Background: Notch EGF repeats are glycosylated with xylose containing O-glycans.
Results: We have identified a human gene encoding an enzyme transferring the second xylose to generate the Xyl-Xyl-Glc trisaccharide on Notch EGF repeats.
Conclusion: Genes encoding all glycosyltransferase activities involved in the O-glucose-linked modification are now known.
Significance: Identification of the responsible genes allows elucidation of the biological role of Notch xylosylation.
Keywords: Endoplasmic Reticulum (ER), Glycobiology, Glycosyltransferases, Notch, Post-translational Modification, O-Glycosylation, Xylosyltransferase
Abstract
The extracellular domain of Notch contains epidermal growth factor (EGF) repeats that are extensively modified with different O-linked glycans. O-Fucosylation is essential for receptor function, and elongation with N-acetylglucosamine, catalyzed by members of the Fringe family, modulates Notch activity. Only recently, genes encoding enzymes involved in the O-glucosylation pathway have been cloned. In the Drosophila mutant rumi, characterized by a mutation in the protein O-glucosyltransferase, Notch signaling is impaired in a temperature-dependent manner, and a mouse knock-out leads to embryonic lethality. We have previously identified two human genes, GXYLT1 and GXYLT2, encoding glucoside xylosyltransferases responsible for the transfer of xylose to O-linked glucose. The identity of the enzyme further elongating the glycan to generate the final trisaccharide xylose-xylose-glucose, however, remained unknown. Here, we describe that the human gene C3ORF21 encodes a UDP-xylose:α-xyloside α1,3-xylosyltransferase, acting on xylose-α1,3-glucoseβ1-containing acceptor structures. We have, therefore, renamed it XXYLT1 (xyloside xylosyltransferase 1). XXYLT1 cannot act on a synthetic acceptor containing an α-linked xylose alone, but requires the presence of the underlying glucose. Activity on Notch EGF repeats was proven by in vitro xylosylation of a mouse Notch1 fragment recombinantly produced in Sf9 insect cells, a bacterially expressed EGF repeat from mouse Notch2 modified in vitro by Rumi and Gxylt2 and in vivo by co-expression of the enzyme with the Notch1 fragment. The enzyme was shown to be a typical type II membrane-bound glycosyltransferase localized in the endoplasmic reticulum.
Introduction
Notch signaling is based on the interaction between transmembrane ligands of the Delta and Jagged/Serrate family and the extracellular domain of Notch receptors. This interaction enables two subsequent proteolytic cleavages that release the intracellular domain of the Notch receptor. After relocation to the nucleus, it binds to nuclear factors, thereby promoting target gene expression. The extracellular domains of Notch receptors and their ligands are composed of tandem epidermal growth factor-like (EGF) repeats (reviewed in Ref. 1) that are modified by unusual O-linked carbohydrates. The EGF repeats harbor consensus sequences containing serine or threonine residues to which O-linked fucose (Fuc), glucose (Glc), or N-acetylglucosamine (GlcNAc) are transferred (2, 3). Modification of O-linked GlcNAc has not been identified yet (3). In contrast, both fucose and glucose are further elongated to form the final tetrasaccharide Siaα2,3/6Galβ1,4GlcNAcβ1,3Fucα1-O-Ser/Thr (2) and trisaccharide Xylα1,3Xylα1,3Glcβ1-O-Ser (2, 4–6), respectively. These O-glycans are found on a variety of other proteins including coagulation factor VII and IX and thrombospondin (7, 8), but are functionally most renowned as modulators of Notch signaling (9–11).
Mice and flies lacking the protein O-fucosyltransferase-1 (Pofut1)2 exhibit Notch loss-of-function phenotypes (12, 13). Although initial fucosylation seems to be a prerequisite for Notch receptor function, transfer of GlcNAc catalyzed by enzymes of the Fringe family results in altered receptor-ligand interactions. Signaling induced by the Notch ligand Delta is increased, whereas signaling induced by Jagged/Serrate is decreased (14–18) as a result of Fringe activity.
In contrast, the importance of the trisaccharide Xyl-Xyl-Glc-O-Ser is less well understood. The initiating protein O-glucosyltransferase (Poglut) has been identified only recently through the Drosophila mutant rumi (19). Rumi-deficient flies show a temperature-sensitive phenotype, which is most severe and equivalent to the complete loss of Notch signaling in flies grown at 28–30 °C. Experiments performed by Acar et al. (19) provide evidence that the proteolytic cleavage of the Notch receptor is impaired in rumi flies but that interaction with the ligand Delta is not influenced. Thus, O-glucosylation appears to play a role for conformational stabilization of Notch at high temperatures to enable the proteolytic cleavage necessary for signal transmission (9, 11, 19, 20). A mammalian ortholog of Rumi has been demonstrated to encode an active Poglut and results in an early lethal phenotype when knocked out in mice (21). In contrast to phenotypes of other global regulators of Notch (13, 22, 23), animals lacking Rumi showed more severe and complex phenotypes, suggesting that other essential proteins are targets for O-glucosylation (21). Very recently, Rumi was shown to function as a protein O-xylosyltransferase (Poxylt, utilizing UDP-Xyl) as well as a Poglut, and it was shown that the O-xylose can be extended to a trisaccharide (Xyl-Xyl-Xyl-O-Ser) on mouse Notch2 (24). Enzymatic activity of the two α1,3-xylosyltransferases involved in extension of O-glucose had been detected in bovine liver cells (25–27), but the identity of the encoding genes long remained unknown.
Recently, we have shown (28) that two members of the human glycosyltransferase 8 family (GT8) (29), GXYLT1 and GXYLT2 (glucoside-xylosyltransferase 1/2), are able to transfer the first α1,3-linked xylose to O-glucosylated mammalian Notch EGF repeats. The enzymes exhibit about 50% identity at amino acid sequence level with differences most apparent in the stem region, but no differences in their acceptor specificity could be observed so far. GXYLT1 and GXYLT2 are, however, not able to elongate substrates containing the disaccharide Xyl-Glc- (28), indicating the requirement for an additional xylosyltransferase. Here, we describe the identification of another human member of the GT8 family, only distantly related to the previously analyzed GT8 members, encoding a xyloside-xylosyltransferase (XXYLT), which further elongates the Xyl-Glc-O-Ser disaccharide on Notch EGF repeats.
EXPERIMENTAL PROCEDURES
Constructs for Baculoviral Expression in Sf9 Cells
Protein A fusion constructs of GXYLT1 and GLT8D1 for recombinant expression via the pFast Bac system (Invitrogen) were reported previously (28). To generate the equivalent plasmid of xyloside xylosyltransferase 1 (XXYLT1), the putative luminal C-terminal domain, starting from Ser-43, was amplified by PCR from human prostate Marathon-Ready cDNA (Clontech) using the primers 5′-atctgaattcaggccgggagaccttctc-3′ and 5′-atctgaattcctagtcctccgggatggga-3′. After EcoRI digestion, the sequence was cloned into the pFast Bac1 vector (Invitrogen), containing the honeybee melittin secretion sequence followed by the protein A coding sequence from pProtA (ProtA-XXYLT1). C-terminal Myc/His-tagged mouse Notch EGF1–5, encoding the first five EGF domains of mouse Notch1 cloned into pFast Bac1, was described previously (28, 30).
