Abstract
The serum and glucocorticoid-inducible kinase SGK1 increases the activity of Orai1, the pore forming unit of store-operated Ca2+ entry, and thus influences Ca2+-dependent cellular functions such as migration. SGK1 further regulates transcription factor nuclear factor κB (NF-κB). This study explored whether SGK1 influences transcription of Orai1 and/or STIM1, the Orai1-activating Ca2+ sensor. Orai1 and STIM1 transcript levels were decreased in mast cells from SGK1 knock-out mice and increased in HEK293 cells transfected with active S422DSGK1 but not with inactive K127NSGK1 or in S422DSGK1-transfected cells treated with the NF-κB inhibitor Wogonin (100 μm). Treatment with the stem cell factor enhanced transcript levels of STIM1 and Orai1 in sgk1+/+ but not in sgk1−/− mast cells and not in sgk1+/+ cells treated with Wogonin. Orai1 and STIM1 transcript levels were further increased in sgk1+/+ and sgk1−/− mast cells by transfection with active NF-κB subunit p65 as well as in HEK293 cells by transfection with NF-κB subunits p65/p50 or p65/p52. They were decreased by silencing of NF-κB subunits p65, p50, or p52 or by NF-κB inhibitor Wogonin (100 μm). Luciferase assay and chromatin immunoprecipitation defined NF-κB-binding sites in promoter regions accounting for NF-κB sensitive genomic regulation of STIM1 and Orai1. Store-operated Ca2+ entry was similarly increased by overexpression of p65/p50 or p65/p52 and decreased by treatment with Wogonin. Transfection of HEK293 cells with p65/p50 or p65/p52 further augmented migration. The present observations reveal powerful genomic regulation of Orai1/STIM1 by SGK1-dependent NF-κB signaling.
Keywords: Calcium Channels, Cell Signaling, Gene Expression, NF-κB Transcription Factor, Phosphatidylinositol 3-Kinase, Protein Kinases, Serine Threonine Protein Kinase, Transcription Factors, Transcription Regulation
Introduction
Alterations of cytosolic Ca2+ activity participate in the regulation of a wide variety of cellular functions including excitation-contraction coupling, exocytosis, migration, cell proliferation, and cell death (1–4). Cytosolic Ca2+ is increased by release of Ca2+ from intracellular stores and/or Ca2+ entry across the cell membrane (5). Ca2+ release from intracellular stores results in the stimulation of Ca2+ release-activated Ca2+ channel (CRAC)2 (6, 7), which consists of the pore forming units Orai1, -2, and/or -3 (8–10) and the endoplasmic reticulum-located regulatory subunit STIM1 or -2 (11–13). The stimulation of the channel leads to the inward current ICRAC and the store-operated Ca2+ entry (SOCE).
Recent observations uncovered the powerful stimulation of ICRAC and SOCE by the serum and glucocorticoid-inducible kinase SGK1 (14), a kinase stimulated by growth factors and involved in stress response (15) and regulation of cell survival (16). SGK1 is partially effective through phosphorylation of the ubiquitin ligase Nedd4-2 (neuronal precursor cells expressed developmentally down-regulated). Nedd4-2 ubiquitinates Orai1, thus preparing the channel protein for degradation (14). The effect of Nedd4-2 on Orai1 parallels that of Nedd4-2 on the epithelial Na+ channel ENaC (16, 17). The phosphorylation of Nedd4-2 leads to binding of the ubiquitin ligase to the protein 14-3-3, which prevents the interaction with the channel protein (18). Accordingly, SGK1 enhances Orai1 protein abundance in the cell membrane (14). STIM is similarly regulated by ubiquitination (19). However, the effect of SGK1 on Orai1 protein abundance is only in part explained by Nedd4-2-dependent protein degradation. Therefore, further experiments were performed to explore whether SGK1, in addition, stimulates Orai1 and/or STIM1 expression. As a matter of fact, RT-PCR revealed an increase of Orai1 and STIM1 transcript levels after expression of constitutively active SGK1. Thus, further experiments were performed to uncover the transcription factor involved. Previously, SGK1 has been shown to foster nuclear translocation and activation of nuclear factor κB (NF-κB) (20–22). Accordingly, this study explored the putative involvement of NF-κB subunits p65 (RELA), p50 (NFKB1), and p52 (NFKB2) in the regulation of Orai1 and STIM1 expression.
EXPERIMENTAL PROCEDURES
Transfection of Cells and NF-κB Inhibition
HEK293 cells were transfected with the constitutively active mutant SGK1S422D (hSGK1SD in pIRES-EGFP), the inactive mutant SGK1K127N (hSGK1KN in pIRES-EGFP) (23), or the NF-κB subunits p65 (24), p50, and p52 (both obtained from imaGenes GmbH) using FuGENE HD transfection reagent (Roche Diagnostics) according to the manufacturer's instructions. For luciferase assay, pGL3-Luciferase Enhancer constructs were cotransfected with 50 ng of pRL-TK (Promega) and p65 using FuGENE HD transfection reagent. Where indicated, NF-κB was inhibited by treatment of the cells for 24 h with 100 μm Wogonin (25, 26). Additionally, knockdown of NF-κB subunits p65, p50, and p52 was performed with 10 nm siRNA (Life Technology Inc., Ambion) according to the manufacturer's instructions using Polyplus Interferin (PeqLab, Germany). Mast cells were transfected with 2 μg of S276Dp65 according to the manufacturer's instructions using solution V and an Amaxa nucleofector device (Köln, Germany; set to program T-020).
Site-directed Mutagenesis
To introduce an aspartic acid for a serine at position 276, QuikChange II site-directed mutagenesis kit (Stratagene, Agilent Technologies) was utilized using the following primer pair (5′-3′ orientation, the mutation is in bold): forward (CAGCTACGGCGGCCTGATGATCGCGAGCTCAGT) and reverse (ACTGAGCTCGCGATCATCAGGCCGCCGTAGCTG). Sequencing was performed to verify the mutation.
Real-time PCR Analysis
To determine STIM1/Orai1 mRNA abundance in HEK293 cells, RNA was extracted using Trifast reagent (Invitrogen) according to the manufacturer's instructions. Subsequently 2 μg of total RNA was reverse-transcribed to cDNA using random hexamer primers (0.1 mm, Roche Diagnostics), 1st strand buffer (Invitrogen), DTT (10 mm, Invitrogen), and SuperScript II reverse transcriptase (200 units, Invitrogen) for 1 h at 42 °C. For inactivation, the samples were incubated for 10 min at 70 °C. Quantitative real-time PCR was applied on the CFX96 Real-Time System® C1000 Thermal Cycler (Bio-Rad) using the following primer pairs (5′-3′ orientation): STIM1 forward (CCTGTGGAAGGCATGGAAGT), STIM1 reverse (CTGAGGCAGCTCCACATATGT), Orai1 forward (CACCTGTTTGCGCTCATGAT), and Orai1 reverse (GGGACTCCTTGACCGAGTTG). The transcript levels of the housekeeping gene TBP were determined for each sample using the following primers (5′-3′ orientation): forward (GCCCGAAACGCCGAATAT) and reverse (CCGTGGTTCGTGGCTCTCT). Amplification of the housekeeping gene TBP was performed to standardize the amount of sample RNA. Knockdown or up-regulation of NF-κB subunits after transfection was examined using the following primer pairs (5′-3′ orientation): p65 forward (ATACCACCAAGACCCACCCC), p65 reverse (TGAGGAGGGTCCTTGGTGAC), p50 forward (ATAATGCCTTCCGGCTGAGTC), p50 reverse (TTGTCTCAGGGCCTCCACC), p52 forward (GAAGGGCCGAAAGACCTATCC), and p52 reverse (GAGCACGAGGTGGGTCACTG).
