Background: IbpA and IbpB, the Escherichia coli sHsps, deoligomerize during heat shock to prevent irreversible protein aggregation.
Results: We analyzed the importance of N and C termini, conserved IEI motif, and arginine 133 for IbpA chaperone function.
Conclusion: All analyzed elements are required for IbpA chaperone function.
Significance: A new structural element important for chaperone activity, localized in the C terminus of sHsp, is suggested.
Keywords: Chaperone Chaperonin, Heat Shock Protein, Peptide Interactions, Protein Aggregation, Protein Folding, Protein-Protein Interactions, Protein Disaggregation
Abstract
Small heat shock proteins are ubiquitous molecular chaperones that, during cellular stress, bind to misfolded proteins and maintain them in a refolding competent state. Two members of the small heat shock protein family, IbpA and IbpB, are present in Escherichia coli. Despite 48% sequence identity, the proteins have distinct activities in promoting protein disaggregation. Cooperation between IbpA and IbpB is crucial for prevention of the irreversible aggregation of proteins. In this study, we investigated the importance of the N- and C-terminal regions of IbpA for self-oligomerization and chaperone functions. Deletion of either the N- or C-terminal region of IbpA resulted in a defect in the IbpA fibril formation process. The deletions also impaired IbpA chaperone function, defined as the ability to stabilize, in cooperation with IbpB, protein aggregates in a disaggregation-competent state. Our results show that the defect in chaperone function, observed in truncated versions of IbpA, is due to the inability of these proteins to interact with substrate proteins and consequently to change the properties of aggregates. At the same time, these versions of IbpA interact with IbpB similarly to the wild type protein. Competition experiments performed with the pC peptide, which corresponds to the IbpA C terminus, suggested the importance of IbpA intermolecular interactions in the stabilization of aggregates in a state competent for disaggregation. Our results suggest that these interactions are not only dependent on the universally conserved IEI motif but also on arginine 133 neighboring the IEI motif. IbpA mutated at arginine 133 to alanine lacked chaperone activity.
Introduction
When a cell is challenged by temperatures above the physiological level, it launches a heat shock response, increasing the synthesis of several chaperone proteins to adapt to the stressful conditions. These heat stress conditions also cause intracellular protein aggregation. The protein aggregates are eliminated by chaperone proteins when the temperature decreases. The disaggregation and refolding of polypeptides trapped in these aggregates depend on the collaborative action of chaperones from different families (1–6). These include small heat shock proteins (sHsps)3 (in Escherichia coli, IbpA and IbpB), the Hsp70 chaperone system (in E. coli, DnaK and its cochaperones DnaJ and GrpE), and the Hsp100 chaperones (in E. coli, ClpB). It has been proposed that sHsps associate with the aggregating polypeptides, thus changing their biochemical properties so that the subsequent Hsp100-Hsp70-mediated disaggregation process becomes much more efficient (1, 6–9).
Small Hsps are widely distributed in both prokaryotes and eukaryotes. Members of this diverse protein family are characterized by a low molecular mass (15- 43 kDa) and a conserved stretch of ∼100 amino acid residues. This domain displays sequence similarity to the vertebrate protein α-crystallin and therefore is called the α-crystallin domain (reviewed in Refs. 10–13).
The E. coli small heat shock proteins IbpA and IbpB were initially identified as proteins associated with inclusion bodies (inclusion body-associated protein) (14) and later shown to be present in aggregates formed by heat shock (15). The deletion of the ibpA and ibpB genes results in decreased viability manifested during prolonged growth at high temperatures and correlates with the increase in intracellular protein aggregation (16).
IbpB has been purified and shown to form 2–3-MDa polydisperse oligomers, which dissociate into smaller structures upon exposure to high temperatures (17, 18). The incubation of IbpB at a high temperature stimulates its chaperone activity, defined as protection against aggregation of insulin in reducing conditions (19). Recent studies determined that the N and C termini of IbpB are important for the association of IbpB into oligomeric structures and for its chaperone activity (18). Other studies showed that the presence of IbpB during substrate denaturation results in a higher efficiency of reactivation of polypeptides trapped in aggregates by the Hsp100-Hsp70 bi-chaperone system (4). However, these results were obtained at relatively low temperatures of substrate denaturation and high concentrations of IbpB.
The second E. coli sHsp, IbpA, has been less studied. It was shown to form a high molecular weight species and exhibit a limited protective effect on several tested substrate proteins subjected to thermal or oxidative stress (19). Recent studies showed that purified IbpA forms fibril-like structures (20).
