently shown that also in Huntington disease (HD) DNA repair dysfunction is a critical factor of the pathological process (9). DNA repair dysfunction in HD is not genetically determined but is caused by the accumulation of neurotoxic species of mutant huntingtin protein (Htt). Indeed, mutant Htt interacts with Ku70 and diminishes DNA-PK kinase activity both in vitro and in vivo. The impairment of DNA damage repair contributes to the increase of DSBs in HD pathology.
In conclusion, based on these data, it is tempting to speculate that protein misfolding and aggregation of amyloidogenic proteins, such as Aβ in AD and Htt in HD, contribute to the DSB accumulation observed in neurodegenerative diseases via inhibition of NHEJ and eventually to neurodegeneration.
Acknowledgment
We thank Silvia Paradisi for helping with oligomeric preparations of Aβ(1–42).
This work was supported by the Italian Ministry of Health.
- AD
- Alzheimer disease
- ROS
- reactive oxygen species
- DSB
- double strand break
- NHEJ
- nonhomologous end joining
- Aβ
- β-amyloid
- HD
- Huntington disease
- DOX
- doxorubicin
- WCE
- whole cell extract
- NAC
- N-acetyl-l-cysteine
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DHE
- dihydroethidium
- LDH
- lactate dehydrogenase
- cs
- catalytic subunit.
REFERENCES
- 1. Robison S. H., Munzer J. S., Tandan R., Bradley W. G. (1987) Alzheimer disease cells exhibit defective repair of alkylating agent-induced DNA damage. Ann. Neurol. 21, 250–258 [DOI] [PubMed] [Google Scholar]
- 2. Mullaart E., Boerrigter M. E., Ravid R., Swaab D. F., Vijg J. (1990) Increased levels of DNA breaks in cerebral cortex of Alzheimer disease patients. Neurobiol. Aging 11, 169–173 [DOI] [PubMed] [Google Scholar]
- 3. Adamec E., Vonsattel J. P., Nixon R. A. (1999) DNA strand breaks in Alzheimer disease. Brain Res. 849, 67–77 [DOI] [PubMed] [Google Scholar]
- 4. Shackelford D. A. (2006) DNA end joining activity is reduced in Alzheimer disease. Neurobiol. Aging 27, 596–605 [DOI] [PubMed] [Google Scholar]
- 5. Weissman L., Jo D. G., Sørensen M. M., de Souza-Pinto N. C., Markesbery W. R., Mattson M. P., Bohr V. A. (2007) Defective DNA base excision repair in brain from individuals with Alzheimer disease and amnestic mild cognitive impairment. Nucleic Acids Res. 35, 5545–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giuliano P., De Cristofaro T., Affaitati A., Pizzulo G. M., Feliciello A., Criscuolo C., De Michele G., Filla A., Avvedimento E. V., Varrone S. (2003) DNA damage induced by polyglutamine-expanded proteins. Hum. Mol. Genet. 12, 2301–2309 [DOI] [PubMed] [Google Scholar]
- 7. Kovtun I. V., Liu Y., Bjoras M., Klungland A., Wilson S. H., McMurray C. T. (2007) OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qi M. L., Tagawa K., Enokido Y., Yoshimura N., Wada Y., Watase K., Ishiura S., Kanazawa I., Botas J., Saitoe M., Wanker E. E., Okazawa H. (2007) Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat. Cell Biol. 9, 402–414 [DOI] [PubMed] [Google Scholar]
- 9. Enokido Y., Tamura T., Ito H., Arumughan A., Komuro A., Shiwaku H., Sone M., Foulle R., Sawada H., Ishiguro H., Ono T., Murata M., Kanazawa I., Tomilin N., Tagawa K., Wanker E. E., Okazawa H. (2010) Mutant huntingtin impairs Ku70-mediated DNA repair. J. Cell Biol. 189, 425–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robbins J. H., Otsuka F., Tarone R. E., Polinsky R. J., Brumback R. A., Nee L. E. (1985) Parkinson disease and Alzheimer disease. Hypersensitivity to x-rays in cultured cell lines. J. Neurol. Neurosurg. Psychiatr. 48, 916–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimura-Miura H., Hattori N., Kang D., Miyako K., Nakabeppu Y., Mizuno Y. (1999) Increased 8-oxo-dGTPase in the mitochondria of substantia nigral neurons in Parkinson disease. Ann. Neurol. 46, 920–924 [PubMed] [Google Scholar]