Bone marrow-derived mast cells from sgk1−/− and wild type mice were analyzed as described above using the following primer pairs (5′-3′ orientation): STIM1 forward (GGAAGGCGTGGAAGTCATCA), STIM1 reverse (TCATACTGTGGCAGCTCCACA), Orai1 forward (GCCAAGCTCAAAGCTTCCA), Orai1 reverse (CATGGTCTGTGTCCAGCTGG), SGK1 forward (GGCTATCTGCACTCCCTAAACA), SGK1 reverse (CCAAAGTCAGTGAGGACGATGT), TBP forward (CACTCCTGCCACACCAGCTT), and TBP reverse (TGGTCTTTAGGTCAAGTTTACAGCC).
Primer pairs were designed binding to two different exons, creating a product of about 90 bp. Relative quantification of gene expression was performed using the 2−ΔΔct method as described earlier (27).
Identification of Putative κB-binding Sites
The genomic sequences of STIM1 and Orai1 promoter were checked for the 5′-GGGRNNYYCC-3′ κB-consensus sequence. DNA sequence of the promoter region (3000 bp upstream of transcription start) was used from ensemble.org, and each possible consensus sequence was tested by self-made sequence alignment.
Luciferase Assay
Sequences containing putative κB-binding sites in the STIM1 and Orai1 promoter region were amplified by PCR from genomic template DNA. PCR was performed using MaximaTM Hot Start TaqDNA polymerase (Fermentas, Germany) with primers containing a KpnI site 5′ and a HindIII site 3′: O1 forward (GTAGGTACCGGAAACAAAGCCAGTAG), O1 reverse (CGTAAGCTTTTCCAGACCAGCCTA), O2 forward (CGTAGGTACCCCAGAGACTTCTTGGG), O2 reverse (GCTAAGCTTCAGGACGGCGAGG), S1 forward (CCTCGGTACCATCCATGTTGTAGCA), and S1 reverse (GCGAAAGCTTACGCTAAAATGGTGTCT). The separate STIM1/Orai1 promoter fragments were cloned into the pGL3-Luciferase Enhancer Vector (Promega). To ensure that the fragments were inserted in the correct orientation and to confirm the correctness of the sequence control restrictions and DNA sequencing was performed (Delphi Test).
Forty-eight hours after transfection, Dual-Luciferase Reporter Assays (Promega) were performed according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla activity. Renilla luciferase was constitutively expressed by the cotransfected vector pRL-TK and thus served as an internal control for the transfection rate.
Animals
Bone marrow was obtained from 6–8-week-old female and male SGK1 knock-out (sgk1−/−) mice and their wild type (sgk1+/+) littermates. Generation, breeding, and genotyping of the mice has been described earlier (28). Animal experiments were conducted according to German law for the welfare of animals and were approved by local authorities.
Culture of Bone Marrow-derived Mast Cells (BMMCs)
Femoral bone marrow cells from 6–8-week-old sgk1+/+ and sgk1−/− mice were cultured for 4 weeks in RPMI 1640 containing 10% fetal calf serum (both from Invitrogen), 1% penicillin/streptomycin, 20 ng/ml interleukin-3 (RD Systems), and 100 ng/ml concentrations of the c-kit ligand stem cell factor (SCF, Peprotech, Tebu-bio). BMMC maturation was confirmed by flow cytometry (FACSCalibur, BD Biosciences) (29).
Treatment of BMMCs
Mast cells were plated in 24-well plates (1 × 106 cells/well). The cells were either treated with Wogonin (100 μm) for 24 h or incubated in the medium deprived of growth factors (SCF, IL-3) in the presence or in the absence of Wogonin (100 μm) for 20 h and after that treated with SCF (100 μg/ml) in the presence or in the absence of Wogonin, respectively, for a further 4 h. Then the cells were collected and washed with PBS.
Immunofluorescence
HEK293 cells transfected with p65/p50 or p65/p52, respectively, were cultured in 24-well plates with a coverslip inside, washed, and fixed with 4% paraformaldehyde. For blocking, unspecific binding HEK293 cells were incubated with 5% normal goat serum, 1× PBS, 0.1% Triton for 1 h at room temperature. Then the cells were exposed to rabbit anti-Orai1 (1:100, Millipore) or rabbit anti-STIM1 (1:100, Abnova) at 4 °C overnight. The cells were rinsed three times with PBS and incubated with secondary FITC goat anti-rabbit antibody (1:1000; Invitrogen) or with Alexa Fluor® 488-conjugated goat anti-mouse antibody (1:1000, Invitrogen) for 1.5 h at room temperature. After three washing steps the nuclei were stained with DRAQ-5 dye (1:1000; Biostatus) for 10 min at room temperature. The slides and coverslips were mounted with ProLong Gold antifade reagent (Invitrogen). Images were taken on a LSM 5 EXCITER confocal laser scanning microscope (Zeiss, Germany) with a water-immersion Plan-Neofluar 40×/1.3 NA differential interference contrast and analyzed with the instrument's software.
Nuclear translocation of p65 was analyzed by incubating HEK293 cells transfected with either SGK1S422D or SGK1K127N or EGFP-containing empty vector or BMMCs derived from sgk1−/− and sgk1+/+ mice with anti-p65 (1:200, Santa Cruz) and Cy3 donkey anti-rabbit antibody (1:1000; Millipore). For a positive control, the translocation of NF-κB subunit p65 in BMMCs was induced by treatment of the HEK293 cells with TNFα (100 ng/ml, 1h, R&D Systems).
Chromatin Immunoprecipitation
For chromatin immunoprecipitation (ChIP), HEK293 cells (80–90% confluent) were incubated at room temperature for 10 min with 1% paraformaldehyde and PBS. The reaction was stopped by adding 125 mm glycine for 5 min. After washing 3 times the cells were incubated with swelling buffer containing 25 m HEPES, pH 7.8, 1.5 mm MgCl2, 10 mm KCl, 0.1% Nonidet P-40, 1 mm DTT, 0.5 mm PMSF, complete EDTA-free Protease Inhibitor Mixture tablets (Roche Diagnostics) for 20 min at 4 °C. The cells were lysed with 1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 7.8, and sonicated 3 times for 30 s at 30% (Sonifer 250, Branson) to shear DNA. Preclearing was performed in 16.7 mm Tris-HCl, pH 8.1, 1.1% Triton X-100, 1.2 mm EDTA, 0.01% SDS, 300 mm NaCl, complete EDTA-free Protease Inhibitor Mixture tablets (Roche Diagnostics) using salmon sperm DNA/protein G-agarose slurry. After centrifugation, 20 μl (1%) of the supernatant were removed and saved. Precleared cell lysates were incubated with the following antibodies at 4 °C overnight; 2 μg of anti-p65 (C20, Santa Cruz) was used for pull down, and anti-rabbit IgG antibody (1:1000, Abcam) was used as the negative control. Immune complexes were recovered with salmon sperm DNA/protein A-agarose slurry. After washing and elution in 1% SDS, 100 mm NaHCO3 overnight at 65 °C, genomic DNA was extracted with phenol/chloroform, and PCR was performed with the following primer pairs (5′-3′ orientation): O1 forward (GTAGGTACCGGAAACAAAGCCAGTAG), O1 reverse (CGTAAGCTTTTCCAGACCAGCCTA), O2 forward (CGTAGGTACCCCAGAGACTTCTTGGG), O2 reverse (GCTAAGCTTCAGGACGGCGAGG), STIM1 forward (CCTCGGTACCATCCATGTTGTAGCA), and STIM1 reverse (GCGAAAGCTTACGCTAAAATGGTGTCT).
Whole Cell Lysates
For total protein analysis cells were harvested with lysis buffer (50 mm Tris, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.4% β-mercaptoethanol, Proteinase-Inhibitor Mixture, Roche Diagnostics) 48–72 h after transfection. Clarified protein lysate was applied to a polyacrylamide gel and analyzed by Western blot.
Western Blot Analysis
100 μg of protein of whole cell lysates were used for Western blot analysis and incubated with primary antibody against Orai1 (1:500, ProteinTech Group), STIM1 (1:500, Abnova, Taiwan), NF-κB p65 (1:1000, Cell Signaling), NF-κB p50 (E-10, 1:500, Santa Cruz), NF-κB p100/p52 (1:1000, Cell Signaling), or tubulin (1:1000, Cell Signaling). For detection, a secondary anti-rabbit IgG antibody conjugated with horseradish peroxidase (HRP) (1:3000, Cell Signaling) or secondary anti-mouse IgG antibody conjugated with HRP (1:3000, Amersham Biosciences) was used. The blots were stripped and reprobed with tubulin to verify equal loading. Antibody binding was detected with ECL detection reagent (Amersham Biosciences). Bands were quantified with ImageJ Software.
