Abstract
The repair of reactive oxygen species-induced base lesions and single strand breaks (SSBs) in the nuclear genome via the base excision (BER) and SSB repair (SSBR) pathways, respectively, is well characterize, and important for maintaining genomic integrity. However, the role of mitochondrial (mt) BER and SSBR proteins in mt genome maintenance is not completely clear. Here we show the presence of the oxidized base-specific DNA glycosylase Nei-like 2 (NEIL2) and the DNA end-processing enzyme polynucleotide kinase 3′-phosphatase (PNKP) in purified human mitochondrial extracts (MEs). Confocal microscopy revealed co-localization of PNKP and NEIL2 with the mitochondrion-specific protein cytochrome c oxidase subunit 2 (MT-CO2). Further, chromatin immunoprecipitation analysis showed association of NEIL2 and PNKP with the mitochondrial genes MT-CO2 and MT-CO3 (cytochrome c oxidase subunit 3); importantly, both enzymes also associated with the mitochondrion-specific DNA polymerase γ. In cell association of NEIL2 and PNKP with polymerase γ was further confirmed by proximity ligation assays. PNKP-depleted ME showed a significant decrease in both BER and SSBR activities, and PNKP was found to be the major 3′-phosphatase in human ME. Furthermore, individual depletion of NEIL2 and PNKP in human HEK293 cells caused increased levels of oxidized bases and SSBs in the mt genome, respectively. Taken together, these studies demonstrate the critical role of NEIL2 and PNKP in maintenance of the mammalian mitochondrial genome.
Keywords: Base Excision Repair, DNA, DNA-binding Protein, DNA Damage, DNA Repair, DNA-Protein Interaction, Mitochondrial DNA
Introduction
Reactive oxygen species-induced genomic damage includes a plethora of oxidized bases, apurinic/apyrimidinic (AP)3 sites, and DNA single strand breaks (SSBs) that are often mutagenic and are etiologically linked to various pathophysiologies, including sporadic cancer, and a multitude of age-related degenerative diseases (1, 2). All of these DNA base lesions are primarily repaired by the evolutionarily conserved base excision repair (BER) pathway in both the nucleus and mitochondria (3). BER for oxidized bases is initiated by the recognition and cleavage of the base lesion from DNA by a DNA glycosylase; the resulting AP site is then cleaved by the intrinsic lyase activity of the glycosylase. In mammalian cells, five oxidized base-specific DNA glycosylases have been identified; they belong to two families based on their reaction mechanisms and structural conservation of their catalytic motifs. OGG1 and NTH1 of the Nth family remove base lesions only from duplex DNA and have β-elimination activity, generating 3′-deoxyribose phosphate and 5′-phosphate (5′-P) groups at the resulting SSB (4). In contrast, the recently discovered NEILs 1–3 of the Nei family are active with duplex as well as single-stranded DNA (occurring transiently during DNA replication and transcription). Our recent studies now show that NEIL2 preferentially repairs oxidative damage from the transcribed genes, and NEIL1 primarily associates with the replication-associated proteins, thus indicating its involvement in repair during DNA replication (5, 6). NEILs 1 and 2 have β,δ-elimination activity, generating 3′-phosphate (3′-P) and 5′-P groups at the SSB site. In the ensuing step, the 3′-deoxyribose phosphate and 3′-P blocking groups are removed by AP endonuclease 1 or polynucleotide kinase 3′-phosphatase (PNKP), respectively, in the two distinct subpathways (7–9). In the mammalian cell nucleus, DNA polymerase β (Polβ) or DNA polymerase δ/ϵ (Polδ/ϵ) then fills in the gap before nick sealing by DNA ligases (ligase IIIα/ligase I) (10).
Besides being generated as intermediates of oxidized base repair, SSBs are also generated in the genome by oxidative stress, ionizing radiation, and various chemotherapeutic agents, which generate strand breaks with a variety of nonligatable “dirty” ends (11–13). Such DNA ends are among the most toxic and mutagenic lesions in mammalian genomes because they are refractory to DNA polymerases and DNA ligases; the conventional 3′-OH and 5′-P ends must be restored for gap filling and DNA ligation to occur. Most SSBs are repaired via a SSBR pathway consisting of four basic steps: SSB detection, DNA end processing, DNA gap filling, and DNA ligation (3, 14, 15). A number of enzymes are available to process the SSBs depending on the nature of the blocking groups at both ends (14, 15). Those with 3′-phosphate termini are noteworthy because they are one of the major SSBs induced by oxidative stress and are also generated as intermediates of NEIL-mediated BER (7, 8). PNKP with dual 3′-phosphatase and 5′-kinase activities is required for processing 3′-P and 5′-OH at strand breaks (16, 17). 5′-OH is generated by some nucleases as intermediates of topoisomerase cleavage and also during DNA replication (16, 18, 19). Persistent SSBs caused by the deficiency of 5′- and/or 3′-end-cleaning enzymes may result in a severe phenotype, including cell death (15). PNKP is thus a key enzyme for processing both 3′ and 5′ termini in SSBR.
Mammalian mitochondria contain their own 16.5-kb circular DNA molecule, which is subjected to continuous attack by endogenous reactive oxygen species because of its proximity to the site of reactive oxygen species generation via mt electron transport system complexes. Furthermore, unlike nuclear DNA, mtDNA lacks protective histones and hence is more susceptible to oxidative damage (20–22). Oxidative damage and SSBs in the mt genome have been implicated in various human degenerative diseases and in aging (23–25). Hence, mtDNA repair is critical for normal cellular functioning. Mitochondria have their own DNA repair systems, and repair of oxidized bases via BER has already been demonstrated for a number of cell types (26). Several mt BER proteins have recently been identified; they are either identical to those found in the nucleus or are nuclear BER protein isoforms that arise from variant RNA splicing. Among the DNA glycosylases for oxidized bases, OGG1 and NTH1 have been shown to localize in mitochondria. Bohr and co-workers (27) recently identified NEIL1 in mitochondria from mouse liver. Although OGG1/NTH1-initiated BER involving AP endonuclease 1 has been fairly well characterized, NEIL-initiated BER in mitochondria and the protein components involved in this pathway have not been established (28). More importantly, little is known about the SSBR in mitochondria. DNA polymerase γ is the only DNA polymerase in mammalian mitochondria and is thus essential for both mt genome replication and repair (29).
Here we provide evidence for the presence of NEIL2 and PNKP in mitochondria isolated from human cells. We also demonstrate a role for PNKP in NEIL-mediated BER of oxidized bases as well as in mt SSBR. The accumulation of oxidized bases and SSBs in the mt genome of NEIL2- and PNKP-depleted cells, respectively, indicated the critical roles of these proteins in maintaining mt genomic integrity.
EXPERIMENTAL PROCEDURES
Cell Culture and Generation of Stable NEIL2-FLAG-expressing Human Embryonic Kidney (HEK293) Cells
HEK293 cells were grown at 37 °C and 5% CO2 in DMEM containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 units/ml streptomycin. HEK293 cells stably expressing NEIL2-FLAG at low levels comparable with the endogenous NEIL2 protein levels were generated as described before (5). Human neuroblastoma SH-SY5Y cells were grown in DMEM:F-12 (1:1) (30).