Constructs for Enzyme Expression in Mammalian Cells
The cDNAs encoding the luminal domains of mouse Gxylt2, starting from Arg-26, or Xxylt1, starting from Ser-43, were subcloned into a pSecTag2 vector (Invitrogen) so that the recombinant proteins were expressed with a C-terminal Myc/His tag in mammalian cells and secreted into the culture media.
Constructs for Immunofluorescence and Cleavage Analysis
The full-length open reading frame of XXYLT1 (National Center for Biotechnology Information (NCBI) reference sequence NM_152531) was cloned into the pcDNA3 vector (Invitrogen) and modified with protein tags to generate the single-tagged XXYLT1-HA (XXYLT1-SRYPYDVPDYASL) and FLAG-XXYLT1 (MDYKDDDDKEF-XXYLT1) and the double-tagged FLAG-XXYLT1-HA (MDYKDDDDKEF-XXYLT1-SRYPYDVPDYASL) plasmids. Similar constructs (B4GALT1-HA and FLAG-B4GALT1-HA) were generated for the control gene, the human β1,4-galactosyltransferase 1 (B4GALT1; NCBI reference sequence NM_001497). To generate an appropriate ER marker, the Golgi-located pAcGFP1-Golgi Vector (Clontech), encoding a GFP fusion protein located in the Golgi apparatus, was modified by insertion of a C-terminal KDEL signal sequence. After PCR on the original plasmid, the modified insert was recloned via BamHI and XbaI restriction sites into the vector. The new construct GFP1-KDEL was tested by immunofluorescence studies in CHO cells. DNA sequences of the constructs were confirmed by sequencing. All the primers are available upon request.
Enzyme Expression and Purification
Protein A fusion constructs of soluble secreted human GLT8D1, GXYLT1, and XXYLT1 were expressed in Sf9 cells by baculovirus infection (Bac-to-Bac®; Invitrogen) and purified by IgG-Sepharose-6 Fast Flow beads (GE Healthcare) as described (28). Soluble secreted mouse Gxylt2 and Xxylt1 were expressed in HEK293T cells and purified from the culture media by nickel-nitrilotriacetic acid affinity chromatography as described previously (24). Protein concentration was determined by Coomassie Blue staining using BSA or protein A as standard.
In Vitro Xylosyltransferase Activity Assays
Assays were performed in a total volume of 50 μl in 100 mm MOPS, pH 7.5, 10 mm MnCl2, 10 mm ATP as described before (28). IgG bead-coupled protein A fused enzymes were incubated for 1 h at 37 °C in the presence of radiolabeled donor sugars UDP-[6-3H]Gal, UDP-[1-3H]Glc (GE Healthcare), or UDP-[U-14C]Xyl (PerkinElmer Life Sciences) at 5 μm with specific activity 4 kBq/nmol for 3H-sugars and 0.75 kBq/nmol for [14C]Xyl, obtained by dilution with cold nucleotide sugars (Sigma and CarboSource Services) and a number of synthetic acceptors (Xyl-Xyl-Glc-R; Xyl-Glc-R, Glc-R (31) or para-nitrophenol (pNP)-linked carbohydrates) at a final concentration of 100 μm. To determine the amount of transferred radiolabeled sugars, donor and acceptor were separated via C18 columns (Sep-PakR Vac 3cc; Waters Corp.), and samples were counted by liquid scintillation (LS 6500; Beckman Coulter). The acquired enzymatic activity was expressed as nmol of xylose transferred per nmol of protein per hour, calculated from a Coomassie Blue staining using a protein A standard as reference. Note that this assay was set up to show specificity but does not allow drawing conclusions about enzymatic efficiency due to the approximately equimolar amount of UDP-Xyl and enzyme in the assay.
A single 16th EGF repeat (EGF16) from mouse Notch2 was prepared and modified with Glc-O or Xyl-O by Rumi as described previously (24). In vitro xylosylation of the Glc-EGF repeat was performed using mouse Gxylt2 that was produced and purified by nickel affinity from medium of HEK293T cells. Typically, a 5-ml reaction mixture contained 50 mm HEPES, pH 7.0, 10 mm MnCl2, 5 μm Glc-EGF repeat, 200 μm UDP-Xyl, 0.5% Nonidet P-40, and ∼10 μg of Gxylt2. Samples were incubated at 37 °C overnight, and the product was purified by reversed phase high performance liquid chromatography (HPLC) as described previously (24). Substrate specificity of XXYLT1 on these differently glycosylated EGF16 acceptors was tested using radiolabeled UDP-Xyl in a reaction volume of 10 μl containing 50 mm HEPES, pH 7.0, 10 mm MnCl2, 10 μm acceptor substrate, 10 μm UDP-[14C]Xyl (5 kBq/nmol; American Radiolabeled Chemicals), 0.5% Nonidet P-40, and 10 ng of the mouse Xxylt1 protein, purified as described above for Gxylt2. After 20 min of incubation at 37 °C, the reaction was stopped by adding 900 μl of 100 mm EDTA, pH 8.0. Samples were loaded onto C18 cartridges (100 mg, Agilent). After washing with 5 ml of H2O, the EGF repeats were eluted with 1 ml of 80% methanol. Incorporation of [14C]Xyl into the EGF repeats was determined by liquid scintillation. Reactions without substrates were used as background controls.
Reverse Phase Chromatography and NMR Analysis after Enzymatic Xylosylation of Xyl-Glc-R
1 mg of synthetic acceptor Xyl-Glc-R was xylosylated in 500 μl of reaction buffer by 100 μl of bead-coupled XXYLT1 in the presence of equimolar amounts of cold UDP-Xyl under standard in vitro activity assay conditions. The sample was taken up in 100 μl of 25% aqueous acetonitrile and analyzed (20 μl) by reverse phase HPLC on a column (25 cm × 4.6 mm) with LC-8 (5 μm) sorbent with elution by 25% aqueous acetonitrile at a flow rate of 0.8 ml/min. The elution profile was compared with profiles of reference trisaccharide Xyl-Xyl-Glc-R and disaccharide Xyl-Glc-R acceptor (31).
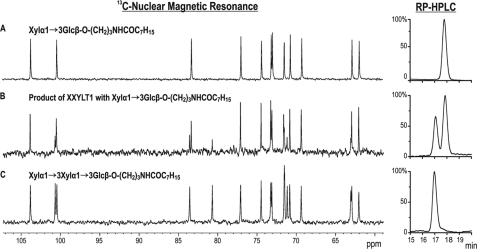
For linkage confirmation in the product of in vitro xylosylation, 1H nuclear magnetic resonance (NMR) (600 MHz, 303 K, D2O, Bruker Avance 600), natural abundance 13C NMR (125 MHz, 303 K, D2O, Bruker Avance 600), and two-dimensional heteronuclear single quantum correlation (HSQC) NMR (600 MHz, 303 K, D2O, Bruker Avance 600) of the reaction mixture were acquired and compared with spectral data for the synthetic reference compounds (see Fig. 3 and supplemental Figs. S1 and S2 and Table S1).