- 12. Bender A., Krishnan K. J., Morris C. M., Taylor G. A., Reeve A. K., Perry R. H., Jaros E., Hersheson J. S., Betts J., Klopstock T., Taylor R. W., Turnbull D. M. (2006) High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 38, 515–517 [DOI] [PubMed] [Google Scholar]
- 13. Kraytsberg Y., Kudryavtseva E., McKee A. C., Geula C., Kowall N. W., Khrapko K. (2006) Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 38, 518–520 [DOI] [PubMed] [Google Scholar]
- 14. Kisby G. E., Milne J., Sweatt C. (1997) Evidence of reduced DNA repair in amyotrophic lateral sclerosis brain tissue. Neuroreport 8, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 15. Ferrante R. J., Browne S. E., Shinobu L. A., Bowling A. C., Baik M. J., MacGarvey U., Kowall N. W., Brown R. H., Jr., Beal M. F. (1997) Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J. Neurochem. 69, 2064–2074 [DOI] [PubMed] [Google Scholar]
- 16. Bogdanov M., Brown R. H., Matson W., Smart R., Hayden D., O'Donnell H., Flint Beal M., Cudkowicz M., (2000) Increased oxidative damage to DNA in ALS patients. Free Radic. Biol. Med. 29, 652–658 [DOI] [PubMed] [Google Scholar]
- 17. Kikuchi H., Furuta A., Nishioka K., Suzuki S. O., Nakabeppu Y., Iwaki T. (2002) Impairment of mitochondrial DNA repair enzymes against accumulation of 8-oxo-guanine in the spinal motor neurons of amyotrophic lateral sclerosis. Acta Neuropathol. 103, 408–414 [DOI] [PubMed] [Google Scholar]
- 18. Merlo D., Di Stasi A. M., Bonini P., Mollinari C., Cardinale A., Cozzolino F., Wisden W., Garaci E. (2005) DNA repair in post-mitotic neurons. A gene-trapping strategy. Cell Death Differ. 12, 307–309 [DOI] [PubMed] [Google Scholar]
- 19. Weissman L., de Souza-Pinto N. C., Stevnsner T., Bohr V. A. (2007) DNA repair, mitochondria, and neurodegeneration. Neuroscience 145, 1318–1329 [DOI] [PubMed] [Google Scholar]
- 20. Butterfield D. A., Drake J., Pocernich C., Castegna A. (2001) Evidence of oxidative damage in Alzheimer disease brain. Central role for amyloid β-peptide. Trends Mol. Med. 7, 548–554 [DOI] [PubMed] [Google Scholar]
- 21. Rashidian J., Iyirhiaro G., Aleyasin H., Rios M., Vincent I., Callaghan S., Bland R. J., Slack R. S., During M. J., Park D. S. (2005) Multiple cyclin-dependent kinases signals are critical mediators of ischemia/hypoxic neuronal death in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 14080–14085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kraemer K. H., Patronas N. J., Schiffmann R., Brooks B. P., Tamura D., DiGiovanna J. J. (2007) Xeroderma pigmentosum, trichothiodystrophy, and Cockayne syndrome. A complex genotype-phenotype relationship. Neuroscience 145, 1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trushina E., McMurray C. T. (2007) Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience 145, 1233–1248 [DOI] [PubMed] [Google Scholar]
- 24. Chiti F., Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 25. Querfurth H. W., LaFerla F. M. (2010) Alzheimer disease. N. Engl. J. Med. 362, 329–344 [DOI] [PubMed] [Google Scholar]
- 26. Walsh D. M., Selkoe D. J. (2007) Aβ oligomers. A decade of discovery. J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 27. Haass C., Selkoe D. J. (2007) Soluble protein oligomers in neurodegeneration. Lessons from the Alzheimer amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 28. Behl C., Davis J. B., Lesley R., Schubert D. (1994) Hydrogen peroxide mediates amyloid β protein toxicity. Cell 77, 817–827 [DOI] [PubMed] [Google Scholar]