Depletion of PNKP and NEIL2 in HEK293 Cells
We have developed microRNA-adapted shRNAmir constructs for targeting endogenous PNKP. Five different single-stranded 97-mer template oligonucleotides containing the human miR30 loop sequences and 5′- and 3′-flanking and antisense sequences targeting human PNKP transcripts were designed using the “Oligo Retriever” program as described previously (31–33). Each of the single-stranded oligonucleotides was PCR-amplified with common 5′ and 3′ PCR primers: 5′-CAGAAGGCTCGAGAAGGTATATTGCTGTTGACAGTGAGCG-3′ and 5′-CTAAAGTAGCCCCTTGAATTCCGAGGCAGTAGGCA-3′. The 128-bp PCR products were purified from an agarose gel, digested with XhoI and EcoRI, and then ligated with the retroviral vector LMP (Open Biosystem). Each individual shRNAmir construct for PNKP was transiently transfected, and the targeting efficiency of each clone was verified by Western blotting and real time (RT)-PCR analyses. Targeted inactivation resulted in extensive cell death, so shRNAmir constructs that down-regulated PNKP by ∼60% were used to target PNKP in the experiments described here.
NEIL2 was depleted in HEK293 cells using siRNA (Sigma; GAAUGAACCUAGAGCGGUG). Cells were harvested 48 h after transfection, and depletion of NEIL2 was confirmed by Western blotting.
Isolation and Purification of Mitochondria
Mitochondria were isolated using the Mitochondria Isolation kit (Pierce protein research product, Thermo Scientific, catalog number 89874) according to the manufacturer's protocol with an optimized Dounce homogenization procedure. Isolated mitochondria were washed with PBS, treated with trypsin (1 mg/ml in PBS) for 15 min at room temperature to remove contaminating proteins adhered to the outer surface of mitochondria, and then extensively washed with PBS. The washed mitochondria were lysed in 50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, and 1% Triton X-100. Different fractions (cytosolic, nuclear, and mitochondrial) were analyzed by 10% SDS-PAGE, tested for the presence of NEIL2 (with rabbit polyclonal antibody (Ab); see Ref. 5) and PNKP (rabbit polyclonal Ab; a gift from Dr. Michael Weinfeld), and tested for the purity of the fractions by successive Western blotting using nucleus- (RNA Pol II, Santa Cruz Biotechnology), cytoplasm- (lactate dehydrogenase, Santa Cruz Biotechnology), and mitochondrion (70-kDa subunit of complex II, Molecular Probes)-specific antibodies.
Immunofluorescence
Human neuroblastoma SH-SY5Y cells were grown on microscope coverslips, fixed in 4% paraformaldehyde in PBS, blocked with 10% goat serum for 60 min, and incubated with primary Ab for NEIL2, PNKP (rabbit polyclonal), or mt-specific MT-CO2 (mouse monoclonal; Santa Cruz Biotechnology) at 4 °C overnight. After incubation with Alexa Fluor 488 (green)- or 568 (red)-conjugated secondary Ab, the coverslips were mounted in ProLong Gold antifade reagent with DAPI (Invitrogen). Images were taken using a Nikon Plan Fluor 60× 0.5–1.25 oil objective mounted on a Nikon Eclipse TE2000 confocal microscope equipped with a Himamatsu EM-CCD camera. The image processing was limited to contrast enhancement.
Chromatin Immunoprecipitation (ChIP) and Re-ChIP Assay
Cells were cultured in DMEM containing 10% FBS. ChIP analysis was performed using a chromatin immunoprecipitation assay kit (Upstate Cell Signaling Solutions, Millipore, Temecula, CA) according to the manufacturer's protocol, and re-ChIP assays were performed as described (5, 34). Briefly, the cells (∼106 cells) were treated with formaldehyde (1% final concentration) for 10 min for cross-linking; then washed twice with PBS; lysed in 200 μl of 50 mm Tris-HCl, pH 8.0, 10 mm EDTA, and 1% SDS with a protease inhibitor mixture; and sonicated to generate 32 ∼400-bp-long DNA fragments. The supernatants were diluted with 20 mm Tris-HCl, pH 8.0, 1.0 mm EDTA, 150 mm NaCl, 1% Triton X-100, 0.01% SDS, and protease inhibitors. The precleared supernatant was then incubated overnight at 4 °C with Ab to FLAG, PNKP, or RNA polymerase II (RNAP II) (Santa Cruz Biotechnology) as indicated in Fig. 3. The immunocomplexes were precipitated with salmon sperm DNA/protein G-agarose, and the agarose beads washed sequentially in a low salt wash buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Triton-X-100, and 0.1% SDS); a high salt wash buffer (same as low salt wash buffer except containing 500 mm NaCl); LiCl wash buffer; and a buffer containing 20 mm Tris-HCl, pH 8.0 and 1 mm EDTA. The immunocomplexes were extracted from the beads with elution buffer (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, 1.0% SDS, and 100 mm NaHCO3).
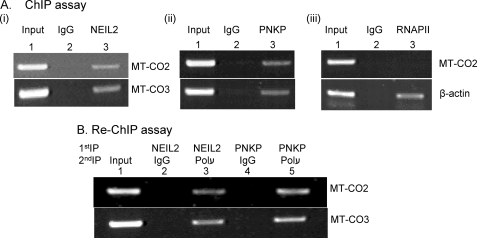
FIGURE 3.
A, ChIP assay. After sonication and immunoprecipitation of cross-linked chromatin separately with Ab for FLAG (panel i), PNKP (panel ii), or RNAP II (panel iii), the IPs were washed, the bound protein-DNA complexes were eluted, and the precipitated DNA was amplified by PCR using mitochondrial (MT-CO2 and MT-CO3) or nuclear (β-actin) gene-specific primers. B, re-ChIP assay. The bound fractions from the first IP were eluted, divided into two aliquots, and subjected to a second IP with IgG (as control) or with a specific Ab. PCR amplifications were performed using specific primers as shown in Table 1B.
The re-ChIP assays were performed as described (5, 34). Briefly, the eluant of the primary immunocomplex obtained with the first Ab was diluted 10-fold with dilution buffer (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, 150 mm NaCl, 1% Triton X-100, and protease inhibitors) and then subjected to further immunoprecipitation with the second Ab. Immunocomplexes were again extracted from the beads with elution buffer. The cross-linking of the eluted immunocomplexes was then reversed with 500 mm NaCl at 65 °C for 4 h, and DNA was isolated from the eluates with proteinase K treatment followed by phenol extraction and ethanol precipitation. The purified DNA was subjected to PCR (30 cycles), and the PCR products were then resolved in a 2.0% agarose gel and visualized by ethidium bromide staining.
In Situ Proximity Ligation Assay (PLA)
HEK293 cells were grown overnight in 16-well chamber slides, fixed with 4% paraformaldehyde, permeabilized with 0.2% Tween 20, and incubated with primary Abs for NEIL2 (mouse monoclonal; Abnova) or PNKP (mouse monoclonal; a gift from Dr. Michael Weinfeld) and Polγ (anti-POLG1; rabbit polyclonal; Agrisera AB, Sweden) and subjected to PLAs using the Duolink PLA kit from OLink Bioscience (Uppsala, Sweden) performed according to the manufacturer's instructions. The nuclei were counterstained with DAPI, and the PLA signals were visualized in a fluorescence microscope (Olympus) at 20× magnification.
Expression and Purification of Recombinant Proteins
Recombinant NEIL2, PNKP, LigIIIα, and Polγ were purified as described previously (7, 35–39), and the purified proteins were stored in 50% glycerol-containing PBS at −20 °C.