FIGURE 3.
Analysis of reaction product of XXYLT1 with synthetic Xyl-Glc-R acceptor. Shown are 125-MHz natural abundance 13C nuclear magnetic resonance spectra (left column) and reverse phase (RP) chromatography profiles (right column) of acceptors and reaction products. A, the pure synthetic reference disaccharide Xyl-Glc-R (Xylα1,3Glcβ-O-(CH2)3NHCOC7H15), B, the reaction product after incubation of 1 mg of Xyl-Glc-R with XXYLT1. C, the synthetic trisaccharide Xyl-Xyl-Glc-R (Xylα1,3Xylα1,3Glcβ-O-(CH2)3NHCOC7H15). Product analysis by HPLC revealed the conversion of more than 30% of Xyl-Glc-R to a product running at the position of Xyl-Xyl-Glc-R. 13C NMR spectra confirmed the identity of signals between the reaction/substrate mixture and signals obtained from the pure synthetic standard of the expected product Xyl-Xyl-Glc-R. Additionally, 1H NMR and HSQC NMR are presented in supplemental Figs. S1 and S2; chemical shifts are listed in supplemental Table S1.
Notch EGF1–5 Expression, Purification, and in Vitro Xylosylation
C-terminal Myc/His-tagged mouse Notch EGF1–5 was expressed in 300 ml of Sf9 insect cells cultured in Insect-Xpress medium (Lonza) by baculovirus infection. After 72 h, the secreted protein was purified by nickel affinity chromatography (HisTrap HP, 1 ml; GE Healthcare) as described (28). Protein fractions, reactive with anti-Myc, were pooled and concentrated/desalted to 150 μl using an Amicon Ultra-4 centrifugal devise (Millipore) and stored in 100 mm MOPS, pH 7, at −20 °C. SDS-Page followed by Coomassie Blue staining and immunoblotting with monoclonal antibody 9E10 (anti-Myc) confirmed the protein purification. Under standard activity assay conditions, 10 μl of purified Notch EGF1–5 was incubated in the presence of 100 μm cold UDP-Xyl as donor and 10 μl of bead-coupled enzyme for 4 h, separated by SDS-PAGE, and analyzed by mass spectrometry (MS).
LC-MS/MS
Protein bands of Notch EGF1–5 were excised from gel and trypsin-digested, and peptides were recovered as described (32). Reverse phase chromatography using acetonitrile as eluent was performed on a Waters nanoACQUITY UPLC device equipped with an analytical column (Waters, BEH130 C18, 100 μm × 100 mm, 1.7-μm particle size) coupled online to an ESI Q-TOF Ultima (Waters). Spectra were recorded in positive ion mode, and peptides were automatically subjected to fragmentation (tandem mass spectrometry (MS/MS)). Unglycosylated or glycosylated versions of EGF16 from mouse Notch2 were analyzed by nano-LC-MS/MS as described previously (24).
In Vivo Enzymatic Activity Assay
Notch EGF1–5 was co-expressed with XXYLT1, or as control, the inactive enzyme GLT8D1 (28) in Sf9 cells by baculovirus infection for 72 h. Purification and analysis of peptide glycosylation of the Notch fragment were performed as specified above.
Immunostaining
Localization studies were carried out in CHO cells. Cell transfection was performed on glass coverslips in 24-well plates using METAFECTENE (Biontex). After 24 h, cells were fixed using 4% PFA in PBS, permeabilized with 0.1% saponin in PBS, and stained with respective primary (rabbit anti-HA (Sigma), mouse anti-FLAG M5 (Sigma), or rabbit α-mannosidase II (gift of Dr. K. Moremen, University of Georgia, Athens, GA)) and secondary antibodies (anti-mouse IgG-Cy3 (Sigma) and anti-rabbit IgG-Alexa Fluor 488 (Invitrogen)) using standard staining conditions. As an ER marker, the GFP1-KDEL construct was co-transfected. Images were acquired with a Zeiss Axiovert 200m.
Cleavage Analysis
CHO cells were grown in 75-cm2 flasks and transfected using METAFECTENE (Biontex) with XXYLT1-HA, FLAG-XXYLT1-HA, B4GALT1-HA, or FLAG-B4GALT1-HA. After 24 h, cells were harvested and lysed by 2 mm EDTA, 1 mm MgCl2, 1% Nonidet P-40 in 50 mm Tris/HCl, pH 8.0, buffer, including protease inhibitor (Roche Applied Science). Proteins were immunoprecipitated using ∼25 μg of mouse anti-HA 12CA5 antibody coupled to 50 μl of protein A-SepharoseTM CL-4B (GE Healthcare). Beads were washed according to the manufacturer's instructions and applied to SDS-PAGE. Protein expression was analyzed by Western blotting using primary antibodies rabbit anti-HA and rabbit anti-FLAG (both from Sigma) followed by visualization with rabbit IRDye 800CW (LI-COR) using the LI-COR Odyssey imager.
RESULTS
Selection of a Potential XXYLT
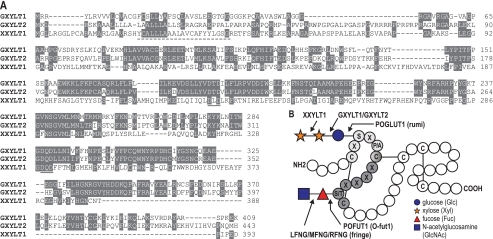
The human genes GXYLT1 and GXYLT2 transfer xylose in α1,3 to O-linked glucose (Fig. 1B). The gene responsible for the transfer of the second xylose remained unknown. Considering that both enzymatic reactions result in the addition of xylose in α1,3 linkage, the genes were anticipated to be homologous. Still, none of the four initially identified members of the GT8 family (GXYLT1, GXYLT2, GLT8D1, and GLT8D2) that were selected based on homology with UDP-Glc:glycoprotein glucosyltransferase (genes UGGT1 and UGGT2) had xylosyltransferase activity toward Xylα1,3Glcβ1 terminated acceptors (28). Using GXYLT1 in a position-specific iterated (PSI)-Blast, a fifth gene was identified, showing less than 20% overall identity at the amino acid level (Fig. 1A). This gene, named C3ORF21 (chromosome 3 open reading frame 21), possessed two conserved DXD motifs and an N-terminal signal sequence or membrane anchor. Therefore, its potential involvement in the synthesis of the Xylα1,3Xylα1,3Glcβ1-O oligosaccharide was investigated.
FIGURE 1.