Measurement of Intracellular Ca2+ Concentration
Fura-2/AM fluorescence was utilized to determine intracellular Ca2+ (29). Cells were excited alternately at 340 and 380 nm through an objective (Fluar 40×/1.30 oil) built in an inverted phase-contrast microscope (Axiovert 100, Zeiss, Oberkochen, Germany). Emitted fluorescence intensity was recorded at 505 nm. Data were acquired using specialized computer software (Metafluor, Universal Imaging). Cytosolic Ca2+ activity was estimated from the 340/380-nm ratio. HEK293 cells were loaded with Fura-2/AM (2.5 μm, Molecular Probes) for 30 min at 37 °C. SOCE was determined by extracellular Ca2+ removal and subsequent Ca2+ readdition in the presence of thapsigargin (1 μm) (30). For quantification of the Ca2+ entry, slope (Δ ratio/s) and peak (Δ ratio) were calculated after readdition of Ca2+.
Experiments were performed with Ringer solution containing 125 mm NaCl, 5 mm KCl, 1.2 mm MgSO4, 2 mm CaCl2, 2 mm Na2HPO4, 32 mm HEPES, 5 mm glucose, pH 7.4. To reach nominally Ca2+-free conditions, experiments were performed using Ca2+-free Ringer solution containing 125 mm NaCl, 5 mm KCl, 1.2 mm MgSO4, 2 mm Na2HPO4, 32 mm HEPES, 0.5 mm EGTA, 5 mm glucose, pH 7.4.
Migration Assay
For migration assays, transwell inserts (BD Falcon 353097) and BD BioCoatTM MatrigelTM Invasion Chambers (BD Biosciences 354480) were used with a pore diameter size of 8 μm. The transwells were placed in a 24-well cell culture plate containing cell culture medium (750 μl). The upper chambers were filled with 500 μl of cell culture medium containing HEK293 cells in a concentration of 5 × 104 cells/ml. After an incubation time of 24 h at 37 °C, migrated cells were analyzed by staining the cell nuclei with DAPI. Before that non-migrated cells were removed by scrubbing with a cotton-tipped swab two times and washing with PBS. After a 15 min fixation in 4% paraformaldehyde, the membrane was removed with a scalpel. After removal from the inserts, membranes were mounted on slides with ProLong Gold antifade reagent (Invitrogen). To determine the total number of migrated cells, the slides were then viewed under the microscope, and the number of cells/field in representative areas was counted. Experiments were performed in triplicate.
Statistics
Data are provided as the mean ± S.E.; n represents the number of independent experiments. All data were tested for significance using Student's unpaired two-tailed t test or ANOVA (Tukey's test or Dunnett's test), where applicable. Results with p < 0.05 were considered statistically significant.
RESULTS
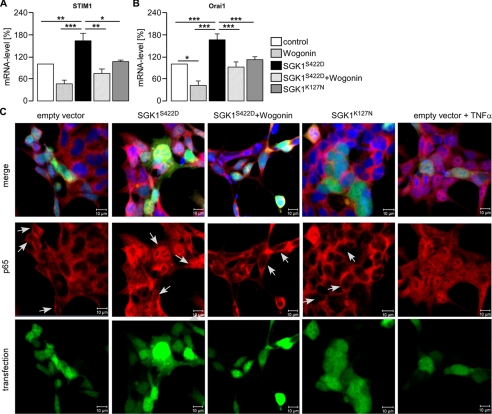
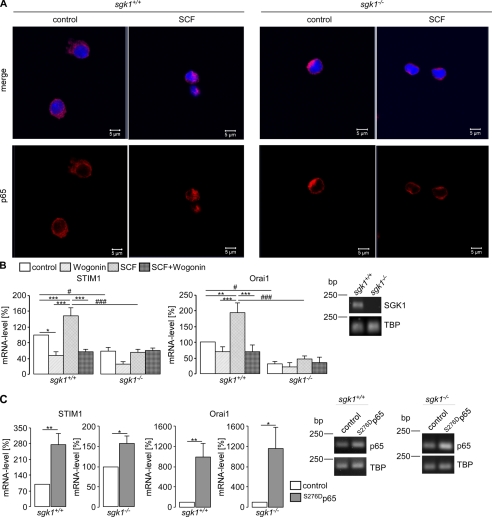
As illustrated in Fig. 1, A and B, transfection of HEK293 with the constitutively active mutant S422DSGK1 but not with the inactive mutant K127NSGK1 was followed by a significant increase of mRNA encoding STIM1 and Orai1. Accordingly, the transcript levels of both genes were enhanced by SGK1. Previous studies revealed that SGK1 activates NF-κB (20–22). Thus, S422DSGK1-transfected and non-transfected HEK293 cells were treated with the NF-κB inhibitor Wogonin (100 μm). The treatment significantly reduced the mRNA levels of STIM1 and Orai1 in both S422DSGK1-overexpressing and non-transfected HEK293 cells (Fig. 1, A and B). Transfection of HEK293 cells with S422DSGK1 but not with K127NSGK1 led to nuclear translocation of the NF-κB subunit p65 (Fig. 1C). Again, the effect of S224DSGK could be reversed by treatment of the cells with Wogonin. For the positive control for p65 translocation, HEK293 cells were incubated with TNFα (100 ng/ml, 1 h). Το elucidate whether the stimulating effect of SGK1 is apparent under endogenous levels of SGK1, the transcript levels of Orai1 and STIM1 were determined in BMMCs isolated from gene targeted mice lacking functional SGK1 (sgk1−/−) and their wild type (sgk1+/+) littermates. As shown in Fig. 2, the Orai1 and STIM1 transcript levels were both significantly higher in sgk1+/+ BMMCs compared with sgk1−/− BMMCs. Stimulation of sgk1+/+, but not of sgk1−/− BMMCs, with SCF (20 ng/ml, 4 h) resulted in a translocation of the NF-κB subunit p65 to the nucleus (Fig. 2A) and accordingly led to significant increase of the STIM1 and Orai1 transcript levels in sgk1+/+ BMMCs but not in sgk1−/− BMMCs (Fig. 2B), an effect abrogated by treatment of sgk1+/+ cells with Wogonin (Fig. 2B). Treatment of BMMCs with Wogonin alone tended to decrease Orai1 mRNA levels, an effect, however, not reaching statistical significance. To further evaluate whether NF-κB-sensitive transcription of STIM1 and Orai1 was downstream of SGK1, BMMCs from sgk1−/− and sgk1+/+ mice were transfected with a constitutively active mutant of p65 (S276Dp65). As shown in Fig. 2C, transfection with S276Dp65 led to a significant increase in transcript levels of STIM1 and Orai1 in BMMCs from both sgk1+/+ and sgk−/− mice.
FIGURE 1.
Effect of SGK1 on STIM1 and Orai1 transcript levels in HEK293 cells. Shown are the arithmetic means (±S.E., n = 13) of the abundance of mRNA encoding STIM1 (A) and Orai1 (B) in HEK293 cells after transfection with constitutively active SGK1S422D or inactive SGK1K127N with or without treatment with Wogonin (100 μm). * (p < 0.05), ** (p < 0.01), and *** (p < 0.001) indicate statistically significant differences (ANOVA). C, confocal microscopy of p65 (red) translocation in HEK293 cells transfected with active SGK1S422D (green) or inactive SGK1K127N (green) or EGFP-empty vector (green) with or without treatment with Wogonin (100 μm) is shown. For a positive control of p65 translocation, HEK293 cells were incubated with TNFα (100 ng/ml, 1h). Nuclei were stained with DRAQ5 (blue). Arrows indicate transfected cells. The experiment was performed three times.