Analysis of DNA Glycosylase Activity of NEIL2 and 3′-Phosphatase Activity of PNKP
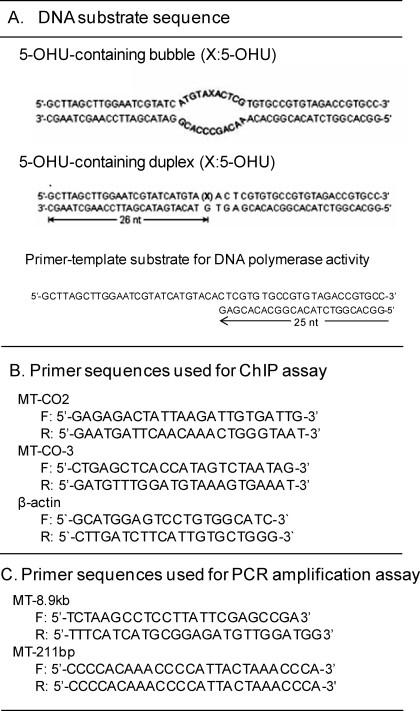
Base excision and strand cleavage activities of DNA glycosylases in ME or using purified NEIL2 were measured using a 5′-32P-labeled 51-mer oligo with 5-OHU in an 11-nt-long bubble oligo (5-OHU·B11; Table 1) in 10 μl of BER buffer containing 25 mm Hepes-KOH, pH 7.6, 50 mm NaCl, 0.5 mm EDTA, 0.5 mm DTT, 100 μg/ml bovine serum albumin (BSA), and 5% glycerol at 37 °C for 15 min followed by 20% urea-PAGE, and the radioactive bands were analyzed in a PhosphorImager. The 3′-phosphatase activity of PNKP was assayed as we described previously (7).
TABLE 1.
DNA substrate and primer sequences used in the study
F, forward; R, reverse.
DNA Trapping Assay
DNA trapping reactions were performed for 30 min at 37 °C by incubating 10 μg of ME or the purified proteins with 32P-labeled bubble substrate oligo (5-OHU·B11) in a reaction mixture containing 25 mm Hepes, pH 7.5, 50 mm KCl, 1 mm EDTA, and 50 mm NaCNBH3 (40). The trapped complexes were separated by 12% SDS-PAGE, and the gels were dried on DE81 paper for PhosphorImager analysis of radioactivity.
Repair of Oxidized Base or SSB Using ME
Repair of the oxidized base lesion 5-OHU was measured in ME (from control or PNKP-depleted cells) using 2 pmol of lesion-containing duplex oligo (5-OHU·G; Table 1). The 20-μl reaction mixture also contained 1 mm ATP, 25 μm unlabeled dNTPs, and 10 μm [α-32P]dNTPs (the concentration of the corresponding cold dNTP was lowered to 15 μm unless specified otherwise) in BER buffer (7), and the reaction mixture was then incubated for 30 min at 37 °C. For measuring repair of an SSB, we generated a circular plasmid substrate containing a single SSB with 3′-P and 5′-P at the strand break (see Fig. 6B). Briefly, pUC19CPD plasmid, which contains two recognition sequences for the restriction enzyme N.BstNBI 32 nt apart (41), was completely digested with N.BstNBI (New England Biolabs). The plasmid was partially denatured by heating at 65 °C for 10 min to remove the 32-nt oligo (5′-GCG GAT ATT AAT GTG ACG GTA GCG AGT CGC TC-3′) and mixed with a biotinylated complementary oligo. The annealed, biotinylated 32-nt duplex was removed from the plasmid using streptavidin-agarose Dynabeads (Invitrogen). The resulting gapped plasmid was extracted with phenol/chloroform and ethanol-precipitated to remove the N.BstNBI. The plasmid was redissolved in Tris-EDTA buffer containing 50 mm NaCl, annealed with U-containing 5′-phosphorylated 32-nt oligo (5′-pGCG GAT ATT AAT GTG ACG GUA GCG AGT CGC TC-3′), and then ligated using T4 DNA ligase. The covalently closed Form-I plasmid containing U was then purified by CsCl gradient centrifugation and converted to a 1-nt gapped plasmid with 3′-P/5′P ends (pUC19CPD-SSB) by treatment with Udg and Fpg. The SSB repair was measured using 200 ng of plasmid substrate as described for 5-OHU repair.
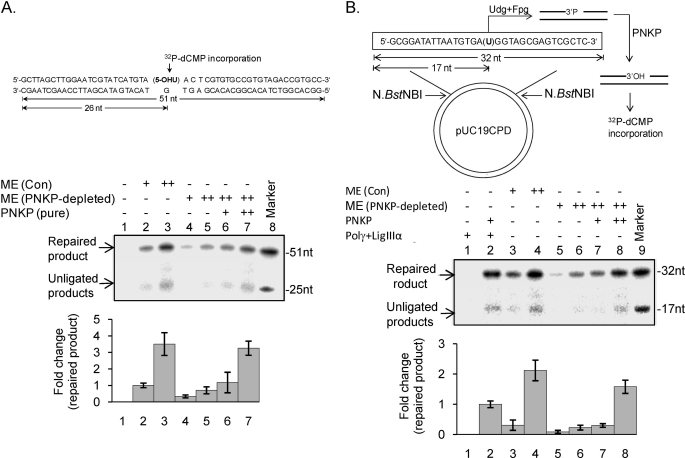
FIGURE 6.
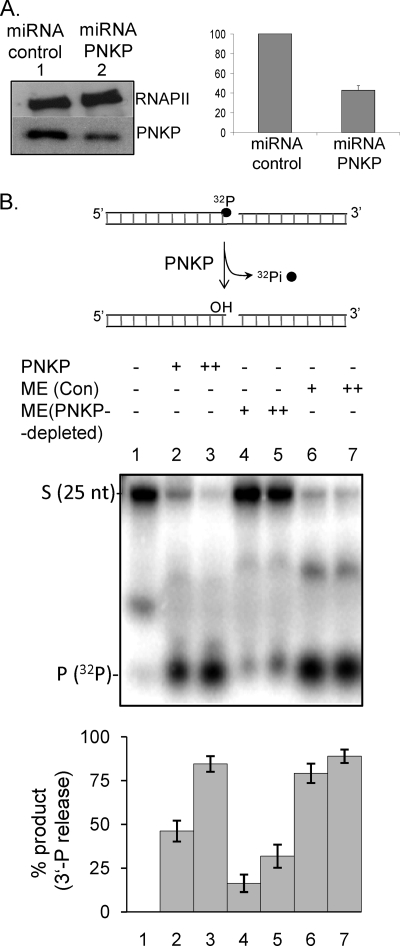
Representative gel showing BER and SSBR using mt extract. A, efficient repair of a 5-OHU lesion in a duplex oligo (top) measured by incorporation of [α-32P]dCMP after excision of 5-OHU. Lane 1, no protein; lanes 2 and 3, ME from control (Con) cells (5 and 10 μg); lanes 4 and 5, PNKP-depleted ME (5 and 10 μg). Addition of purified PNKP (50 and 100 fmol) restored efficient repair (lanes 6 and 7). Lane 8, size markers. The histogram (bottom) represents quantitation of the repair products with lane 2 arbitrarily set as 1. B, a plasmid DNA containing a single U was treated with Udg/Fpg to generate 3′-P and 5′-P with a one nucleotide gap to assess the 3′-phosphatase activity of PNKP. Repair was then monitored by incorporation of [α-32P]dCMP in the presence of Polγ and LigIIIα. The repaired product was digested with N.BstNBI and analyzed by denaturing gel electrophoresis. Other details are provided under “Experimental Procedures.” Lane 1, purified Polγ and LigIIIα (50 fmol); lane 2, reconstitution of SSBR using purified PNKP, Polγ, and LigIII (50 fmol each); lanes 3 and 4, ME (5 and 10 μg); lanes 5 and 6, PNKP-depleted ME (5 and 10 μg); lanes 7 and 8, PNKP-depleted ME plus purified PNKP (50 and 100 fmol); lane 9, size markers. Quantitation of the radioactive bands (lanes 2–8) is represented in a histogram (bottom) with lane 3 arbitrarily set as 1. A schematic representation of the generation a 3′-P-containing plasmid DNA substrate is shown (top). The standard error of the mean was calculated from at least three independent experiments.