- 29. Huang X., Cuajungco M. P., Atwood C. S., Hartshorn M. A., Tyndall J. D., Hanson G. R., Stokes K. C., Leopold M., Multhaup G., Goldstein L. E., Scarpa R. C., Saunders A. J., Lim J., Moir R. D., Glabe C., Bowden E. F., Masters C. L., Fairlie D. P., Tanzi R. E., Bush A. I. (1999) Cu(II) potentiation of Alzheimer Aβ neurotoxicity. Correlation with cell-free hydrogen peroxide production and metal reduction. J. Biol. Chem. 274, 37111–37116 [DOI] [PubMed] [Google Scholar]
- 30. Huang X., Atwood C. S., Hartshorn M. A., Multhaup G., Goldstein L. E., Scarpa R. C., Cuajungco M. P., Gray D. N., Lim J., Moir R. D., Tanzi R. E., Bush A. I. (1999) The Aβ peptide of Alzheimer disease directly produces hydrogen peroxide through metal ion reduction. Biochemistry 38, 7609–7616 [DOI] [PubMed] [Google Scholar]
- 31. Lucassen P. J., Chung W. C., Kamphorst W., Swaab D. F. (1997) DNA damage distribution in the human brain as shown by in situ end labeling; area-specific differences in aging and Alzheimer disease in the absence of apoptotic morphology. J. Neuropathol. Exp. Neurol. 56, 887–900 [DOI] [PubMed] [Google Scholar]
- 32. Stadelmann C., Brück W., Bancher C., Jellinger K., Lassmann H. (1998) Alzheimer disease. DNA fragmentation indicates increased neuronal vulnerability, but not apoptosis. J. Neuropathol. Exp. Neurol. 57, 456–464 [DOI] [PubMed] [Google Scholar]
- 33. Boerrigter M. E., Wei J. Y., Vijg J. (1992) DNA repair and Alzheimer disease. J. Gerontol. 47, B177–B184 [DOI] [PubMed] [Google Scholar]
- 34. Critchlow S. E., Jackson S. P. (1998) DNA end-joining. From yeast to man. Trends Biochem. Sci. 23, 394–398 [DOI] [PubMed] [Google Scholar]
- 35. West S. C. (2003) Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 36. Thompson L. H., Schild D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477, 131–153 [DOI] [PubMed] [Google Scholar]
- 37. Shrivastav M., De Haro L. P., Nickoloff J. A. (2008) Regulation of DNA double strand break repair pathway choice. Cell Res. 18, 134–147 [DOI] [PubMed] [Google Scholar]
- 38. Lieber M. R., Ma Y., Pannicke U., Schwarz K. (2003) Mechanism and regulation of human nonhomologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4, 712–720 [DOI] [PubMed] [Google Scholar]
- 39. Mills K. D., Ferguson D. O., Alt F. W. (2003) The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194, 77–95 [DOI] [PubMed] [Google Scholar]
- 40. Smith G. C., Jackson S. P. (1999) The DNA-dependent protein kinase. Genes Dev. 13, 916–934 [DOI] [PubMed] [Google Scholar]
- 41. Weterings E., Chen D. J. (2008) The endless tale of nonhomologous end-joining. Cell Res. 18, 114–124 [DOI] [PubMed] [Google Scholar]
- 42. Simpson J. E., Ince P. G., Haynes L. J., Theaker R., Gelsthorpe C., Baxter L., Forster G., Lace G. L., Shaw P. J., Matthews F. E., Savva G. M., Brayne C., Wharton S. B. (2010) Population variation in oxidative stress and astrocyte DNA damage in relation to Alzheimer-type pathology in the ageing brain. Neuropathol. Appl. Neurobiol. 36, 25–40 [DOI] [PubMed] [Google Scholar]
- 43. Stine W. B., Jr., Dahlgren K. N., Krafft G. A., LaDu M. J. (2003) In vitro characterization of conditions for amyloid-β peptide oligomerization and fibrillogenesis. J. Biol. Chem. 278, 11612–11622 [DOI] [PubMed] [Google Scholar]
- 44. Willmore E., de Caux S., Sunter N. J., Tilby M. J., Jackson G. H., Austin C. A., Durkacz B. W. (2004) A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood 103, 4659–4665 [DOI] [PubMed] [Google Scholar]
- 45. Veuger S. J., Curtin N. J., Richardson C. J., Smith G. C., Durkacz B. W. (2003) Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 63, 6008–6015 [PubMed] [Google Scholar]