Sequence alignment of Notch xylosyltransferases and overview of enzymes involved in Notch glycosylation. A, amino acid sequence alignment of XXYLT1 with GXYLT1 and GXYLT2. Sequence conservation in at least two of the three aligned enzymes is indicated in black. The predicted transmembrane domain in XXYLT1 is underlined (dashed underline), as well as the conserved DXD motifs (solid underline). B, overview of enzymes involved in O-fucosylation and O-glucosylation of specific consensus sequences on EGF repeats. Shown is one EGF repeat and the enzymes acting on EGF repeats exclusively. GlcNAc-Fuc-O can be further extended by nonspecific galactosyl- and sialyltransferases. Indicated are the official human gene name abbreviations (HUGO Gene Nomenclature Committee (HGNC)) and, in brackets, the Drosophila names or abbreviations.
Xylosyltransferase Activity with Synthetic Acceptors
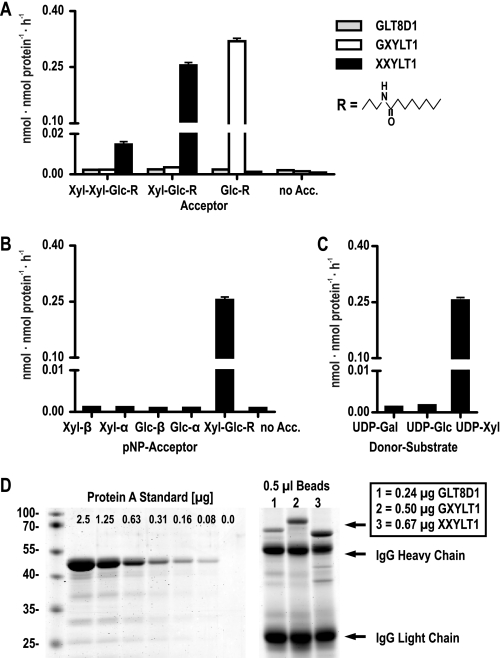
To test the enzymatic activity of the putative xylosyltransferase, renamed XXYLT1, the protein, lacking the predicted membrane-bound domain, was fused N-terminally to a protein A tag and cloned in a baculoviral vector with a signal sequence to promote secretion of the fusion protein. After expression in Sf9 insect cells, the protein was isolated from the culture medium by capture with IgG beads, binding the N-terminal protein A tag. Bead-coupled enzyme was used for in vitro glycosyltransferase activity assays with artificial synthetic compounds that mimic the natural acceptor structures (28, 31). Assays carried out with the radiolabeled donor substrate UDP-[14C]Xyl revealed xylosyltransferase activity of XXYLT1 with the Xyl-Glc-R synthetic acceptor (Fig. 2A). Marginal increased values, when compared with the negative control, were observed for the Xyl-Xyl-Glc-R acceptor, which can be interpreted as due to the presence of degradation products within the acceptor compound, rather than as resulting from generation of a product with three xylose residues. This conclusion is supported by the lack of any signal in the HPLC profile of Fig. 3B that could represent such a product. XXYLT1 showed a clear donor substrate specificity for UDP-Xyl (Fig. 2C). Interestingly, no enzymatic activity could be observed with a panel of sugars linked to pNP as acceptors, (Fig. 2B) including α-linked Xyl-pNP, suggesting that the enzyme requires the disaccharide Xylα1,3Glcβ1 as minimal acceptor structure.
FIGURE 2.
In vitro enzymatic activity of XXYLT1. A, activity of XXYLT1 when compared with GXYLT1 and GLT8D1 with artificial, synthetic acceptors mimicking the naturally occurring carbohydrate substrates on Notch EGF repeats. B, activity of XXYLT1 with different monosaccharides linked to pNP when compared with Xyl-Glc-R. C, UDP-sugar specificity of XXYLT1 using Xyl-Glc-R as acceptor. D, quantification of 0.5 μl of IgG bead-coupled enzymes by a protein A standard stained with Coomassie Blue. For each assay of A–C, 10 μl of enzyme beads was used; enzymatic activity is presented as nmol of transferred sugar per nmol of enzyme per hour, and values are the mean ± S.E. of three independent measurements.
Analysis of the Reaction Product after in Vitro Xylosylation by XXYLT1
The reaction product generated by XXYLT1 was investigated by HPLC and NMR studies to confirm the structure of the formed trisaccharide, especially of the direction and configuration of the linkage between the two xylosyl units (Fig. 3). HPLC data of Xyl-Glc-R modified by XXYLT1 in the presence of UDP-Xyl, shown in Fig. 3B, indicated that the enzyme generated a product migrating at the same position as the synthetic reference trisaccharide Xyl-Xyl-Glc-R (Fig. 3C). Moreover, acquired 13C and 1H NMR spectra of the enzymatically modified disaccharide (Fig. 3B; supplemental Fig. S1B) are identical to the spectra of the synthetic reference trisaccharide (Fig. 3C; supplemental Fig. S1C). In particular, the characteristic low field (80.3 ppm) location of C3′ signal in the 13C NMR spectrum unequivocally shows that the xylosyl residue is linked through an α1,3 linkage, whereas the α-configuration of the terminal xylose unit is indicated by the coupling constant JH1,H2 of 3.7 Hz (supplemental Fig. S1; supplemental Table S1). Finally, the precise nature of the product was also confirmed by a two-dimensional HSQC NMR spectrum (supplemental Fig. S2).
Xylosylation of Notch EGF Repeats by XXYLT1
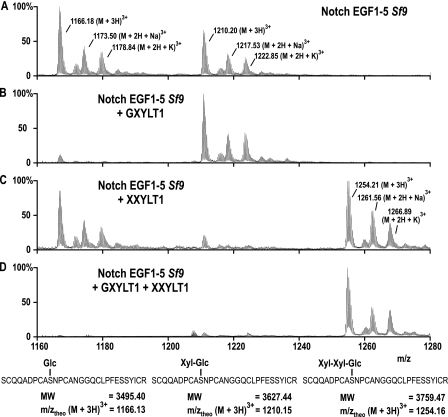
To determine whether Notch is modified by XXYLT1, an in vitro assay was carried out to test a naturally occurring EGF acceptor. A mouse Notch fragment containing five EGF repeats with consensus sequences for O-glucosylation in repeats 2 and 4 (Notch EGF1–5) was expressed in Sf9 insect cells and purified by nickel-affinity chromatography. The purified acceptor presented both Glc-O and Xyl-Glc-O modifications in about a one-to-one ratio as shown by mass spectral analysis of the glycopeptide fragment from EGF4 (m/z 1166 and 1210, Fig. 4A). Incubation of this acceptor with GXYLT1 resulted in the expected conversion of Glc-O into Xyl-Glc-O, visible by the disappearance of the peak at m/z 1166 (Fig. 4B). In contrast, incubation of Notch EGF1–5 with XXYLT1 resulted in the de novo generation of a product at m/z 1254 (Fig. 4C) and loss of the Xyl-Glc-O peak at m/z 1210. The new peak at m/z 1254 demonstrated the addition of one pentose residue to the Xyl-Glc-O-containing peptide. Modification of Notch EGF1–5 with both enzymes, GXYLT1 and XXYLT1, resulted in a complete shift toward the formation of the trisaccharide Xyl-Xyl-Glc-O on EGF repeat 4 (Fig. 4D). The same was observed for the peptide of EGF repeat 2, but overlapping signals from another peptide troubled the picture (data not shown).