Analysis of Oxidized Bases and SSBs in mt Genome by PCR Amplification Assay
Mitochondrial genome-specific semiquantitative PCR assays of long DNA fragments for measuring DNA damage were performed as described earlier (42) using LongAmp Taq DNA polymerase (New England Biolabs) and amplifying an 8.9-kb region of mt DNA. Preliminary assays were carried out to ensure the linearity of PCR amplification with respect to the number of cycles and DNA concentration. Damage to mt DNA was normalized to mt genome copy number determined by amplification of a 211-bp fragment using specific primers (Table 1). Unrepaired oxidized bases in DNA from NEIL2-depleted cells were measured by digestion with Fpg/endonuclease III to generate strand breaks before PCR analysis (43).
RESULTS
Presence of NEIL2 and PNKP in Mammalian Mitochondria
We previously reported the unusual activity of NEIL1 and NEIL2 in excising lesions from DNA bubble structures (unlike OGG/NTH1, which are active only with duplex DNA (44)). Interestingly, we also found a similar DNA glycosylase activity in the purified ME from HEK293 cells (Fig. 1A, lane 2). To test the presence of NEILs in mitochondria, we first analyzed trapped complex formation using ME from HEK293 cells with a 5-OHU-containing bubble substrate (5-OHU·B11). These glycosylases form a transient Schiff's base at the AP site (after excision of the base lesion) that can be trapped with NaCNBH3 or NaBH4. Fig. 1B shows the formation of two distinct trapped complexes with the ME (lane 4). The presence of similarly sized trapped products in parallel trapping assays with recombinant NEIL1 (lane 2) and NEIL2 (lane 3) suggested that both NEILs are present in mitochondria. The presence of NEIL1 in mitochondria has already been reported without detailed characterization (27). To provide evidence that NEIL2 also contributes to the repair of oxidative damage in the mitochondrial genome, we compared DNA glycosylase/AP lyase activity (on 5-OHU·B11) with the ME prepared from control versus NEIL2-depleted cells (siRNA-mediated; Fig. 1C). Fig. 1D shows an ∼50% decrease in activity with ME from NEIL2-depleted cells compared with control (lane 2 versus lane 3). The mitochondrial presence of NEIL2 was further confirmed by Western analysis of the ME from HEK293 cells using anti-NEIL2 Ab (Fig. 1E and Ref. 5). We have shown previously that NEIL-initiated repair in the nucleus utilizes PNKP, not AP endonuclease 1, for processing the β,δ-elimination product 3′-P at the strand break (7, 8). We thus postulated that PNKP should be present in the mitochondria; indeed, it was found to be present in the ME (Fig. 1E, lane 3). Lane 4 contained recombinant NEIL2 and PNKP (10 ng). Quantitation of the band intensities on the blots indicated that 30 μg of ME contained ∼20 ng of PNKP and ∼4 ng of NEIL2. Our data thus suggest that PNKP is a relatively abundant DNA repair protein in mitochondria. PNKP is known to be involved in multiple repair pathways (BER, SSBR, and double strand break repair), so its abundance may be a requirement for the cells.
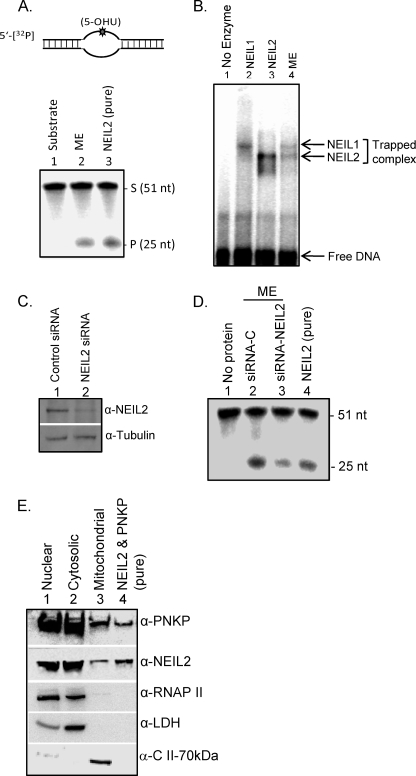
FIGURE 1.
Identification of NEIL2 and PNKP in mitochondria. A, a 5′-32P-labeled 51-mer oligo (5-OHU·B11; Table 1A) was used for a DNA glycosylase/AP lyase assay with purified ME (10 μg; lane 2). Lane 1, no protein; lane 3, purified NEIL2 (20 fmol) as a positive control. B, trapping assay of purified NEIL1 (20 fmol; lane 2), NEIL2 (lane 3), and ME (10 μg; lane 4) with 5′-32P-labeled 5-OHU·B11 oligo. Trapped complexes of the NEILs and free DNA are indicated. C, Western analysis of NEIL2 depletion in HEK293 cell extracts (10 μg) by NEIL2-specific siRNA; tubulin was used as a loading control. D, DNA glycosylase/AP lyase activity (with 5′-32P-labeled 5-OHU·B11) in the ME (5 μg) prepared from control (lane 2) versus NEIL2-depleted (lane 3) cells. E, Western blot analysis of cytosolic, nuclear, and mt fractions from HEK293 cells. Abs specific for RNAP II, lactate dehydrogenase (LDH), and the 70-kDa subunit of the electron transport chain complex II (C II-70kDa) were used as nuclear, cytosolic, and mt markers, respectively, to show the purity of the mt preparation. Cytosolic, nuclear, and mitochondrial fractions were loaded in equal amounts (30 μg). Purified NEIL2 and PNKP (10 ng) were used as references. S, 32P-labeled 3′-P-containing oligo substrate; P, released phosphate; siRNA-C, siRNA control.
One inherent challenge in studying mitochondrial proteins is the difficulty of removing nuclear and cytosolic contaminants from purified mitochondria. Some proteins originally shown to be present in the mitochondria could in fact be adventitiously associated with the mitochondrial outer membrane. However, such extraneous proteins are susceptible to trypsin, which does not degrade the matrix proteins (45, 46). In this study, we treated the mitochondrial pellet with trypsin before extraction in mt lysis buffer. To confirm the purity of the mitochondrial fraction, we subjected the mitochondrial, cytosolic, and nuclear fractions to Western analysis using Abs specific for each (Fig. 1E). Lactate dehydrogenase, specific for the cytosolic fraction, was absent in the mitochondrial and nuclear fractions. An RNAP II Ab was used to check for contamination by the nuclear fraction; we observed that isolated mitochondria were free from contamination by the cytosolic or nuclear protein, and both NEIL2 and PNKP were indeed present in the ME. Finally, the identity of the mitochondrial fraction was confirmed by the presence of the 70-kDa subunit of complex II.