FIGURE 2.
Effect of SGK1 and NF-κB on STIM1 and Orai1 transcript levels in BMMCs. A, confocal microscopy of p65 (red) translocation in BMMCs from gene- targeted mice lacking functional SGK1 (sgk1−/−) and their wild type (sgk1+/+) littermates with or without (control) exposure to SCF (100 ng/ml) is shown. Nuclei were stained with DRAQ5 (blue). B, arithmetic means (±S.E., n = 14) of relative mRNA levels of STIM1 (left panel) and Orai1 (right panel) in sgk1+/+ and sgk1−/− BMMCs with or without exposure to SCF (100 ng/ml) and with or without the presence of Wogonin (100 μm) are shown. Knock out of SGK1 was determined utilizing RT-PCR and agarose gel electrophoresis. ** (p < 0.01) and *** (p < 0.001) indicate statistically significant differences in one group, and # (p < 0.05) and ### (p < 0.001) indicate statistically significant differences between groups (ANOVA). C, arithmetic means (±S.E., n = 6) of relative mRNA levels for STIM1 (left and middle left panel) and Orai1 (right and middle right panel) in BMMCs from sgk1−/− and sgk1+/+ mice without (control) or with transfection with a constitutively active p65 mutant are shown. The success of transfection was determined utilizing RT-PCR and agarose gel electrophoresis. * (p < 0.05) and **(p < 0.01) indicate statistically significant differences (Student's unpaired t test).
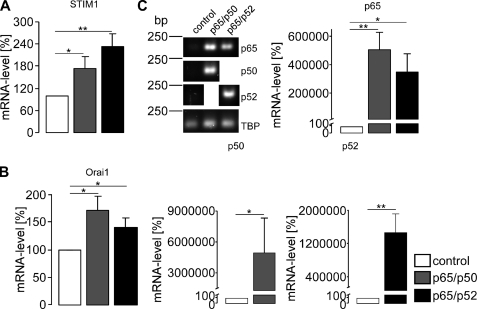
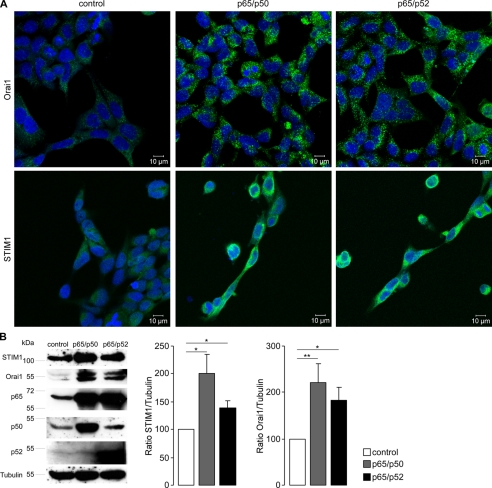
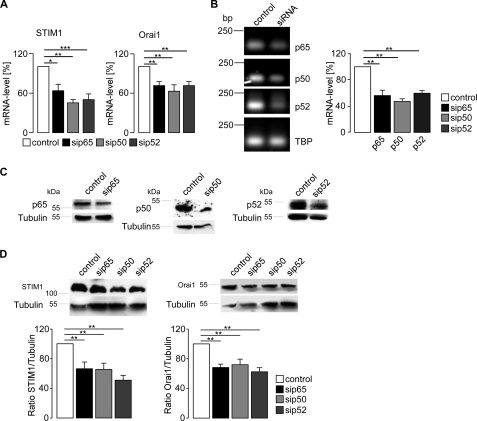
Further experiments explored whether transfection of HEK293 cells with NF-κB influences the transcript levels of STIM1 and Orai1. As shown in Fig. 3, transfection with the NF-κB subunits p65/p50 or p65/p52 indeed significantly increased the STIM1 (Fig. 3A) and Orai1 (Fig. 3B) transcript levels. The success of transfection was verified by specific primer pairs for p65, p50, and p52 using RT-PCR (Fig. 3C). TBP was used as an internal control for RT-PCR. Further experiments explored whether STIM1 and Orai1 protein could also be increased by transfection with the NF-κB subunits. As a result, fluorescence imaging (Fig. 4A) and Western blot analysis (Fig. 4B) revealed significant increases in STIM1 and Orai1 protein after transfection with p65/p50 or p65/p52. Tubulin was used as loading control for Western blot analysis. The success of transfection was controlled with specific antibodies against p65, p50, and p52, respectively.
FIGURE 3.
Effect of the NF-κB subunits p65, p50, and p52 on STIM1 and Orai1 transcript levels in HEK293 cells. Arithmetic means (±S.E., n = 14–17) of the abundance of mRNA encoding STIM1 (A) and Orai1 (B) in HEK293 cells expressing the NF-κB subunits p65/p50 or p65/p52. C, the success of NF-κB subunit transfection was determined utilizing RT-PCR and agarose gel electrophoresis. Upper left panel, shown is a representative agarose gel of cDNA after PCR extracted from HEK293 cells transfected with p65/p50 or p65/p52. TBP was used as an internal control. Upper right and lower panel, shown are the arithmetic means (±S.E., n = 14–17) of the mRNA abundance encoding p65 (upper right panel), p50 (lower left panel), or p52 (lower right panel) in HEK293 expressing p65/p50 or p65/p52 as analyzed by the 2−ΔΔCT method (27). * (p < 0.05) and ** (p < 0.01) indicate statistically significant differences (Student's unpaired t test or ANOVA).
FIGURE 4.
Effect of NF-κB on STIM1 and Orai1 protein abundance in HEK293 cells. A, confocal microscopy of STIM1 and Orai1 protein abundance in HEK293 cells is shown. Shown are HEK293 cells transfected with control plasmid (control), p65 and p50, or p65 and p52. The nuclei were stained with DRAQ5 (blue), Orai1 with a FITC-conjugated antibody (green, upper panel), and STIM1 with an Alexa Fluor® 546-conjugated antibody (green, lower panel). The experiment was completed three times. B, shown is a Western blot analysis of whole cell lysate in p65/p50- or p65/p52-transfected HEK293 cells showing STIM1 and Orai1 protein abundance. The blot was stripped and reprobed with tubulin to ensure equal loading and with p65, p50, and p52 to control transfection. Left, representative blot; middle, arithmetic means of STIM1/tubulin ratio (±S.E., n = 6); right, arithmetic means of Orai1/tubulin ratio (±S.E., n = 6). * (p < 0.05) and ** (p < 0.01) indicate statistically significant differences (Student's unpaired t test).
In a next step, NF-κB was silenced with siRNA in HEK293 cells. As illustrated in Fig. 5A, silencing of p65, p50, or p52 each led to a significant decrease of both STIM1 and Orai1 transcript levels. The success of the knockdown was verified by RT-PCR (Fig. 5B) and Western blotting (Fig. 5C). Silencing of NF-κB subunits also decreased protein levels of STIM1 and Orai1 (Fig. 5D).
FIGURE 5.
Effect of NF-κB subunit silencing on STIM1 and Orai1 transcript levels. A, arithmetic means (±S.E., n = 4–8) of the abundance of mRNA encoding STIM1 and Orai1 in HEK293 cells without and with silencing of p65, p50, and p52. * (p < 0.05), ** (p < 0.01), and *** (p < 0.001) indicate statistically significant differences (ANOVA). B, left panel, shown is a representative agarose gel of cDNA after RT-PCR extracted from HEK293 cells silenced with siRNA for p65, p50, or p52 to control the success of knock down. TBP was used as an internal control. Right panel, arithmetic means (±S.E., n = 3–8) of the mRNA abundance encoding p65 (black bar), p50 (light gray bar), or p52 (dark gray bar) in HEK293 cells compared with cells transfected with scramble siRNA (white bar) are shown. ** (p < 0.01) indicate statistically significant differences (ANOVA). C, Western blot analysis of whole cell lysate in HEK293 cells silenced with siRNA for p65, p50, or p52 is shown. As control, scrambled siRNA was utilized. The success of knockdown of p65, p50, or p52 was tested with specific antibodies. The blot was stripped and reprobed with tubulin to control equal loading. The experiment was completed five times. D, Western blot analysis of whole cell lysate in HEK293 cells silenced with siRNA for p65, p50, or p52 is shown. Upper left panel, shown is a representative blot of STIM1. Upper right panel, shown is a representative blot of Orai1. Lower left panel, shown is the arithmetic means of STIM1/tubulin ratio (±S.E., n = 5). Lower right panel, shown is the arithmetic means of Orai1/tubulin (±S.E., n = 5). ** (p < 0.01) indicate statistically significant differences (ANOVA).