FIGURE 4.
In vitro xylosyltransferase activity on Notch EGF1–5. LC-ESI-MS of mouse Notch EGF1–5, produced in Sf9 insect cells and after in vitro incubation with the indicated enzymes. A, a peptide of EGF4 showed Glc-O and Xyl-Glc-O modifications (m/z 1166 and 1210). B, after incubation with GXYLT1 in the presence of UDP-Xyl, the Glc-O signal is strongly reduced. C, XXYLT1 extended the Xyl-Glc-O disaccharide by one xylose residue, indicated by the appearing signal at m/z 1254. D, incubation of Notch EGF1–5 with both enzymes GXYLT1 and XXYLT1 almost completely shifted the carbohydrate modification to Xyl-Xyl-Glc-O. Peptide sequencing was done by MS/MS, verifying that all signals were derived from the indicated peptide (supplemental Fig. S3). MW, molecular weight.
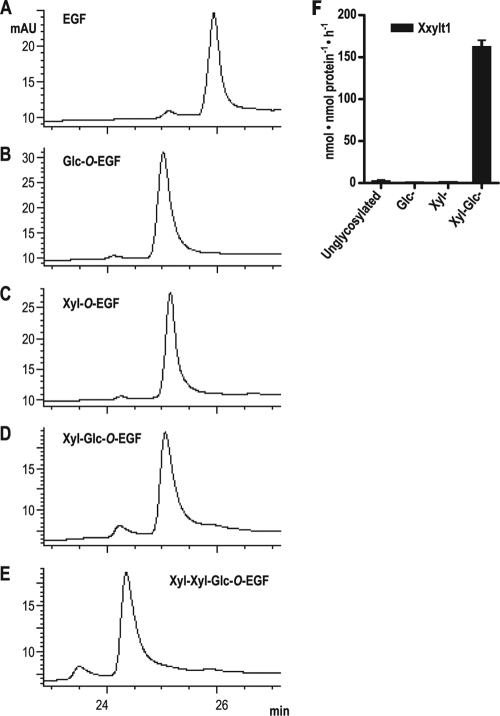
Because Rumi was recently shown to be capable of transferring either xylose or glucose to certain EGF repeats (e.g. EGF16 from mouse Notch2), and Xyl-O can be further elongated to a Xyl-Xyl-Xyl-O trisaccharide (24), we wanted to test whether XXYLT1 can add xylose to an O-xylosylated EGF repeat. Bacterially expressed EGF16 (unglycosylated) was incubated with Rumi in the presence of UDP-Glc or UDP-Xyl to generate the O-glucosylated and O-xylosylated forms, respectively. Reverse phase HPLC analysis showed that the glycosylated EGF repeats eluted slightly earlier than the unglycosylated form (Fig. 5, A–C). Glc-O-EGF16 was converted to Xyl-Glc-O-EGF16 by incubation with mouse Gxylt2 and UDP-Xyl (Fig. 5D; all structures were confirmed by mass spectrometry, supplemental Fig. S4). Each of these EGF repeats was tested as a potential acceptor substrate for mouse Xxylt1, but only Xyl-Glc-O-EGF16 was converted by the enzyme (Fig. 5, E and F), showing that Xxylt1 is not able to elongate Xyl-O-EGF16.
FIGURE 5.
Xyl-O-EGF is not modified by Xxylt. A–C, analysis of EGF16 from mouse Notch2 by reverse phase HPLC before modification (A) or after incubation with Rumi and UDP-Glc (B) or with Rumi and UDP-Xyl (C). The product from B (Glc-O-EGF) was purified and incubated with mouse Gxylt2 and UDP-xylose to generate Xyl-Glc-O-EGF. D and E, Xyl-Glc-O-EGF was incubated with UDP-xylose in the absence (D) or the presence (E) of mouse Xxylt1 and analyzed by reverse phase HPLC. The presence of the glycans on all forms of EGF16 were confirmed by mass spectrometry (supplemental Fig. S4). F, each of the purified EGF repeats generated in A–D were incubated with mouse Xxylt1 and UDP-[14C]xylose. The values indicate the mean ± S.E. of three independent measurements. mAU, milliabsorbance units.
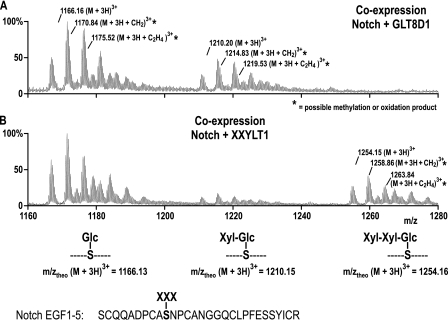
In vivo enzymatic activity of XXYLT1 was tested by a co-expression approach. The MS data shown in Fig. 4A already indicated that insect cells endogenously express a glucoside xylosyltransferase, which partially xylosylates O-glucose of mouse Notch EGF repeats. However, so far, no trisaccharide modification has been detected in Sf9 cells. Upon co-transfection of Notch EGF1–5 and XXYLT1 in Sf9 cells, we could detect the trisaccharide modification at m/z 1254 (Fig. 6B). In comparison, samples obtained from co-expression of Notch EGF1–5 and the inactive enzyme GLT8D1 only showed Glc-O and Xyl-Glc-O modifications (Fig. 6A).
FIGURE 6.
XXYLT1 shows in vivo activity on Notch EGF repeats. A, mass spectrometric analysis of Notch EGF1–5 produced in Sf9 cells that were co-infected with the enzyme GLT8D1 as a negative control showed a mixture of Glc-O and Xyl-Glc-O modifications. B, co-infection with XXYLT1 resulted in de novo generation of the complete trisaccharide at m/z 1254 at the expense of the disaccharide at m/z 1210. There was no further elongation than the trisaccharide observed. Similar to Fig. 4, peptide identity was confirmed by MS/MS (data not shown).