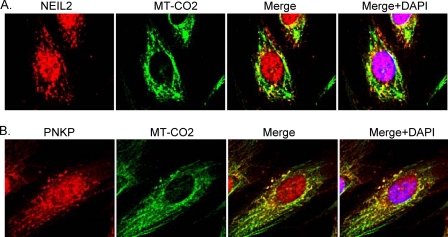
To further confirm that NEIL2 and PNKP localize to the mitochondria in human cells, we co-stained SH-SY5Y cells with the mitochondrion-specific MT-CO2 Ab and Ab for NEIL2 or PNKP. Upon immunofluorescence microscopy, we observed significant co-localization of NEIL2 (Fig. 2A) or PNKP (Fig. 2B) with MT-CO2, indicating the presence of a substantial fraction of NEIL2 and PNKP in mitochondria. Taken together, these results provide the first evidence for the presence of both NEIL2 and PNKP in mammalian mitochondria.
FIGURE 2.
Colocalization of NEIL2 and PNKP in mitochondria. The mitochondrial presence of NEIL2 (A) and PNKP (B) was investigated in human neuroblastoma SH-SY5Y cells using Abs for NEIL2 (rabbit polyclonal; see Ref. 5), PNKP (rabbit polyclonal), and mitochondrion-specific MT-CO2 (mouse monoclonal; Santa Cruz Biotechnology). The nuclei are counterstained with DAPI (blue). Red, NEIL2 and PNKP; green, Mitochondria; yellow, overlap of red and green showing significant co-localization of NEIL2 and PNKP with MT-CO2. Other details are under “Experimental Procedures.”
NEIL2 and PNKP Associate with Mitochondrial Genes
To further confirm the association of NEIL2 and PNKP with the mtDNA, we carried out ChIP (Fig. 3A) followed by a second ChIP (re-ChIP; Fig. 3B) in NEIL2-FLAG-expressing HEK293 cells to resolve whether the proteins were enriched on the same region of the mtDNA. We first confirmed the presence of NEIL2-FLAG in mt extract using anti-FLAG Ab (Sigma) (supplemental Fig. S1), then immunoprecipitated the cross-linked protein-DNA complexes separately with anti-FLAG Ab and PNKP Ab, washed the IPs, eluted the bound immune-DNA complexes, and amplified the precipitated DNA by PCR using mt gene-specific (MT-CO2 and MT-CO3) primers. Amplification of these mt-specific genes was observed for both NEIL2 and PNKP (Fig. 3A, panels i and ii). Lack of any amplification of mt genes in IP using anti-RNAP II Ab or IgG served as controls (Fig. 3A, panel iii, upper panel). As expected, amplification of nuclear β-actin gene fragment was observed in the RNAP II IP (Fig. 3A, panel iii, lower panel). In the re-ChIP assay, we subjected the cross-linked chromatin fraction to a first IP with anti-FLAG and PNKP Ab separately and prepared them for a second immunopulldown (re-ChIP) with anti-Polγ Ab or IgG. Amplification was observed for NEIL2-Polγ sequential immunoprecipitation (lane 3) but not in the case of IgG (lane 2), indicating the specific association of NEIL2 and Polγ on the mt genome (Fig. 3B). Similarly, we demonstrated an association between PNKP and Polγ on the mt genome (Fig. 3B, lane 5).
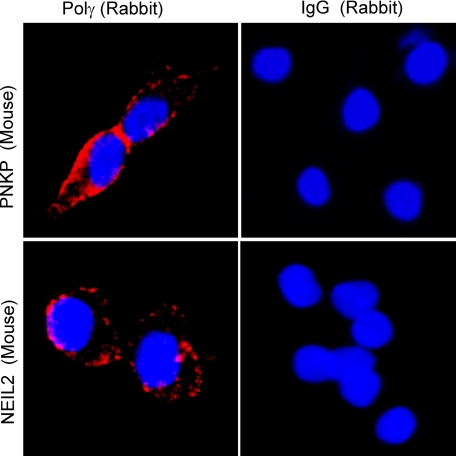
To further confirm the association of NEIL2 or PNKP with Polγ, we used an in situ PLA in which the close physical association of two proteins is visualized by a fluorescent signal (Olink Bioscience). This is a relatively new technique to study the interaction of endogenous proteins. In this assay, two proteins were immunostained with two primary Abs that were raised in two different host species, such as one in mouse (in this case NEIL2 and PNKP) and the other in rabbit Ab (Polγ). A species-specific second Ab, each containing a short oligo (PLA probe), was then allowed to bind to the primary Ab. When the two Abs are in close proximity (<40 nm), the oligos in the PLA probes can be amplified and visualized with a fluorescent probe as distinct foci. The assay has been shown to be highly specific for physically interacting endogenous proteins in a complex (47–49). We detected fluorescent signals for both NEIL2-Polγ and PNKP-Polγ (Fig. 4). The interactions between NEIL2-Polγ and PNKP-Polγ were observed in the perinuclear compartments as expected. No signals were detected when control IgGs were used in place of specific primary Abs. Taken together, these data clearly demonstrated the co-association of NEIL2 and PNKP with Polγ on the mitochondrial genome.
FIGURE 4.
Detection of NEIL2 and PNKP (mouse Ab) interaction with Polγ (rabbit Ab) in HEK293 cells by proximity ligation assays. Upper panel, PNKP (mouse monoclonal; a gift from Dr. Michael Weinfeld) with Polγ (anti-POLG1; Agrisera AB) or IgG (rabbit Ab). Lower panel, NEIL2 (mouse monoclonal; Abnova) with Polγ (rabbit Ab) or IgG (rabbit Ab).
PNKP Is Major 3′-Phosphatase in Mitochondria
To provide direct evidence for the role of PNKP in repairing mtDNA, we have depleted PNKP using microRNA in the HEK293 cell line, which showed a ∼60% reduction in its PNKP level as determined by Western analysis as well as by quantitative reverse transcription PCR (Fig. 5A, right panel, lane 2). We then examined the 3′-phosphatase activity in MEs from control and PNKP-depleted cells using an oligo substrate containing an SSB with 3′-32P as described under “Experimental Procedures” (Fig. 5B). The 3′-phosphatase activity of PNKP would release the 3′-32P, which was analyzed by separation by denaturing PAGE. Robust 3′-phosphatase activity was observed in ME from cells expressing control miRNA (Fig. 5B, lanes 6 and 7); however, ME from PNKP-depleted cells had only a residual 20–30% 3′-phosphatase activity (lanes 4 and 5) compared with that from control miRNA-treated cells, clearly indicating that PNKP was the major 3′-phosphatase activity in the ME.
FIGURE 5.
Representative gel showing 3′-phosphatase activity of PNKP in mitochondria. The substrate for the PNKP 3′-phosphatase assays was generated as we have described previously (7). A, Western analysis of PNKP in control (lane 1) and PNKP-specific miRNA-expressing cells (lane 2). Upper panel, RNAP II as control. PNKP transcript levels in control versus miRNA-treated cells were quantitated by quantitative PCR (right panel). B, a 32P-labeled 3′-phosphate-containing oligo (0.5 pmol) was used to measure the 3′-phosphatase activity of PNKP in MEs (5 and 10 μg) prepared from control (Con) miRNA-expressing (lanes 6 and 7) and PNKP-depleted cells (lanes 4 and 5). Lane 1, substrate alone; lanes 2 and 3, purified PNKP (50 and 100 fmol) as a positive control. S, 32P-labeled 3′-P-containing oligo substrate; P, released phosphate. The histogram shows quantitation of the percentage (%) of product (32Pi) released. The standard error of the mean was calculated from at least three independent experiments. The radioactive bands were quantitated using Quantity One software from Bio-Rad. S.E. was calculated using Microsoft Excel 7.0.