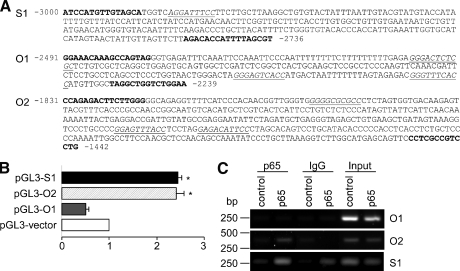
Analysis of the STIM1 and Orai1 promoter revealed putative NF-κB-binding sites in the region 2980 bp upstream of STIM1 and 2420 bp upstream of Orai1 transcription start (Fig. 6A). We, therefore, explored the possibility of transcriptional activation of STIM1 and Orai1 by NF-κB utilizing ChIP and luciferase assay. For STIM1 promoter (S1) and two regions of Orai1 promoter (O1 and O2), putative κB-like binding sites were subcloned into pGL3-vector cotransfected with p65 and pRL-TK and analyzed for luciferase activity. For control of basal luciferase activity cells were transfected with a control plasmid (pcDNA3.1+) instead of p65. Firefly luciferase activity was normalized to Renilla luciferase activity. As shown in Fig. 6B, cotransfection of pGL3-S1 with the integrated STIM1 promoter and p65 resulted in a 2.3 ± 0.2 (n = 14)-fold increase of luciferase activity. Cotransfection with pGL3-O2 (contains the promoter region O2 of Orai1) and p65 resulted in a 2.4 ± 0.2 (n = 21)-fold increase of luciferase activity. In contrast, cotransfection with pGL3-O1 containing the promoter region O1 of Orai1 and p65 did not activate luciferase activity. In ChIP experiments the binding of p65 to putative NF-κB response elements was further investigated. This was achieved using anti-p65 (2 μg, C20, Santa Cruz) or anti-rabbit IgG (1:1000, Abcam) antibodies to pull down p65-bound DNA extracted from HEK293 cells transiently transfected with p65. For comparison, the cells were transfected with a control plasmid. PCR indicated p65 binding to the STIM1 and Orai1 promoter sequence using primer pairs flanking the putative binding sites. As illustrated in Fig. 6C, we could observe pulldown of one putative binding site in the STIM1 promoter as well as in the Orai1 promoter (O2). There was no pulldown detectable in a second putative κB-binding site in Orai1 promoter (O1) and when using rabbit IgG as negative control.
FIGURE 6.
NF-κB-sensitive transcription of Orai1 and STIM1. A, analysis of the DNA sequence of 11p15.5 in the region adjacent to the STIM1 transcriptional start site (S1) and of 12q24.31 in the region adjacent to the Orai1 transcriptional start site (O1 and O2) revealed putative κB-like binding sites (italic, underlined) is shown. For ChIP experiments DNA was amplified by PCR and subcloned into pGL3-vector for luciferase reporter assay using the primers depicted in bold. B, a luciferase reporter assay was applied to study the ability of NF-κB to activate the target promoters of STIM1 and Orai1. HEK293 cells were transfected with luciferase reporter plasmids containing the putative κB-binding sites and additionally transfected with p65 or control plasmid. The results are presented as -fold increase in the activity of luciferase activity (±S.E., n = 11–21) compared with luciferase activity transfected with the control plasmid. * (p < 0.05) indicates statistically significant differences (ANOVA). C, binding of NF-κB to DNA was explored utilizing ChIP of genomic DNA extracted from HEK293 cells transfected with p65 or a control plasmid. Protein-chromatin complexes were immunoprecipitated with anti-p65 or anti-rabbit IgG. The input served as a positive control, IgG as a negative control. The experiment was carried out five times.
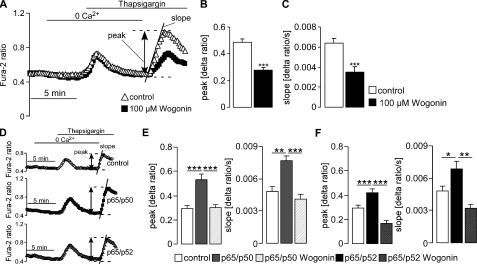
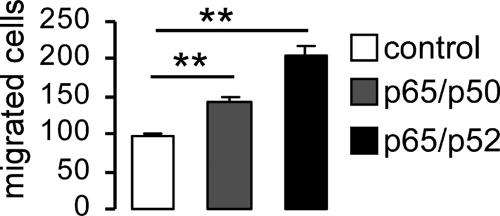
A further series of experiments explored whether NF-κB-sensitive transcription of STIM1 and Orai1 influenced SOCE. To this end, the fura-2 fluorescence-ratio was determined by fluorescence spectrometry. SOCE was measured upon store depletion by inhibition of the vesicular Ca2+-ATPase with thapsigargin in HEK293 cells in the presence and absence of Wogonin (100 μm). Stores were depleted by thapsigargin (1 μm) first added to the Ca2+-free medium. Thapsigargin triggered release of Ca2+ from intracellular stores, which was not modified by Wogonin treatment (Fig. 7A). Re-addition of Ca2+ (1 mm) to the medium in the continued presence of thapsigargin led to an entry of Ca2+ through plasma membrane Ca2+ channels and to a subsequent increase of the fluorescence ratio (Fig. 7A). The fluorescence Δratio (peak) and the slope of the ratio (Δratio/time) were analyzed upon Ca2+ re-addition. Both, peak, and slope values of SOCE were significantly decreased by the NF-κB inhibitor Wogonin (Fig. 7, A–C). Conversely, both peak and slope were significantly increased after transfecting the cells with NF-κB subunits p65/p50 or p65/p52 (Fig. 7, D–F). When p65/p50- and p65/p52-transfected cells were treated with Wogonin, the stimulating effect of p65/p50 or p65/p52 overexpression was abrogated (Fig. 7, E and F). Finally, the impact of NF-κB-sensitive transcriptional regulation of STIM1 and Orai1 on HEK293 cell migration was estimated in a transwell migration assay from translocation of cells from one chamber to another across a membrane with a pore diameter size of 8 μm. As a result, migratory activity was significantly higher in p65/p50- and p65/p52-transfected HEK293 cells than in cells transfected with an empty vector (Fig. 8).
FIGURE 7.
Effect of NF-κB on SOCE in HEK293 cells. A, shown are representative tracings of the fura-2 fluorescence ratio in fluorescence spectrometry during and after Ca2+ depletion and thapsigargin (1 μm) addition in HEK293 cells with or without treatment with Wogonin (100 μm). Arithmetic means (±S.E., n = 3, each experiment 14–21 cells) of peak (B) and slope (C) increase of the fura-2-fluorescence ratio in cells without (white bars) or with (black bars) treatment with Wogonin. D, shown are representative tracings of the fura-2 fluorescence ratio in fluorescence spectrometry during and after Ca2+ depletion and thapsigargin (1 μm) addition in HEK293 cells transfected with control plasmid (upper panel), p65/p50 (middle panel), or p65/p52 (lower panel). E, shown are the arithmetic means (±S.E., n = 3, each experiment 14–33 cells) of peak (Δ ratio) and slope (Δ ratio/s) of fura-2 fluorescence increase after readmission of Ca2+ in HEK293 cells transfected with control plasmid or p65/p50 with or without treatment with Wogonin (100 μm). F, shown are the arithmetic means (±S.E., n = 3, each experiment 14–33 cells) of peak (Δ ratio) and slope (Δ ratio/s) of fura-2 fluorescence increase after readmission of Ca2+ in HEK293 cells transfected with control plasmid or p65/p52 with or without treatment with Wogonin (100 μm). * (p < 0.05), ** (p < 0.001), *** (p < 0.001) indicate statistically significant differences (ANOVA).