Intracellular Location and Cleavage Analysis of XXYLT1
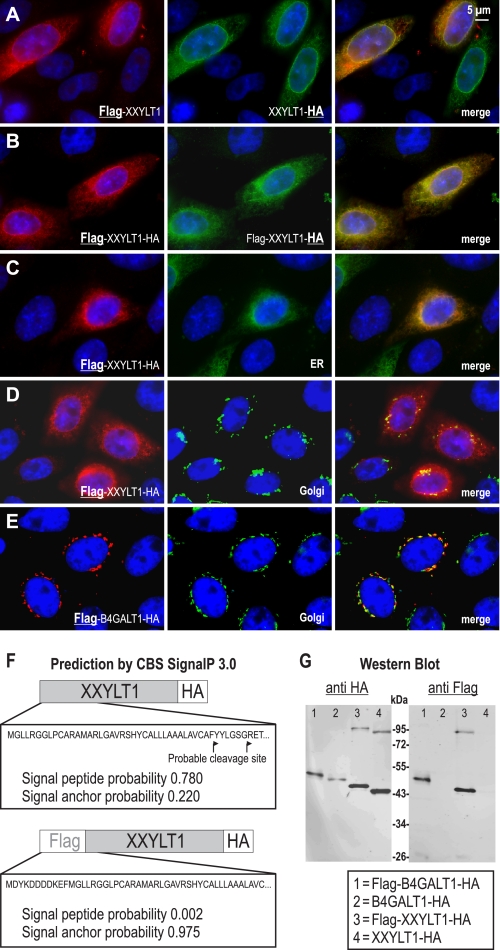
To determine the intracellular localization of the newly identified glycosyltransferase, we transiently expressed N-terminal FLAG-tagged and/or C-terminal HA-tagged XXYLT1 in CHO cells followed by immunofluorescence staining. Expression of the three constructs resulted in identical subcellular expression patterns, and signals acquired from the double-tagged FLAG-XXYLT1-HA construct clearly matched (Fig. 7, A and B). Co-localization of FLAG-XXYLT1-HA and an ER-retained GFP construct indicated that XXYLT1 is located in the ER (Fig. 7C). In contrast, no merge between the Golgi marker mannosidase II and XXYLT1 was found (Fig. 7D). The same construct of β4-galactosyltransferase (B4GALT1), which was used as a control, was always located, as expected, in the Golgi apparatus (Fig. 7E) (33).
FIGURE 7.
Intracellular localization and cleavage analysis of XXYLT1. Images A–E show immunostainings of CHO cells that were transiently transfected with tagged plasmids. A, co-localization of the single-tagged constructs FLAG-XXYLT1 and XXYLT1-HA. B, overlaying signals from the N-terminal FLAG tag and the C-terminal HA tag of the FLAG-XXYLT1-HA construct. C, co-localization of XXYLT1 visualized by the FLAG tag and a co-transfected GFP1-KDEL as ER marker. D, no merge was observed after staining of tagged XXYLT1 and the Golgi marker mannosidase II. E, as control, the established Golgi-located β1,4-galactosyltransferase (FLAG-B4GALT1-HA) was investigated in parallel. F, signal sequence prediction (SignalP 3.0) indicated a high probability for XXYLT1 to have a cleaved signal sequence, which is not predicted anymore in the FLAG-tagged construct. G, lysates of cells transiently expressing FLAG-XXYLT1-HA, the control protein FLAG-B4GALT1-HA, as well as C-terminally HA-tagged enzymes were analyzed by Western blotting using either anti-HA or anti-FLAG antibody, showing no indication of cleavage of the protein. XXYLT1 partly runs as an SDS-resistant dimer.
Both Poglut/Rumi and Pofut1 are soluble enzymes in the ER (19, 34), whereas most other glycosyltransferases are type II membrane proteins in the Golgi (35). The question, therefore, arose whether XXYLT1 is a typical type II glycosyltransferase with a signal anchor or is cleaved after a signal sequence has directed the protein into the ER. Moreover, a prediction program (SignalP 3.0) (36) predicted the protein to have a cleaved signal sequence with a total probability of 0.78 (Fig. 7F). To answer this question, the double-tagged FLAG-XXYLT1-HA and C-terminally tagged XXYLT1-HA proteins were analyzed by Western blotting. As control, B4GALT1, which is an established non cleaved type II transmembrane protein (33), was investigated in parallel. Two identical blots were generated and developed with either an anti-FLAG or an anti-HA antibody (Fig. 7G). In all cases, XXYLT1 behaved like B4GALT1. The double-tagged protein was visible as a single band, and no smaller products reacting only with the anti-HA antibody were observed. As the N-terminal fused FLAG tag might shield a possible cleavage signal at the N terminus, the XXYLT1-HA was investigated too. The absence of the FLAG tag reduced the molecular mass by ∼1.4 kDa. For both B4GALT1 and XXYLT1, a slightly shortened protein, corresponding to this difference, was now visible in the blot developed with the HA antibody. However, again, no smaller, potentially cleaved, products were observed. Protein bands running at higher molecular weight most likely represented dimers of XXYLT1. These data indicated that XXYLT1 most likely is a typical type II transmembrane protein, but localized in the ER.
DISCUSSION
This study describes the identification of an α1,3-xylosyltransferase acting on the Xylα1,3Glc-O-linked glycan of Notch EGF domains and was accordingly named XXYLT1. Like the α1,3-xylosyltransferases GXYLT1 and GXYLT2 involved in the preceding glycosylation step, XXYLT1 belongs to the glycosyltransferase family 8. However, although it catalyzes the transfer of a xylose residue in α1,3 linkage as the GXYLTs, XXYLT1 uses a different acceptor substrate and only presents a low identity of less than 20% to these enzymes. XXYLT1 has all the properties to encode the xylosyltransferase activity previously detected in HepG2 cells (26) and to be responsible for xylosylation of Notch and other EGF repeat-containing proteins. Formal proof that this is the only enzyme in the human genome with this activity, however, still has to be provided.
The glycosyltransferase family 8 comprises a broad range of glycosyltransferases, from both bacteria and eukaryotes, involved in the transfer of a variety of sugars. Mammalian members of this family have only been shown to transfer either glucose or xylose. Glucose is the donor sugar for glycogenin (37) (the autocatalytic precursor for glycogen) and for UDP-Glc:glycoprotein glucosyltransferase (38). GXYLT1, GXYLT2, and the newly identified XXYLT1 have now been shown to encode xylosyltransferases acting on O-glucosylated EGF repeats. However, the enzymatic activity of the remaining enzymes LARGE1, LARGE2, GLT8D1, and GLT8D2 is still unknown. LARGE proteins comprise two predicted catalytic domains of glycosyltransferases, one of the GT8 family and one of family 49 (39), and are known to act on dystroglycan (40).
Both GXYLT and XXYLT seem to have emerged at the same time with the appearance of metazoans. As for GXYLT1 and GXYLT2, there is no ortholog of XXYLT in the unicellular Monosiga brevicollis, which actually has a LARGE-like glycosyltransferase. However, clear orthologs of both GXYLT and XXYLT are found in most metazoans, including Drosophila melanogaster. The latter is rather surprising because we have not observed the product of XXYLT on Notch produced in Sf9 insect cells (28). Similarly, in O-linked glycans isolated from Drosophila, only the Xyl-Glc disaccharide was detected (41). The expression and activity of Drosophila XXYLT still have to be confirmed.
In contrast to GXYLT1 and GXYLT2, XXYLT1 showed no activity with xylose linked to pNP or with Xyl-O-EGF16, a structure recently demonstrated to be formed by Poglut/Rumi (24). The GXYLTs were able to use Glcβ-pNP as substrate, but XXYLT1 was only active with an acceptor containing Xylα1,3Glcβ1. This is actually a logical requirement to prevent the enzyme from using its transferred xylose in α1,3 linkage again as acceptor substrate. Indeed, no further extension beyond the trisaccharide is observed both naturally and in our in vitro assays. Whether the xylosyltransferases recognize the underlying EGF repeat is not known. Recognizing EGF repeats would be required to differentially xylosylate EGF repeats. At least in Drosophila, xylosylation appears to be EGF repeat-dependent (42), indicating that the Drosophila xylosyltransferase shows site specificity. On the other hand, mouse Notch1 overexpressed in tissue culture cells is mainly uniformly modified with the trisaccharide Xyl-Xyl-Glc (43).