PNKP Is Required for Both BER and SSBR in Mitochondria
We next examined the role of PNKP in the repair of oxidized bases and SSBs in MEs from control and PNKP-depleted cells. Repair of 5-OHU was measured as described before by analyzing the incorporation of [α-32P]dCMP after excision of the lesion from a duplex oligo substrate (3, 7). Efficient repair was observed in ME from control cells (Fig. 6A, lanes 2 and 3), but PNKP depletion caused a significant decrease in repair (∼70%; lanes 4 and 5). Addition of recombinant PNKP restored the repair activity (lanes 6 and 7). These data clearly suggest that PNKP plays a major role in repair of oxidized bases in the mitochondrial genome.
We next investigated the role of PNKP in mitochondrial SSBR using a plasmid substrate containing a single SSB with a 3′-P blocking group. SSB repair was measured by incorporation of [α-32P]dCMP after processing of the 3′-P by PNKP. We first reconstituted SSB repair using the recombinant mt BER components, PNKP, Polγ, and LigIII (Fig. 6B, lane 2). As for oxidized BER, ME from control cells showed robust SSB repair (Fig. 6B, lanes 3 and 4), and a marked decrease in repair was observed with ME from PNKP-depleted cells (lanes 5 and 6). Addition of recombinant PNKP (50 and 100 fmol) restored the SSB repair in PNKP-depleted ME (lanes 7 and 8). Together, these data show that PNKP plays a critical role in both BER and SSBR in mitochondria.
Evidence for Role of NEIL2 and PNKP in mt Genome Maintenance
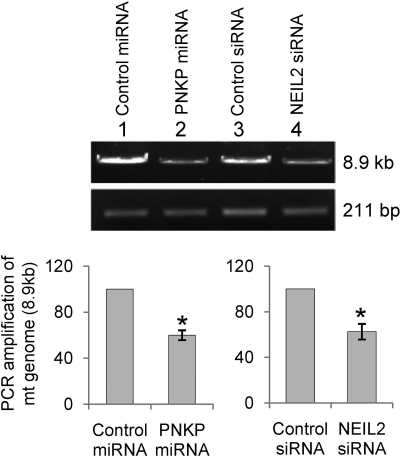
To provide direct evidence for the role of NEIL2 and PNKP in repairing mitochondrial genomes, the relative levels of base damage in the mitochondrial genes were measured in NEIL2- and PNKP-depleted and control cells using a semiquantitative long amplification PCR assay (42). To measure unrepaired oxidized bases in NEIL2-depleted cells (Fig. 7), the isolated DNA was digested with Fpg and Nth (Escherichia coli) to generate SSBs at the sites of base lesions. However, genomic DNA from PNKP-depleted cells did not need to be treated with Fpg/Nth because these cells accumulate SSBs even without the enzymatic treatment. Fig. 7 shows that there was a 37.5 ± 6.5% decrease in PCR amplification due to unrepaired oxidized base damage in the mitochondrial DNA of NEIL2-depleted cells. Similarly, a 40 ± 4.2% decrease in amplified product was observed with PNKP-depleted cells, indicating a substantial increase in unrepaired SSBs in mtDNA of PNKP-depleted cells (Fig. 7) compared with their respective controls. Before analysis, the DNA was normalized for mt copy number by PCR of a 211-bp region of the mt genome (43). Taken together, these results strongly suggest an important role for NEIL2 and PNKP in the maintenance of mt genomic integrity.
FIGURE 7.
Quantitation of mitochondrial DNA damage in NEIL2- and PNKP-depleted cells. Upper panel, representative gel showing PCR-amplified fragments of the long amplicon (8.9-kb region) and 211-bp region. The relative levels of endogenous DNA damage were calculated by quantitating the long amplicon (8.9-kb region) PCR product after normalizing for mt copy number by PCR of a 211-bp region of mt genome. Bottom panel, quantitation of the amplified products is represented in histograms with the amplicon from control siRNA- or miRNA-treated cells arbitrarily set as 100. The DNA from NEIL2-depleted cells was digested with Fpg/endonuclease III before analysis. Other details are provided under “Experimental Procedures.” *, p < 0.01. The standard error of the mean was calculated from at least three independent experiments.
DISCUSSION
PNKP was cloned and characterized many years ago (16, 17); however, the functional significance of its presence in mitochondria and more specifically its role in mt SSBR and NEIL-initiated BER have not been examined. To our knowledge, our studies here showed for the first time the presence of NEIL2 and PNKP in mitochondria and provided evidence for their critical roles in the repair of mt genomic damage. The mt forms of NEIL2 and PNKP identified in this study are identical to their nuclear forms. Analysis of the protein sequences of NEIL2 and PNKP showed no canonical mitochondrial targeting sequences. However, this should not be troubling as more than 50% of mitochondrial proteins do not use the classical import pathway that requires the recognition of a specific sequence (50). In addition, many nuclear proteins (for example, NF-κB, p53, BRCA1, and AP-1) have been detected in mitochondria despite lacking canonical mitochondrial targeting sequences, indicating the existence of still unknown mechanisms of intracellular trafficking (51–55).
It is important to note that there is a general skepticism about unequivocally establishing the presence of a protein in mitochondria because it is hard to remove protein contaminants associated with the mt outer membrane from the preparation even after repeated banding by equilibrium centrifugation (56). Minimal extramitochondrial contamination was achieved in this study by treating the fractionated, intact mitochondria with trypsin followed by its inactivation with trypsin inhibitor (28, 46). The resulting mitochondrial preparations were meticulously tested for nuclear and cytoplasmic contamination using Abs specific for proteins exclusively present in these compartments (Fig. 1E).
DNA damage repair is a highly regulated and multistep process, and the proteins involved in the pathway act in concert (57). To understand this coordinated series of events, it is thus important to identify the interacting proteins and their functional association to form a repair complex. We have previously identified such a BER complex in the nucleus involving NEIL2, PNKP, Polβ, and LigIIIα (8). In this study, we have shown that NEIL2 and PNKP are in close proximity with the mitochondrial Polγ. Furthermore, PNKP, Polγ, and LigIII are the minimal protein components required for mitochondrial SSBR as determined by in vitro reconstitution of complete SSBR with purified proteins (Fig. 6B). Notably, depletion of NEIL2 or PNKP caused an ∼40% increase in endogenous DNA damage accumulation even though the level of the proteins (NEIL or PNKP) was depleted by only 60–70%. These data suggest that inactivation or complete depletion of either NEIL2 or PNKP would cause severe damage to the mitochondrial genome and hence strongly implicate these proteins in mt genomic maintenance.