- 46. Shearman M. S., Ragan C. I., Iversen L. L., (1994) Inhibition of PC12 cell redox activity is a specific, early indicator of the mechanism of β-amyloid-mediated cell death. Proc. Natl. Acad. Sci. U.S.A. 91, 1470–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diaz J. C., Simakova O., Jacobson K. A., Arispe N., Pollard H. B. (2009) Small molecule blockers of the Alzheimer Aβ calcium channel potently protect neurons from Aβ cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 3348–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yankner B. A., Dawes L. R., Fisher S., Villa-Komaroff L., Oster-Granite M. L., Neve R. L. (1989) Neurotoxicity of a fragment of the amyloid precursor associated with Alzheimer disease. Science 245, 417–420 [DOI] [PubMed] [Google Scholar]
- 49. Del Mar Martínez-Senac M., Villalaín J., Gómez-Fernández J. C. (1999) Structure of the Alzheimer β-amyloid peptide (25–35) and its interaction with negatively charged phospholipid vesicles. Eur. J. Biochem. 265, 744–753 [DOI] [PubMed] [Google Scholar]
- 50. Pike C. J., Burdick D., Walencewicz A. J., Glabe C. G., Cotman C. W. (1993) Neurodegeneration induced by β-amyloid peptides in vitro. The role of peptide assembly state. J. Neurosci. 13, 1676–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pike C. J., Walencewicz-Wasserman A. J., Kosmoski J., Cribbs D. H., Glabe C. G., Cotman C. W. (1995) Structure-activity analyses of β-amyloid peptides. Contributions of the β 25–35 region to aggregation and neurotoxicity. J. Neurochem. 64, 253–265 [DOI] [PubMed] [Google Scholar]
- 52. Iversen L. L., Mortishire-Smith R. J., Pollack S. J., Shearman M. S. (1995) The toxicity in vitro of β-amyloid protein. Biochem. J. 311, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Casciola-Rosen L. A., Anhalt G. J., Rosen A. (1995) DNA-dependent protein kinase is one of a subset of autoantigens specifically cleaved early during apoptosis. J. Exp. Med. 182, 1625–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song Q., Lees-Miller S. P., Kumar S., Zhang Z., Chan D. W., Smith G. C., Jackson S. P., Alnemri E. S., Litwack G., Khanna K. K., Lavin M. F. (1996) DNA-dependent protein kinase catalytic subunit. A target for an ICE-like protease in apoptosis. EMBO J. 15, 3238–3246 [PMC free article] [PubMed] [Google Scholar]
- 55. McConnell K. R., Dynan W. S., Hardin J. A. (1997) The DNA-dependent protein kinase catalytic subunit (p460) is cleaved during Fas-mediated apoptosis in Jurkat cells. J. Immunol. 158, 2083–2089 [PubMed] [Google Scholar]
- 56. Shackelford D. A., Tobaru T., Zhang S., Zivin J. A. (1999) Changes in expression of the DNA repair protein complex DNA-dependent protein kinase after ischemia and reperfusion. J. Neurosci. 19, 4727–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Soldani C., Scovassi A. I. (2002) Poly(ADP-ribose) polymerase-1 cleavage during apoptosis. An update. Apoptosis 7, 321–328 [DOI] [PubMed] [Google Scholar]
- 58. Kruman I. I., Wersto R. P., Cardozo-Pelaez F., Smilenov L., Chan S. L., Chrest F. J., Emokpae R., Jr., Gorospe M., Mattson M. P. (2004) Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 41, 549–561 [DOI] [PubMed] [Google Scholar]
- 59. Banáth J. P., Olive P. L. (2003) Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double strand breaks. Cancer Res. 63, 4347–4350 [PubMed] [Google Scholar]
- 60. Gottlieb T. M., Jackson S. P. (1993) The DNA-dependent protein kinase: Requirement for DNA ends and association with Ku antigen. Cell 15, 131–142 [DOI] [PubMed] [Google Scholar]
- 61. Collis S. J., DeWeese T. L., Jeggo P. A., Parker A. R. (2005) The life and death of DNA-PK. Oncogene 24, 949–961 [DOI] [PubMed] [Google Scholar]
- 62. Burden D. A., Osheroff N. (1998) Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta 1400, 139–154 [DOI] [PubMed] [Google Scholar]