FIGURE 8.
Influence of NF-κB on the migration of HEK293 cells. Arithmetic means ± S.E. (n =) of the number of migrated HEK293 cells transfected with empty vector (control) or with NF-κB subunits p65/p50 or p65/p52 are shown. ** (p < 0.01) indicate significant differences (ANOVA).
DISCUSSION
This study uncovers a novel element in the regulation of Orai1 and STIM1, the components that constitute the CRAC and contribute to the SOCE. We demonstrate that transfection of human embryonic kidney (HEK293) cells with constitutively active S422DSGK1, but not with inactive K127NSGK1, enhances the STIM1 and Orai1 transcript levels. SGK1 has previously been shown to activate the transcription factor NF-κB (20–22). Accordingly, treatment of S422DSGK1-transfected HEK293 cells with the NF-κB inhibitor Wogonin strongly decreased STIM1 or Orai1 transcript levels. Transfection with S422DSGK1 but not with K127NSGK1 or with S422DSGK1 in the presence of Wogonin was further followed by nuclear translocation of p65 in HEK293 cells. STIM1 and Orai1 transcript levels were significantly lower in BMMCs from SGK1 knock-out (sgk1−/−) mice when compared with BMMCs from their corresponding wild type (sgk1+/+) littermates. Along those lines treatment with the stem cell factor SCF, a strong activator of the PI3K pathway (31), enhanced transcript levels of STIM1 and Orai1 in BMMCs from sgk1+/+ but not from sgk1−/− mice. Consequently, active SGK1 is required for up-regulation of STIM1 and Orai1 transcription by growth factor stimulation of mast cells. Simultaneous treatment of sgk1+/+ BMMCs with SCF and the NF-κB inhibitor Wogonin abolished the stimulating effect of SCF on STIM1 and Orai1 transcription. In addition, transfection of sgk1−/− and sgk1+/+ mast cells with a constitutively active p65 mutant (S276Dp65) strongly increased transcript levels of STIM1 and Orai1.
According to this study the SGK1-dependent up-regulation of Orai1 and STIM1 transcription is mimicked in HEK293 cells by overexpressing the NF-κB subunits p65/p50 or p65/p52 and reversed by silencing of the NF-κB subunits p65, p50, and p52 and by treatment with the NF-κB inhibitor Wogonin. Subsequently, overexpression of p65/p50 or p65/p52 also enhanced, whereas silencing of p65, p50, or p52 reduced, STIM1 and Orai1 protein abundance. SOCE was similarly up-regulated by overexpression of p65/p50 and p65/p52 and decreased by Wogonin treatment.
Application of the luciferase reporter assay allowed identifying putative binding regions of NF-κB in the region upstream of STIM1 and Orai1. Two promoter regions, S1 and O2, were activated by p65 transfection. To distinguish between direct and indirect modes of binding, ChIP was performed to determine if any of these putative κB-like binding sites exhibit bona fide p65 binding. Amplification of pulled down DNA with primers flanking κB-like sites revealed binding of p65 to DNA for regions S1 and O2. Region O1 showed no binding of p65. Thus, it appears safe to conclude that NF-κB participates in the SGK1-dependent genomic regulation of STIM1 and Orai1.
NF-κB-dependent STIM1 and Orai1 expression may be relevant for the regulation of migration (32), which can be stimulated by expression of Orai1, STIM1, and constitutively active SGK1 but not by coexpression of inactive SGK1 (14). Migration is stronger in HEK293 cells transfected with p65/p50 or p65/p52 than in nontransfected control cells. NF-κB-dependent STIM1 and Orai1 expression may be also relevant for mast cell degranulation. As shown earlier (33), antigen-induced Ca2+ entry, activation of Ca2+-sensitive K+ channels, and degranulation were significantly decreased in BMMCs from sgk1−/− mice. The blunted Ca2+ entry into mast cells was paralleled by a decreased anaphylactic reaction of sgk1−/− mice (33). Furthermore, mast cells lacking STIM1 had impaired Ca2+ influx after stimulation with the immunoglobulin E receptor FcϵRI as well as decreased degranulation, cytokine production, and anaphylactic reaction (34). In addition, they showed decreased activation of NF-κB after FcϵRI-mediated mast cell stimulation (34). Similarly, mast cells derived from Orai1-deficient mice are defective in degranulation, cytokine secretion, and IgE-mediated passive cutaneous anaphylaxis (35). It has been shown that Ca2+ entry mediated by ICRAC stimulates several transcription factors including NF-κB (36). According to the present observations, the NF-κB activation after Ca2+ entry enhances Orai1 and STIM1 expression and is thus part of a positive feedback further augmenting Ca2+ influx.
STIM1 and Orai1 are expressed in a wide variety of tissues (7, 37–39) and serve a multitude of functions. As SGK1 is stimulated by growth factors (40, 41), the kinase links growth factor receptor-mediated signaling with the store-operated Ca2+ entry. Orai1 (42–45) and SGK1 (46, 47) have been shown to be critically important in the regulation of cell proliferation. SOCE has further been shown to participate in cytokine production (48), migration of tumor (49), and mast cells (50). Also diseases like muscle dystrophy (51) and non-progressive myopathy (52) are associated with SOCE malfunction. NF-κB plays a pivotal role in inflammation (53), immune cell differentiation and maturation (53), secondary lymphoid organogenesis (53), fibrosis (20, 22), autophagy (54), antioxidative defense (55), muscle dystrophy (56, 57), cell survival (58), tumor growth (59, 60), cancer cachexia (61), insulin resistance (62), arteriosclerosis (63), and bone formation (64, 65). It is tempting to speculate that overlapping (patho)physiological significance of SGK1, STIM1/Orai1, and NF-κB is at least partially due to NF-κB-sensitive regulation of STIM1/Orai1 expression.
ICRAC activation is further controlled by Lyn and Syk kinases (66), which, however, other than SGK1, are not effective through regulation of STIM1 or Orai1 expression. In contrast to Lyn and Syk kinases, SGK1 plays a dual role in Orai1 regulation, i.e. inhibition of ubiquitination and stimulation of expression. Both effects increase Orai1 protein abundance. Unlike Lyn and Syk kinases, those effects of SGK1 are slow, rendering the respective cell more sensitive to immediate regulators of Orai1. In conclusion, NF-κB is a powerful regulator of STIM1 and Orai1 expression and thus influences SOCE-dependent functions, including migration.
Acknowledgments
We thank Prof. Dr. Florian Greten (Technische Universität München) for providing the plasmid encoding the NF-κB subunit p65. We acknowledge preparation of the manuscript by S. Rübe and L. Subasic.
This work was supported by the Deutsche Forschungsgemeinschaft (GK 1302 and SFB 773).
- CRAC
- Ca2+ release-activated channel
- SOCE
- store-operated Ca2+ entry
- BMMC
- bone marrow-derived mast cell(s)
- SCF
- stem cell factor
- ANOVA
- analysis of variance.