To examine the role of PNKP in repair of the mt genome, we used PNKP-depleted ME (by ∼60%) to demonstrate that PNKP is the major 3′-phosphatase in mitochondria (Fig. 5B). This is an important finding because 3′-P is one of the major blocked ends as we have discussed previously. The PNKP-depleted ME also showed significantly less efficient total BER and SSBR (Fig. 6), thus highlighting the role of PNKP in both NEIL-mediated BER and SSBR in mitochondria. The importance of SSBR is demonstrated by the observation that two of the proteins involved in this pathway, aprataxin and tyrosyl-DNA phosphodiesterase 1, are mutated in hereditary neurodegenerative diseases (58–60). Recent reports of the presence of tyrosyl-DNA phosphodiesterase 1 and aprataxin in mitochondria further underscore the importance of end-processing activity in mt genome maintenance (53, 61). Recently it has been shown that mutation in or lower levels of PNKP cause an autosomal recessive disease (denoted MCSZ; Ref. 62) characterized by microcephaly, intractable seizures, and developmental delay. Whether mt genomic damage contributes to the pathogenesis of MCSZ should be an important area for future studies. Thus, understanding how mt proteins repair oxidized bases and SSBs is critically important because any decrease of repair capacity leading to the accumulation of mt genomic damage could contribute to the onset of various diseases and/or pathologies.
Supplementary Material
Acknowledgments
We acknowledge the generosity of Drs. Michael Weinfeld and John Hays for giving us PNKP Abs (both mouse monoclonal and rabbit polyclonal) and pUC19CPD plasmid, respectively, and Dr. Chandrasekha Yallampalli for allowing us to use the fluorescence microscope. We thank Dr. David Konkel for carefully editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA102271 and ES017353 (to T. K. H.), CA81063 (to S. M. M.), and ES018948 (to I. B.) from the USPHS.

This article contains supplemental Fig. S1.
- AP
- apurinic/apyrimidinic
- Ab
- antibody
- BER
- base excision repair
- ME
- mitochondrial extract
- mt
- mitochondrial
- MT-CO2
- mitochondrial cytochrome c oxidase subunit 2
- MT-CO3
- mitochondrial cytochrome c oxidase subunit 3
- NTH1
- endonuclease III homolog 1
- OGG1
- 8-oxoguanine DNA glycosylase 1
- NEIL
- Nei-like
- PLA
- proximity ligation assay
- 3′-P
- 3′-phosphate
- 5′-P
- 5′-phosphate
- PNKP
- polynucleotide kinase 3′-phosphatase
- Pol
- DNA polymerase
- SSB
- single strand break
- SSBR
- single strand break repair
- 5-OHU
- 5-hydroxyuridine
- Lig
- ligase
- RNAP II
- RNA polymerase II
- nt
- nucleotide(s)
- IP
- immunoprecipitation
- miRNA
- microRNA
- MCSZ
- microcephaly, infantile-onset seizures, and developmental delay.
REFERENCES
- 1. Breen A. P., Murphy J. A. (1995) Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 18, 1033–1077 [DOI] [PubMed] [Google Scholar]
- 2. Cadet J. L., Brannock C. (1998) Free radicals and the pathobiology of brain dopamine systems. Neurochem. Int. 32, 117–131 [DOI] [PubMed] [Google Scholar]
- 3. Hegde M. L., Hazra T. K., Mitra S. (2008) Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 18, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dodson M. L., Michaels M. L., Lloyd R. S. (1994) Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 269, 32709–32712 [PubMed] [Google Scholar]
- 5. Banerjee D., Mandal S. M., Das A., Hegde M. L., Das S., Bhakat K. K., Boldogh I., Sarkar P. S., Mitra S., Hazra T. K. (2011) Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 286, 6006–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dou H., Theriot C. A., Das A., Hegde M. L., Matsumoto Y., Boldogh I., Hazra T. K., Bhakat K. K., Mitra S. (2008) Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 283, 3130–3140 [DOI] [PubMed] [Google Scholar]
- 7. Wiederhold L., Leppard J. B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A. E., Izumi T., Prasad R., Wilson S. H., Mitra S., Hazra T. K. (2004) AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15, 209–220 [DOI] [PubMed] [Google Scholar]
- 8. Das A., Wiederhold L., Leppard J. B., Kedar P., Prasad R., Wang H., Boldogh I., Karimi-Busheri F., Weinfeld M., Tomkinson A. E., Wilson S. H., Mitra S., Hazra T. K. (2006) NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair 5, 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Das S., Chattopadhyay R., Bhakat K. K., Boldogh I., Kohno K., Prasad R., Wilson S. H., Hazra T. K. (2007) Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J. Biol. Chem. 282, 28474–28484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stucki M., Pascucci B., Parlanti E., Fortini P., Wilson S. H., Hübscher U., Dogliotti E. (1998) Mammalian base excision repair by DNA polymerases δ and ϵ. Oncogene 17, 835–843 [DOI] [PubMed] [Google Scholar]
- 11. Giloni L., Takeshita M., Johnson F., Iden C., Grollman A. P. (1981) Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J. Biol. Chem. 256, 8608–8615 [PubMed] [Google Scholar]
- 12. Henner W. D., Grunberg S. M., Haseltine W. A. (1983) Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J. Biol. Chem. 258, 15198–15205 [PubMed] [Google Scholar]
- 13. Weinfeld M., Mani R. S., Abdou I., Aceytuno R. D., Glover J. N. (2011) Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 36, 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caldecott K. W. (2001) Mammalian DNA single-strand break repair: an X-ra(y)ted affair. BioEssays 23, 447–455 [DOI] [PubMed] [Google Scholar]
- 15. Caldecott K. W. (2008) Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631 [DOI] [PubMed] [Google Scholar]
- 16. Jilani A., Ramotar D., Slack C., Ong C., Yang X. M., Scherer S. W., Lasko D. D. (1999) Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 274, 24176–24186 [DOI] [PubMed] [Google Scholar]
- 17. Karimi-Busheri F., Daly G., Robins P., Canas B., Pappin D. J., Sgouros J., Miller G. G., Fakhrai H., Davis E. M., Le Beau M. M., Weinfeld M. (1999) Molecular characterization of a human DNA kinase. J. Biol. Chem. 274, 24187–24194 [DOI] [PubMed] [Google Scholar]
- 18. Barry M. A., Eastman A. (1993) Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch. Biochem. Biophys. 300, 440–450 [DOI] [PubMed] [Google Scholar]
- 19. Pohjanpelto P., Hölttä E. (1996) Phosphorylation of Okazaki-like DNA fragments in mammalian cells and role of polyamines in the processing of this DNA. EMBO J. 15, 1193–1200 [PMC free article] [PubMed] [Google Scholar]
- 20. Mandavilli B. S., Santos J. H., Van Houten B. (2002) Mitochondrial DNA repair and aging. Mutat. Res. 509, 127–151 [DOI] [PubMed] [Google Scholar]
- 21. Richter C., Park J. W., Ames B. N. (1988) Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. U.S.A. 85, 6465–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaneko M., Inoue F. (1998) The sensitivity to DNA single strand breakage in mitochondria, but not in nuclei, of Chinese hamster V79 and variant cells correlates with their cellular sensitivity to hydrogen peroxide. Toxicol. Lett. 99, 15–22 [DOI] [PubMed] [Google Scholar]
- 23. Wallace D. C. (2002) Animal models for mitochondrial disease. Methods Mol. Biol. 197, 3–54 [DOI] [PubMed] [Google Scholar]
- 24. Copeland W. C. (2010) Understanding heterogeneous diseases in mtDNA maintenance. Methods 51, 363. [DOI] [PubMed] [Google Scholar]
- 25. de Souza-Pinto N. C., Wilson D. M., 3rd, Stevnsner T. V., Bohr V. A. (2008) Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair 7, 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bohr V. A. (2002) Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 32, 804–812 [DOI] [PubMed] [Google Scholar]
- 27. Hu J., de Souza-Pinto N. C., Haraguchi K., Hogue B. A., Jaruga P., Greenberg M. M., Dizdaroglu M., Bohr V. A. (2005) Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 280, 40544–40551 [DOI] [PubMed] [Google Scholar]