- 63. Boldogh I., Roy G., Lee M. S., Bacsi A., Hazra T. K., Bhakat K. K., Das G. C., Mitra S. (2003) Reduced DNA double strand breaks in chlorambucil-resistant cells are related to high DNA-PKcs activity and low oxidative stress. Toxicology 193, 137–152 [DOI] [PubMed] [Google Scholar]
- 64. Lu H. R., Zhu H., Huang M., Chen Y., Cai Y. J., Miao Z. H., Zhang J. S., Ding J. (2005) Reactive oxygen species elicit apoptosis by concurrently disrupting topoisomerase II and DNA-dependent protein kinase. Mol. Pharmacol. 68, 983–994 [DOI] [PubMed] [Google Scholar]
- 65. Chen L., Liu L., Yin J., Luo Y., Huang S. (2009) Hydrogen peroxide-induced neuronal apoptosis is associated with inhibition of protein phosphatase 2A and 5, leading to activation of MAPK pathway. Int. J. Biochem. Cell Biol. 41, 1284–1295 [DOI] [PubMed] [Google Scholar]
- 66. Johnstone E. M., Babbey L. E., Stephenson D., Paul D. C., Santerre R. F., Clemens J. A., Williams D. C., Little S. P. (1996) Nuclear and cytoplasmic localization of the β-amyloid peptide (1–43) in transfected 293 cells. Biochem. Biophys. Res. Commun. 220, 710–718 [DOI] [PubMed] [Google Scholar]
- 67. Bückig A., Tikkanen R., Herzog V., Schmitz A. (2002) Cytosolic and nuclear aggregation of the amyloid β-peptide following its expression in the endoplasmic reticulum. Histochem. Cell Biol. 118, 353–360 [DOI] [PubMed] [Google Scholar]
- 68. Hegde M. L., Anitha S., Latha K. S., Mustak M. S., Stein R., Ravid R., Rao K. S. (2004) First evidence for helical transitions in supercoiled DNA by amyloid β peptide (1–42) and aluminum. A new insight in understanding Alzheimer disease. J. Mol. Neurosci. 22, 19–31 [DOI] [PubMed] [Google Scholar]
- 69. Ohyagi Y., Asahara H., Chui D. H., Tsuruta Y., Sakae N., Miyoshi K., Yamada T., Kikuchi H., Taniwaki T., Murai H., Ikezoe K., Furuya H., Kawarabayashi T., Shoji M., Checler F., Iwaki T., Makifuchi T., Takeda K., Kira J., Tabira T. (2005) Intracellular Aβ42 activates p53 promoter. A pathway to neurodegeneration in Alzheimer disease. FASEB J. 19, 255–257 [DOI] [PubMed] [Google Scholar]
- 70. Zhao Y., Thomas H. D., Batey M. A., Cowell I. G., Richardson C. J., Griffin R. J., Calvert A. H., Newell D. R., Smith G. C., Curtin N. J. (2006) Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 66, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 71. Culmsee C., Bondada S., Mattson M. P. (2001) Hippocampal neurons of mice deficient in DNA-dependent protein kinase exhibit increased vulnerability to DNA damage, oxidative stress, and excitotoxicity. Brain Res. Mol. Brain Res. 87, 257–262 [DOI] [PubMed] [Google Scholar]
- 72. Daido S., Yamamoto A., Fujiwara K., Sawaya R., Kondo S., Kondo Y. (2005) Inhibition of the DNA-dependent protein kinase catalytic subunit radiosensitizes malignant glioma cells by inducing autophagy. Cancer Res. 65, 4368–4375 [DOI] [PubMed] [Google Scholar]
- 73. Neema M., Navarro-Quiroga I., Chechlacz M., Gilliams-Francis K., Liu J., Lamonica K., Lin S. L., Naegele J. R. (2005) DNA damage and nonhomologous end joining in excitotoxicity. Neuroprotective role of DNA-PKcs in kainic acid-induced seizures. Hippocampus 15, 1057–1071 [DOI] [PubMed] [Google Scholar]
- 74. Chakravarthy B. R., Walker T., Rasquinha I., Hill I. E., MacManus J. P. (1999) Activation of DNA-dependent protein kinase may play a role in apoptosis of human neuroblastoma cells. J. Neurochem. 72, 933–942 [DOI] [PubMed] [Google Scholar]
- 75. Shao L., Goronzy J. J., Weyand C. M. (2010) DNA-dependent protein kinase catalytic subunit mediates T-cell loss in rheumatoid arthritis. EMBO Mol. Med. 2, 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Orii K. E., Lee Y., Kondo N., McKinnon P. J. (2006) Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. U.S.A. 103, 10017–10022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McKinnon P. J. (2009) DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 10, 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nouspikel T., Hanawalt P. C. (2003) When parsimony backfires. Neglecting DNA repair may doom neurons in Alzheimer disease. BioEssays 25, 168–173 [DOI] [PubMed] [Google Scholar]