Human XXYLT1 is predicted to have a cleavable signal sequence rather than an N-terminal signal anchor (Fig. 7). This is in fact the case for many glycosyltransferases that are in reality non-cleaved type II transmembrane proteins of the Golgi (35). In contrast, Pofut1 and Poglut/Rumi, the enzymes responsible for initial O-fucosylation and O-glucosylation, are soluble proteins in the ER and have a cleavable signal sequence (19, 34). Our experiments clearly showed that XXYLT1 is not cleaved and behaves like the archetypal type II membrane-bound glycosyltransferase B4GALT1 (33). On the other hand, it does not show the typical Golgi localization of B4GALT1. It is localized primarily to the ER. To avoid influencing the subcellular localization by changing the N- or C-terminal sequence, we have generated both N-terminally and C-terminally tagged constructs. These always showed identical localization as well as a construct that was double-tagged. We, thus, conclude from these experiments that XXYLT1 is a type II transmembrane protein, typical for glycosyltransferases, but is located in the ER. XXYLT1 behaves very similar to COSMC in these respects. COSMC is an ER-localized chaperone specific for the Golgi glycosyltransferase T-synthase and is itself a type II transmembrane protein with homology to the same T-synthase. It resides in the ER due to formation of a homodimer by a disulfide bond within the transmembrane domain that is essential for ER retention (44). Indeed, we observed dimer formation for XXYLT1 (Fig. 7G), and two cysteine residues, potentially responsible for intermolecular disulfide bond formation, are present within the transmembrane domain. There are, therefore, strong indications that XXYLT1 is retained in the ER by the same mechanism as COSMC. Moreover, the xylosyltransferase has an AXXXAXXXA motif within the predicted transmembrane domain. AXXXA or GXXXG motifs in other proteins have been shown to promote membrane helix interactions (45, 46).
With the previous identification of the glucosyltransferase Rumi, the two GXYLTs, and now the cloning of XXYLT1, all genes encoding the enzymes required for the formation of the Xyl-Xyl-Glc- glycotope are currently known. These important steps will now allow investigating the function of these modifications.
Supplementary Material
Acknowledgments
UDP-Xyl isolation by the CarboSource Services at the Complex Carbohydrate Research Center, University of Georgia, Athens GA, was supported in part by National Science Foundation Research Coordination Networks Grant 0090281.
This work was supported by funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) for the Cluster of Excellence REBIRTH (From Regenerative Biology to Reconstructive Therapy) and by National Institutes of Health Grant GM61126 (to R. S. H.).

This article contains supplemental Figs. S1–S4 and Table S1.
- Pofut
- protein O-fucosyltransferase
- Poglut
- protein O-glucosyltransferase
- B4GALT1
- β1,4-galactosyltransferase 1
- GALT
- galactosyltransferase
- GLT8D
- glycosyltransferase 8 family domain
- GT8
- glycosyltransferase 8 (family)
- GXYLT
- glucoside xylosyltransferase
- XXYLT
- xyloside xylosyltransferase
- ER
- endoplasmic reticulum
- HSQC
- heteronuclear single quantum coherence
- pNP
- para-nitrophenol
- COSMC
- core 1 β3-Gal-T-specific molecular chaperone.
REFERENCES
- 1. Kopan R., Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., Haltiwanger R. S. (2000) Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 275, 9604–9611 [DOI] [PubMed] [Google Scholar]
- 3. Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., Okajima T. (2008) O-linked N-acetylglucosamine is present on the extracellular domain of Notch receptors. J. Biol. Chem. 283, 35486–35495 [DOI] [PubMed] [Google Scholar]
- 4. Hase S., Kawabata S., Nishimura H., Takeya H., Sueyoshi T., Miyata T., Iwanaga S., Takao T., Shimonishi Y., Ikenaka T. (1988) A new trisaccharide sugar chain linked to a serine residue in bovine blood coagulation factors VII and IX. J. Biochem. 104, 867–868 [DOI] [PubMed] [Google Scholar]
- 5. Hase S., Nishimura H., Kawabata S., Iwanaga S., Ikenaka T. (1990) The structure of (xylose)2glucose-O-serine 53 found in the first epidermal growth factor-like domain of bovine blood clotting factor IX. J. Biol. Chem. 265, 1858–1861 [PubMed] [Google Scholar]
- 6. Whitworth G. E., Zandberg W. F., Clark T., Vocadlo D. J. (2010) Mammalian Notch is modified by d-Xyl-α1,3-d-Xyl-α1–3-d-Glc-β1-O-Ser: implementation of a method to study O-glucosylation. Glycobiology 20, 287–299 [DOI] [PubMed] [Google Scholar]
- 7. Nishimura H., Kawabata S., Kisiel W., Hase S., Ikenaka T., Takao T., Shimonishi Y., Iwanaga S. (1989) Identification of a disaccharide (Xyl-Glc) and a trisaccharide (Xyl2-Glc) O-glycosidically linked to a serine residue in the first epidermal growth factor-like domain of human factors VII and IX and protein Z and bovine protein Z. J. Biol. Chem. 264, 20320–20325 [PubMed] [Google Scholar]
- 8. Nishimura H., Takao T., Hase S., Shimonishi Y., Iwanaga S. (1992) Human factor IX has a tetrasaccharide O-glycosidically linked to serine 61 through the fucose residue. J. Biol. Chem. 267, 17520–17525 [PubMed] [Google Scholar]
- 9. Jafar-Nejad H., Leonardi J., Fernandez-Valdivia R. (2010) Role of glycans and glycosyltransferases in the regulation of Notch signaling. Glycobiology 20, 931–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stanley P. (2007) Regulation of Notch signaling by glycosylation. Curr. Opin. Struct. Biol. 17, 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takeuchi H., Haltiwanger R. S. (2010) Role of glycosylation of Notch in development. Semin. Cell Dev. Biol. 21, 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Okajima T., Irvine K. D. (2002) Regulation of Notch signaling by O-linked fucose. Cell 111, 893–904 [DOI] [PubMed] [Google Scholar]
- 13. Shi S., Stanley P. (2003) Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 100, 5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hicks C., Johnston S. H., diSibio G., Collazo A., Vogt T. F., Weinmaster G. (2000) Fringe differentially modulates Jagged1 and Delta1 signaling through Notch1 and Notch2. Nat. Cell Biol. 2, 515–520 [DOI] [PubMed] [Google Scholar]
- 15. Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., Vogt T. F. (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 [DOI] [PubMed] [Google Scholar]
- 16. Fleming R. J., Gu Y., Hukriede N. A. (1997) Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development 124, 2973–2981 [DOI] [PubMed] [Google Scholar]