REFERENCES
- 1. Burgoyne R. D. (2007) Neuronal calcium sensor proteins. Generating diversity in neuronal Ca2+ signaling. Nat. Rev. Neurosci. 8, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Orrenius S., Zhivotovsky B., Nicotera P. (2003) Regulation of cell death. The calcium-apoptosis link. Nat. Rev. Mol. Cell. Biol. 4, 552–565 [DOI] [PubMed] [Google Scholar]
- 3. Roderick H. L., Cook S. J. (2008) Ca2+ signaling checkpoints in cancer. Remodeling Ca2+ for cancer cell proliferation and survival. Nat. Rev. Cancer 8, 361–375 [DOI] [PubMed] [Google Scholar]
- 4. Salter R. D., Watkins S. C. (2009) Dendritic cell altered states. What role for calcium? Immunol. Rev. 231, 278–288 [DOI] [PubMed] [Google Scholar]
- 5. Parekh A. B., Penner R. (1997) Store depletion and calcium influx. Physiol. Rev. 77, 901–930 [DOI] [PubMed] [Google Scholar]
- 6. Putney J. W., Jr. (2005) Capacitative calcium entry. Sensing the calcium stores. J. Cell Biol. 169, 381–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cahalan M. D., Zhang S. L., Yeromin A. V., Ohlsen K., Roos J., Stauderman K. A. (2007) Molecular basis of the CRAC channel. Cell Calcium 42, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., Hogan P. G. (2006) Orai1 is an essential pore subunit of the CRAC channel. Nature 443, 230–233 [DOI] [PubMed] [Google Scholar]
- 9. Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang S. L., Kozak J. A., Jiang W., Yeromin A. V., Chen J., Yu Y., Penna A., Shen W., Chi V., Cahalan M. D. (2008) Store-dependent and -independent modes regulating Ca2+ release-activated Ca2+ channel activity of human Orai1 and Orai3. J. Biol. Chem. 283, 17662–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fahrner M., Muik M., Derler I., Schindl R., Fritsch R., Frischauf I., Romanin C. (2009) Mechanistic view on domains mediating STIM1-Orai coupling. Immunol. Rev. 231, 99–112 [DOI] [PubMed] [Google Scholar]
- 12. Peinelt C., Vig M., Koomoa D. L., Beck A., Nadler M. J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J. P. (2006) Amplification of CRAC current by STIM1 and CRACM1 (Orai1). Nat. Cell Biol. 8, 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Penna A., Demuro A., Yeromin A. V., Zhang S. L., Safrina O., Parker I., Cahalan M. D. (2008) The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature 456, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eylenstein A., Gehring E. M., Heise N., Shumilina E., Schmidt S., Szteyn K., Münzer P., Nurbaeva M. K., Eichenmüller M., Tyan L., Regel I., Föller M., Kuhl D., Soboloff J., Penner R., Lang F. (2011) Stimulation of Ca2+-channel Orai1/STIM1 by serum- and glucocorticoid-inducible kinase 1 (SGK1). FASEB J. 25, 2012–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leong M. L., Maiyar A. C., Kim B., O'Keeffe B. A., Firestone G. L. (2003) Expression of the serum- and glucocorticoid-inducible protein kinase, Sgk, is a cell survival response to multiple types of environmental stress stimuli in mammary epithelial cells. J. Biol. Chem. 278, 5871–5882 [DOI] [PubMed] [Google Scholar]
- 16. Lang F., Böhmer C., Palmada M., Seebohm G., Strutz-Seebohm N., Vallon V. (2006) (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol. Rev. 86, 1151–1178 [DOI] [PubMed] [Google Scholar]
- 17. Snyder P. M. (2009) Sci. Signal. 2, e41. [DOI] [PubMed] [Google Scholar]
- 18. Liang X., Butterworth M. B., Peters K. W., Walker W. H., Frizzell R. A. (2008) An obligatory heterodimer of 14-3-3β and 14-3-3ϵ is required for aldosterone regulation of the epithelial sodium channel. J. Biol. Chem. 283, 27418–27425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keil J. M., Shen Z., Briggs S. P., Patrick G. N. (2010) Regulation of STIM1 and SOCE by the ubiquitin-proteasome system (UPS). PLoS One 5, e13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. BelAiba R. S., Djordjevic T., Bonello S., Artunc F., Lang F., Hess J., Görlach A. (2006) The serum- and glucocorticoid-inducible kinase Sgk-1 is involved in pulmonary vascular remodeling. Role in redox-sensitive regulation of tissue factor by thrombin. Circ. Res. 98, 828–836 [DOI] [PubMed] [Google Scholar]
- 21. Tai D. J., Su C. C., Ma Y. L., Lee E. H. (2009) SGK1 phosphorylation of IκB kinase α and p300 up-regulates NF-κB activity and increases N-methyl-d-aspartate receptor NR2A and NR2B expression. J. Biol. Chem. 284, 4073–4089 [DOI] [PubMed] [Google Scholar]
- 22. 22 Vallon V., Wyatt A. W., Klingel K., Huang D. Y., Hussain A., Berchtold S., Friedrich B., Grahammer F., Belaiba R. S., Görlach A., Wulff P., Daut J., Dalton N. D., Ross J., Jr., Flögel U., Schrader J., Osswald H., Kandolf R., Kuhl D., Lang F. (2006) SGK1-dependent cardiac CTGF formation and fibrosis following DOCA treatment. J. Mol. Med. 84, 396–404 [DOI] [PubMed] [Google Scholar]
- 23. Lang F., Klingel K., Wagner C. A., Stegen C., Warntges S., Friedrich B., Lanzendorfer M., Melzig J., Moschen I., Steuer S., Waldegger S., Sauter M., Paulmichl M., Gerke V., Risler T., Gamba G., Capasso G., Kandolf R., Hebert S. C., Massry S. G., Broër S. (2000) Deranged transcriptional regulation of cell volume-sensitive kinase hSGK in diabetic nephropathy. Proc. Natl. Acad. Sci. U.S.A. 97, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanjabi S., Williams K. J., Saccani S., Zhou L., Hoffmann A., Ghosh G., Gerondakis S., Natoli G., Smale S. T. (2005) A c-Rel subdomain responsible for enhanced DNA binding affinity and selective gene activation. Genes Dev. 19, 2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fas S. C., Baumann S., Zhu J. Y., Giaisi M., Treiber M. K., Mahlknecht U., Krammer P. H., Li-Weber M. (2006) Wogonin sensitizes resistant malignant cells to TNFalpha- and TRAIL-induced apoptosis. Blood 108, 3700–3706 [DOI] [PubMed] [Google Scholar]
- 26. Lee S. O., Jeong Y. J., Yu M. H., Lee J. W., Hwangbo M. H., Kim C. H., Lee I. S. (2006) Wogonin suppresses TNFα-induced MMP-9 expression by blocking the NF-κB activation via MAPK signaling pathways in human aortic smooth muscle cells. Biochem. Biophys. Res. Commun. 351, 118–125 [DOI] [PubMed] [Google Scholar]
- 27. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wulff P., Vallon V., Huang D. Y., Völkl H., Yu F., Richter K., Jansen M., Schlünz M., Klingel K., Loffing J., Kauselmann G., Bösl M. R., Lang F., Kuhl D. (2002) Impaired renal Na+ retention in the sgk1-knockout mouse. J. Clin. Invest. 110, 1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shumilina E., Lam R. S., Wölbing F., Matzner N., Zemtsova I. M., Sobiesiak M., Mahmud H., Sausbier U., Biedermann T., Ruth P., Sausbier M., Lang F. (2008) Blunted IgE-mediated activation of mast cells in mice lacking the Ca2+-activated K+ channel KCa3.1. J. Immunol. 180, 8040–8047 [DOI] [PubMed] [Google Scholar]
- 30. Bird G. S., DeHaven W. I., Smyth J. T., Putney J. W., Jr. (2008) Methods for studying store-operated calcium entry. Methods 46, 204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ali K., Bilancio A., Thomas M., Pearce W., Gilfillan A. M., Tkaczyk C., Kuehn N., Gray A., Giddings J., Peskett E., Fox R., Bruce I., Walker C., Sawyer C., Okkenhaug K., Finan P., Vanhaesebroeck B. (2004) Essential role for the p110δ phosphoinositide 3-kinase in the allergic response. Nature 431, 1007–1011 [DOI] [PubMed] [Google Scholar]
- 32. Becchetti A., Arcangeli A. (2010) Integrins and ion channels in cell migration. Implications for neuronal development, wound healing, and metastatic spread. Adv. Exp. Med. Biol. 674, 107–123 [DOI] [PubMed] [Google Scholar]