- 28. Chattopadhyay R., Wiederhold L., Szczesny B., Boldogh I., Hazra T. K., Izumi T., Mitra S. (2006) Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 34, 2067–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Longley M. J., Prasad R., Srivastava D. K., Wilson S. H., Copeland W. C. (1998) Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase γ and its role in mitochondrial base excision repair in vitro. Proc. Natl. Acad. Sci. U.S.A. 95, 12244–12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hegde M. L., Hegde P. M., Holthauzen L. M., Hazra T. K., Rao K. S., Mitra S. (2010) Specific Inhibition of NEIL-initiated repair of oxidized base damage in human genome by copper and iron: potential etiological linkage to neurodegenerative diseases. J. Biol. Chem. 285, 28812–28825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dickins R. A., Hemann M. T., Zilfou J. T., Simpson D. R., Ibarra I., Hannon G. J., Lowe S. W. (2005) Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37, 1289–1295 [DOI] [PubMed] [Google Scholar]
- 32. Patrick M. E., Adcock P. M., Gomez T. M., Altekruse S. F., Holland B. H., Tauxe R. V., Swerdlow D. L. (2004) Salmonella enteritidis infections, United States, 1985–1999. Emerg. Infect. Dis. 10, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silva J. M., Li M. Z., Chang K., Ge W., Golding M. C., Rickles R. J., Siolas D., Hu G., Paddison P. J., Schlabach M. R., Sheth N., Bradshaw J., Burchard J., Kulkarni A., Cavet G., Sachidanandam R., McCombie W. R., Cleary M. A., Elledge S. J., Hannon G. J. (2005) Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 37, 1281–1288 [DOI] [PubMed] [Google Scholar]
- 34. Kumar D., Ray A., Ray B. K. (2009) Transcriptional synergy mediated by SAF-1 and AP-1: critical role of N-terminal polyalanine and two zinc finger domains of SAF-1. J. Biol. Chem. 284, 1853–1862 [DOI] [PubMed] [Google Scholar]
- 35. Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., Gustafsson C. M. (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31, 289–294 [DOI] [PubMed] [Google Scholar]
- 36. Hazra T. K., Kow Y. W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S. K., Mitra S., Izumi T. (2002) Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 277, 30417–30420 [DOI] [PubMed] [Google Scholar]
- 37. Korhonen J. A., Pham X. H., Pellegrini M., Falkenberg M. (2004) Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taylor R. M., Whitehouse C. J., Caldecott K. W. (2000) The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 28, 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitehouse C. J., Taylor R. M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D. D., Weinfeld M., Caldecott K. W. (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104, 107–117 [DOI] [PubMed] [Google Scholar]
- 40. Hazra T. K., Izumi T., Maidt L., Floyd R. A., Mitra S. (1998) The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res. 26, 5116–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang H., Hays J. B. (2003) Mismatch repair in human nuclear extracts: effects of internal DNA-hairpin structures between mismatches and excision-initiation nicks on mismatch correction and mismatch-provoked excision. J. Biol. Chem. 278, 28686–28693 [DOI] [PubMed] [Google Scholar]
- 42. Santos J. H., Meyer J. N., Mandavilli B. S., Van Houten B. (2006) Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 314, 183–199 [DOI] [PubMed] [Google Scholar]
- 43. Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D. F., Demple B., Shen B. (2008) Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 32, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dou H., Mitra S., Hazra T. K. (2003) Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684 [DOI] [PubMed] [Google Scholar]
- 45. Gordon D. M., Wang J., Amutha B., Pain D. (2001) Self-association and precursor protein binding of Saccharomyces cerevisiae Tom40p, the core component of the protein translocation channel of the mitochondrial outer membrane. Biochem. J. 356, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Szczesny B., Tann A. W., Longley M. J., Copeland W. C., Mitra S. (2008) Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 283, 26349–26356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Ostman A., Landegren U. (2002) Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 [DOI] [PubMed] [Google Scholar]
- 48. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 49. Johansson H., Svensson F., Runnberg R., Simonsson T., Simonsson S. (2010) Phosphorylated nucleolin interacts with translationally controlled tumor protein during mitosis and with Oct4 during interphase in ES cells. PLoS One 5, e13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. (2008) Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Psarra A. M., Sekeris C. E. (2008) Nuclear receptors and other nuclear transcription factors in mitochondria: regulatory molecules in a new environment. Biochim. Biophys. Acta 1783, 1–11 [DOI] [PubMed] [Google Scholar]
- 52. Marchenko N. D., Zaika A., Moll U. M. (2000) Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275, 16202–16212 [DOI] [PubMed] [Google Scholar]
- 53. Das B. B., Dexheimer T. S., Maddali K., Pommier Y. (2010) Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 107, 19790–19795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bottero V., Rossi F., Samson M., Mari M., Hofman P., Peyron J. F. (2001) IκB-α, the NF-κB inhibitory subunit, interacts with ANT, the mitochondrial ATP/ADP translocator. J. Biol. Chem. 276, 21317–21324 [DOI] [PubMed] [Google Scholar]
- 55. De Bosscher K., Vanden Berghe W., Haegeman G. (2003) The interplay between the glucocorticoid receptor and nuclear factor-κB or activator protein-1: molecular mechanisms for gene repression. Endocr. Rev. 24, 488–522 [DOI] [PubMed] [Google Scholar]
- 56. Scovassi A. I. (2004) Mitochondrial poly(ADP-ribosylation): from old data to new perspectives. FASEB J. 18, 1487–1488 [DOI] [PubMed] [Google Scholar]
- 57. Mitra S., Izumi T., Boldogh I., Bhakat K. K., Hill J. W., Hazra T. K. (2002) Choreography of oxidative damage repair in mammalian genomes. Free Radic. Biol. Med. 33, 15–28 [DOI] [PubMed] [Google Scholar]
- 58. Date H., Onodera O., Tanaka H., Iwabuchi K., Uekawa K., Igarashi S., Koike R., Hiroi T., Yuasa T., Awaya Y., Sakai T., Takahashi T., Nagatomo H., Sekijima Y., Kawachi I., Takiyama Y., Nishizawa M., Fukuhara N., Saito K., Sugano S., Tsuji S. (2001) Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat. Genet. 29, 184–188 [DOI] [PubMed] [Google Scholar]
- 59. Moreira M. C., Barbot C., Tachi N., Kozuka N., Uchida E., Gibson T., Mendonça P., Costa M., Barros J., Yanagisawa T., Watanabe M., Ikeda Y., Aoki M., Nagata T., Coutinho P., Sequeiros J., Koenig M. (2001) The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat. Genet. 29, 189–193 [DOI] [PubMed] [Google Scholar]
- 60. Takashima H., Boerkoel C. F., John J., Saifi G. M., Salih M. A., Armstrong D., Mao Y., Quiocho F. A., Roa B. B., Nakagawa M., Stockton D. W., Lupski J. R. (2002) Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32, 267–272 [DOI] [PubMed] [Google Scholar]
- 61. Sykora P., Croteau D. L., Bohr V. A., Wilson D. M., 3rd (2011) Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 108, 7437–7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen J., Gilmore E. C., Marshall C. A., Haddadin M., Reynolds J. J., Eyaid W., Bodell A., Barry B., Gleason D., Allen K., Ganesh V. S., Chang B. S., Grix A., Hill R. S., Topcu M., Caldecott K. W., Barkovich A. J., Walsh C. A. (2010) Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 42, 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.