- 17. Panin V. M., Papayannopoulos V., Wilson R., Irvine K. D. (1997) Fringe modulates Notch-ligand interactions. Nature 387, 908–912 [DOI] [PubMed] [Google Scholar]
- 18. Brückner K., Perez L., Clausen H., Cohen S. (2000) Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406, 411–415 [DOI] [PubMed] [Google Scholar]
- 19. Acar M., Jafar-Nejad H., Takeuchi H., Rajan A., Ibrani D., Rana N. A., Pan H., Haltiwanger R. S., Bellen H. J. (2008) Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leonardi J., Fernandez-Valdivia R., Li Y. D., Simcox A. A., Jafar-Nejad H. (2011) Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling. Development 138, 3569–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fernandez-Valdivia R., Takeuchi H., Samarghandi A., Lopez M., Leonardi J., Haltiwanger R. S., Jafar-Nejad H. (2011) Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 138, 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donoviel D. B., Hadjantonakis A. K., Ikeda M., Zheng H., Hyslop P. S., Bernstein A. (1999) Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13, 2801–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oka C., Nakano T., Wakeham A., de la Pompa J. L., Mori C., Sakai T., Okazaki S., Kawaichi M., Shiota K., Mak T. W., Honjo T. (1995) Disruption of the mouse RBP-Jκ gene results in early embryonic death. Development 121, 3291–3301 [DOI] [PubMed] [Google Scholar]
- 24. Takeuchi H., Fernández-Valdivia R. C., Caswell D. S., Nita-Lazar A., Rana N. A., Garner T. P., Weldeghiorghis T. K., Macnaughtan M. A., Jafar-Nejad H., Haltiwanger R. S. (2011) Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 108, 16600–16605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishimizu T., Sano K., Uchida T., Teshima H., Omichi K., Hojo H., Nakahara Y., Hase S. (2007) Purification and substrate specificity of UDP-d-xylose:β-d-glucoside α-1,3-d-xylosyltransferase involved in the biosynthesis of the Xyl α1–3Xyl α1,3Glc β1-O-Ser on epidermal growth factor-like domains. J. Biochem. 141, 593–600 [DOI] [PubMed] [Google Scholar]
- 26. Minamida S., Aoki K., Natsuka S., Omichi K., Fukase K., Kusumoto S., Hase S. (1996) Detection of UDP-d-xylose: α-d-xyloside α-3-xylosyltransferase activity in human hepatoma cell line HepG2. J. Biochem. 120, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 27. Omichi K., Aoki K., Minamida S., Hase S. (1997) Presence of UDP-d-xylose: β-d-glucoside α-1,3-d-xylosyltransferase involved in the biosynthesis of the Xylα1,3Glc β-Ser structure of glycoproteins in the human hepatoma cell line HepG2. Eur. J. Biochem. 245, 143–146 [DOI] [PubMed] [Google Scholar]
- 28. Sethi M. K., Buettner F. F., Krylov V. B., Takeuchi H., Nifantiev N. E., Haltiwanger R. S., Gerardy-Schahn R., Bakker H. (2010) Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated Notch epidermal growth factor repeats. J. Biol. Chem. 285, 1582–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shao L., Moloney D. J., Haltiwanger R. (2003) Fringe modifies O-fucose on mouse Notch1 at epidermal growth factor-like repeats within the ligand-binding site and the Abruptex region. J. Biol. Chem. 278, 7775–7782 [DOI] [PubMed] [Google Scholar]
- 31. Krylov V., Ustyuzhanina N., Grachev A., Bakker H., Nifantiev N. (2007) Stereoselective synthesis of the 3-aminopropyl glycosides of α-d-Xyl-(1→3)β-d-Glc and α-d-Xyl-(1→3)-α-d-Xyl-(1→3)-β-d-Glc and of their corresponding N-octanoyl derivatives. Synthesis 2007, 3147–3154 [Google Scholar]
- 32. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 33. Russo R. N., Shaper N. L., Taatjes D. J., Shaper J. H. (1992) β-1,4-galactosyltransferase: a short NH2-terminal fragment that includes the cytoplasmic and transmembrane domain is sufficient for Golgi retention. J. Biol. Chem. 267, 9241–9247 [PubMed] [Google Scholar]
- 34. Luo Y., Haltiwanger R. S. (2005) O-Fucosylation of Notch occurs in the endoplasmic reticulum. J. Biol. Chem. 280, 11289–11294 [DOI] [PubMed] [Google Scholar]
- 35. Munro S., Freeman M. (2000) The Notch signaling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr. Biol. 10, 813–820 [DOI] [PubMed] [Google Scholar]
- 36. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 37. Pitcher J., Smythe C., Cohen P. (1988) Glycogenin is the priming glucosyltransferase required for the initiation of glycogen biogenesis in rabbit skeletal muscle. Eur. J. Biochem. 176, 391–395 [DOI] [PubMed] [Google Scholar]
- 38. Parker C. G., Fessler L. I., Nelson R. E., Fessler J. H. (1995) Drosophila UDP-glucose:glycoprotein glucosyltransferase: sequence and characterization of an enzyme that distinguishes between denatured and native proteins. EMBO J. 14, 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grewal P. K., McLaughlan J. M., Moore C. J., Browning C. A., Hewitt J. E. (2005) Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology 15, 912–923 [DOI] [PubMed] [Google Scholar]
- 40. Grewal P. K., Holzfeind P. J., Bittner R. E., Hewitt J. E. (2001) Mutant glycosyltransferase and altered glycosylation of α-dystroglycan in the myodystrophy mouse. Nat. Genet. 28, 151–154 [DOI] [PubMed] [Google Scholar]
- 41. Aoki K., Porterfield M., Lee S. S., Dong B., Nguyen K., McGlamry K. H., Tiemeyer M. (2008) The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J. Biol. Chem. 283, 30385–30400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rana N. A., Xu M. L., Moss H., Rab A., Leonardi J., Jafar-Nejad H., Haltiwanger R. S. (2010) Fringe benefits: Analysis of O-glycosylation and fringe elongation on the extracellular domain of Drosophila Notch. Glycobiology 20, 1486 [Google Scholar]
- 43. Rana N. A., Nita-Lazar A., Takeuchi H., Kakuda S., Luther K. B., Haltiwanger R. S. (2011) O-Glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J. Biol. Chem. 286, 31623–31637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun Q., Ju T., Cummings R. D. (2011) The transmembrane domain of the molecular chaperone Cosmc directs its localization to the endoplasmic reticulum. J. Biol. Chem. 286, 11529–11542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Erkens G. B., Berntsson R. P., Fulyani F., Majsnerowska M., Vujičić-Žagar A., Ter Beek J., Poolman B., Slotboom D. J. (2011) The structural basis of modularity in ECF-type ABC transporters. Nat. Struct. Mol. Biol. 18, 755–760 [DOI] [PubMed] [Google Scholar]
- 46. Schneider D., Engelman D. M. (2004) Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J. Mol. Biol. 343, 799–804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.