- 33. Sobiesiak M., Shumilina E., Lam R. S., Wölbing F., Matzner N., Kaesler S., Zemtsova I. M., Lupescu A., Zahir N., Kuhl D., Schaller M., Biedermann T., Lang F. (2009) Impaired mast cell activation in gene-targeted mice lacking the serum- and glucocorticoid-inducible kinase SGK1. J. Immunol. 183, 4395–4402 [DOI] [PubMed] [Google Scholar]
- 34. Baba Y., Nishida K., Fujii Y., Hirano T., Hikida M., Kurosaki T. (2008) Essential function for the calcium sensor STIM1 in mast cell activation and anaphylactic responses. Nat. Immunol. 9, 81–88 [DOI] [PubMed] [Google Scholar]
- 35. Vig M., DeHaven W. I., Bird G. S., Billingsley J. M., Wang H., Rao P. E., Hutchings A. B., Jouvin M. H., Putney J. W., Kinet J. P. (2008) Defective mast cell effector functions in mice lacking the CRACM1 pore subunit of store-operated calcium release-activated calcium channels. Nat. Immunol. 9, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dolmetsch R. E., Xu K., Lewis R. S. (1998) Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392, 933–936 [DOI] [PubMed] [Google Scholar]
- 37. Feske S. (2007) Calcium signaling in lymphocyte activation and disease. Nat. Rev. Immunol. 7, 690–702 [DOI] [PubMed] [Google Scholar]
- 38. Schaff U. Y., Dixit N., Procyk E., Yamayoshi I., Tse T., Simon S. I. (2010) Orai1 regulates intracellular calcium, arrest, and shape polarization during neutrophil recruitment in shear flow. Blood 115, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shin D. M., Muallem S. (2008) Skeletal muscle dressed in SOCs. Nat. Cell Biol. 10, 639–641 [DOI] [PubMed] [Google Scholar]
- 40. Rudd C. E., Taylor A., Schneider H. (2009) CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol. Rev. 229, 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Segal R. A. (2003) Selectivity in neurotrophin signaling. Theme and variations. Annu. Rev. Neurosci. 26, 299–330 [DOI] [PubMed] [Google Scholar]
- 42. Yu F., Sun L., Machaca K. (2010) Constitutive recycling of the store-operated Ca2+ channel Orai1 and its internalization during meiosis. J. Cell Biol. 191, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baryshnikov S. G., Pulina M. V., Zulian A., Linde C. I., Golovina V. A. (2009) Orai1, a critical component of store-operated Ca2+ entry, is functionally associated with Na+/Ca2+ exchanger and plasma membrane Ca2+ pump in proliferating human arterial myocytes. Am. J. Physiol. Cell Physiol. 297, C1103–C1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Faouzi M., Hague F., Potier M., Ahidouch A., Sevestre H., Ouadid-Ahidouch H. (2011) Down-regulation of Orai3 arrests cell-cycle progression and induces apoptosis in breast cancer cells but not in normal breast epithelial cells. J. Cell. Physiol. 226, 542–551 [DOI] [PubMed] [Google Scholar]
- 45. Motiani R. K., Abdullaev I. F., Trebak M. (2010) A novel native store-operated calcium channel encoded by Orai3. Selective requirement of Orai3 versus Orai1 in estrogen receptor-positive versus estrogen receptor-negative breast cancer cells. J. Biol. Chem. 285, 19173–19183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lang F., Perrotti N., Stournaras C. (2010) Colorectal carcinoma cells. Regulation of survival and growth by SGK1. Int. J. Biochem. Cell Biol. 42, 1571–1575 [DOI] [PubMed] [Google Scholar]
- 47. Wang K., Gu S., Nasir O., Föller M., Ackermann T. F., Klingel K., Kandolf R., Kuhl D., Stournaras C., Lang F. (2010) SGK1-dependent intestinal tumor growth in APC-deficient mice. Cell Physiol. Biochem. 25, 271–278 [DOI] [PubMed] [Google Scholar]
- 48. Johnstone L. S., Graham S. J., Dziadek M. A. (2010) STIM proteins. Integrators of signaling pathways in development, differentiation, and disease. J. Cell. Mol. Med. 14, 1890–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang S., Zhang J. J., Huang X. Y. (2009) Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 15, 124–134 [DOI] [PubMed] [Google Scholar]
- 50. Jung I. D., Lee H. S., Lee H. Y., Choi O. H. (2009) FcϵRI-mediated mast cell migration. Signaling pathways and dependence on cytosolic free Ca2+ concentration. Cell. Signal. 21, 1698–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Edwards J. N., Friedrich O., Cully T. R., von Wegner F., Murphy R. M., Launikonis B. S. (2010) Upregulation of store-operated Ca2+ entry in dystrophic mdx mouse muscle. Am. J. Physiol. Cell Physiol. 299, C42–C50 [DOI] [PubMed] [Google Scholar]
- 52. Picard C., McCarl C. A., Papolos A., Khalil S., Lüthy K., Hivroz C., LeDeist F., Rieux-Laucat F., Rechavi G., Rao A., Fischer A., Feske S. (2009) STIM1 mutation associated with a syndrome of immunodeficiency and autoimmunity. N. Engl. J. Med. 360, 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shih V. F., Tsui R., Caldwell A., Hoffmann A. (2011) A single NF-κB system for both canonical and non-canonical signaling. Cell Res. 21, 86–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Criollo A., Senovilla L., Authier H., Maiuri M. C., Morselli E., Vitale I., Kepp O., Tasdemir E., Galluzzi L., Shen S., Tailler M., Delahaye N., Tesniere A., De Stefano D., Younes A. B., Harper F., Pierron G., Lavandero S., Zitvogel L., Israel A., Baud V., Kroemer G. (2010) IKK connects autophagy to major stress pathways. Autophagy 6, 189–191 [DOI] [PubMed] [Google Scholar]
- 55. Lu S. C. (2009) Regulation of glutathione synthesis. Mol. Aspects Med. 30, 42–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Monici M. C., Aguennouz M., Mazzeo A., Messina C., Vita G. (2003) Activation of nuclear factor-κB in inflammatory myopathies and Duchenne muscular dystrophy. Neurology 60, 993–997 [DOI] [PubMed] [Google Scholar]
- 57. Porter J. D., Merriam A. P., Leahy P., Gong B., Khanna S. (2003) Dissection of temporal gene expression signatures of affected and spared muscle groups in dystrophin-deficient (mdx) mice. Hum. Mol. Genet. 12, 1813–1821 [DOI] [PubMed] [Google Scholar]
- 58. Mandrup-Poulsen T., Pickersgill L., Donath M. Y. (2010) Blockade of interleukin 1 in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 6, 158–166 [DOI] [PubMed] [Google Scholar]
- 59. Kasinski A. L., Slack F. J. (2010) Potential microRNA therapies targeting Ras, NF-κB, and p53 signaling. Curr. Opin. Mol. Ther. 12, 147–157 [PubMed] [Google Scholar]
- 60. Shen H. M., Tergaonkar V. (2009) NF-κB signaling in carcinogenesis and as a potential molecular target for cancer therapy. Apoptosis 14, 348–363 [DOI] [PubMed] [Google Scholar]
- 61. Tisdale M. J. (2009) Mechanisms of cancer cachexia. Physiol. Rev. 89, 381–410 [DOI] [PubMed] [Google Scholar]
- 62. Ruan H., Pownall H. J. (2009) The adipocyte IKK/NF-κB pathway. A therapeutic target for insulin resistance. Curr. Opin. Investig. Drugs 10, 346–352 [PubMed] [Google Scholar]
- 63. Jandeleit-Dahm K., Cooper M. E. (2008) The role of AGEs in cardiovascular disease. Curr. Pharm. Des. 14, 979–986 [DOI] [PubMed] [Google Scholar]
- 64. O'Brien C. A. (2010) Control of RANKL gene expression. Bone 46, 911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reid I. R. (2008) Anti-resorptive therapies for osteoporosis. Semin. Cell Dev. Biol. 19, 473–478 [DOI] [PubMed] [Google Scholar]
- 66. Chung S. C., Limnander A., Kurosaki T., Weiss A., Korenbrot J. I. (2007) Coupling Ca2+ store release to ICRAC channel activation in B lymphocytes requires the activity of Lyn and Syk kinases. J. Cell Biol. 177, 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]