- 79. Nyström T. (2005) Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 24, 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grune T., Reinheckel T., Davies K. J. (1997) Degradation of oxidized proteins in mammalian cells. FASEB J. 11, 526–534 [PubMed] [Google Scholar]
- 81. Tseng B. P., Green K. N., Chan J. L., Blurton-Jones M., LaFerla F. M. (2008) Aβ inhibits the proteasome and enhances amyloid and Tau accumulation. Neurobiol. Aging 29, 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cecarini V., Bonfili L., Amici M., Angeletti M., Keller J. N., Eleuteri A. M. (2008) Amyloid peptides in different assembly states and related effects on isolated and cellular proteasomes. Brain Res. 1209, 8–18 [DOI] [PubMed] [Google Scholar]
- 83. Roselli F., Tirard M., Lu J., Hutzler P., Lamberti P., Livrea P., Morabito M., Almeida O. F. (2005) Soluble β-amyloid(1–40) induces NMDA-dependent degradation of postsynaptic density-95 at glutamatergic synapses. J. Neurosci. 25, 11061–11070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cissé M., Halabisky B., Harris J., Devidze N., Dubal D. B., Sun B., Orr A., Lotz G., Kim D. H., Hamto P., Ho K., Yu G. Q., Mucke L. (2011) Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature 469, 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Varadarajan S., Kanski J., Aksenova M., Lauderback C., Butterfield D. A. (2001) Different mechanisms of oxidative stress and neurotoxicity for Alzheimer Aβ(1–42) and Aβ(25–35). J. Am. Chem. Soc. 123, 5625–5631 [DOI] [PubMed] [Google Scholar]
- 86. Butterfield D. A., Kanski J. (2002) Methionine residue 35 is critical for the oxidative stress and neurotoxic properties of Alzheimer amyloid β-peptide(1–42). Peptides 23, 1299–1309 [DOI] [PubMed] [Google Scholar]
- 87. Amador F. C., Henriques A. G., da Cruz E Silva O. A., da Cruz E Silva E. F. (2004) Monitoring protein phosphatase 1 isoform levels as a marker for cellular stress. Neurotoxicol. Teratol. 26, 387–395 [DOI] [PubMed] [Google Scholar]
- 88. Vintém A. P., Henriques A. G., da Cruz E Silva O. A., da Cruz E Silva E. F. (2009) PP1 inhibition by Aβ peptide as a potential pathological mechanism in Alzheimer disease. Neurotoxicol. Teratol. 31, 85–88 [DOI] [PubMed] [Google Scholar]
- 89. Douglas P., Moorhead G. B., Ye R., Lees-Miller S. P. (2001) Protein phosphatases regulate DNA-dependent protein kinase activity. J. Biol. Chem. 276, 18992–18998 [DOI] [PubMed] [Google Scholar]
- 90. Wong R. H., Chang I., Hudak C. S., Hyun S., Kwan H. Y., Sul H. S. (2009) A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell 136, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Olsen B. B., Issinger O. G., Guerra B. (2010) Regulation of DNA-dependent protein kinase by protein kinase CK2 in human glioblastoma cells. Oncogene 29, 6016–6026 [DOI] [PubMed] [Google Scholar]
- 92. Iimoto D. S., Masliah E., DeTeresa R., Terry R. D., Saitoh T. (1990) Aberrant casein kinase II in Alzheimer disease. Brain Res. 507, 273–280 [DOI] [PubMed] [Google Scholar]
- 93. Chauhan A., Chauhan V. P., Murakami N., Brockerhoff H., Wisniewski H. M. (1993) Amyloid β-protein stimulates casein kinase I and casein kinase II activities. Brain Res. 629, 47–52 [DOI] [PubMed] [Google Scholar]
- 94. Barzilai A. (2007) The contribution of the DNA damage response to neuronal viability. Antioxid. Redox. Signal. 9, 211–218 [DOI] [PubMed] [Google Scholar]