IbpA and IbpB proteins are 48% identical to each other at the amino acid level (21), yet their roles in the disaggregation of protein aggregates are quite distinct (9). We reported that the presence of IbpA during heat denaturation of substrate is sufficient to change the macroscopic properties of aggregates, yet this alone does not increase the efficiency of the subsequent Hsp100-Hsp70-dependent reactivation of such aggregated polypeptides. The presence of IbpB is required to increase the efficiency of Hsp100-Hsp70-mediated disaggregation and refolding (9). This observation points out the cooperative mode of action of IbpA and IbpB, in which IbpA associates first with the aggregating substrate and attracts IbpB to the complex. This is in agreement with IbpA and IbpB cellular localization studies (16), which showed that in the presence of IbpA, IbpB is found in cell fractions containing aggregates and membranes, whereas in the absence of IbpA it is found in the cytoplasmic fraction. Also, recent studies of Butland et al. (22) on a protein-protein interaction network in E. coli determined that the pool of interacting partners of IbpA is much bigger than that of IbpB.
Here we analyzed the importance of both the N- and C-terminal regions of IbpA for self-oligomerization and chaperone function. Our results revealed that arginine 133 in the C terminus of IbpA, in addition to the conserved isoleucine motif IEI (amino acids 134–136), is involved in higher order structure formation and plays an important role in IbpA chaperone function.
MATERIALS AND METHODS
Proteins and Peptides
Published protocols were used for the purification of E. coli DnaK, DnaJ, GrpE, ClpB, IbpA, IbpB, and His-IbpB (5, 23, 24). Protein concentrations were determined with the Bio-Rad protein assay (based on the Bradford method) using bovine serum albumin as a standard. Molar concentrations are given assuming a hexameric structure for ClpB and a monomeric structure for the other proteins. Pfu polymerase, restriction enzymes, and other reagents for gene manipulation were obtained from Fermentas. Purified firefly luciferase (QuantiLum Recombinant Luciferase, catalog number E 1702) was purchased from Promega. Malate dehydrogenase (MDH) and anti-luciferase polyclonal rabbit antibodies were purchased from Sigma-Aldrich. Anti-IbpAB polyclonal rabbit antibodies were a kind gift from Dr. Ewa Laskowska (University of Gdańsk). The peptides were synthesized by a standard solid phase method on the 9050 Plus PepSynthesizer (Millipore). The peptides were purified by preparative reverse phase HPLC using a Luna C8 column (Phenomenex). The purity of the peptides was greater than 98%. The identity and purity of the peptides was confirmed by mass spectrometry.
Expression and Purification of IbpAΔN and IbpAΔC
The ibpAΔN11 and ibpAΔC11 genes were created by PCR using a pUC18-derived pCA plasmid (16) bearing the ibpA gene as a template. The amplified DNA fragments were cloned into the pET15 vector (Novagen) at the NcoI/BamHI sites. The obtained constructs were verified by DNA sequencing. IbpAΔN and IbpAΔC proteins were expressed in the E. coli BL21 (DE3) strain. The cells were grown in LB medium supplemented with ampicillin (100 μg/ml) at 37 °C. Protein expression was induced with 1 mm isopropyl β-d-thiogalactopyranoside at A600 = 0.6. The bacterial pellet was resuspended in buffer A (50 mm Tris-HCl, pH 7.4, 100 mm KCl, 10% (v/v) glycerol, 1 mm dithiothreitol) and disrupted by sonication. Further purification was performed according to the protocol for the wild type protein (5) with the following modification: after Q-Sepharose chromatography, fractions containing the IbpAΔN or IbpAΔC were pooled and loaded onto a hydroxyapatite column (Bio-Rad). Proteins bound to the resin were eluted with a linear gradient of 50- 300 mm phosphate buffer, pH 6.8.
Expression and Purification of IbpA R133A and IbpA AEA
The ibpA R133A and ibpA AEA mutations were generated with the QuikChangeTM site-directed mutagenesis kit (Stratagene) using a pET15b plasmid with the ibpA gene cloned in the NcoI/BamHI sites as a template. The obtained constructs were verified by sequencing. The expression and purification were performed as described above. Because both proteins were mainly found in the soluble fractions after lysis, the purification did not require urea in the buffers.
Luciferase Denaturation and Refolding Experiments
Firefly luciferase reactivation was performed as described previously (9). Luciferase (1.5 μm) was inactivated in buffer B (50 mm Tris-HCl, pH 7.4, 150 mm KCl, 20 mm magnesium acetate, 5 mm dithiothreitol) at 48 °C for 10 min in the absence or presence of IbpB (7.8 μm) and IbpA (2 μm) or various amounts of IbpA variants as indicated. For the experiments involving peptides, luciferase was thermally inactivated at 48 °C for 10 min in the absence or presence of IbpA (2 μm), IbpB (7.8 μm), and different concentrations of the peptides pC, pC-AEA, pC-R/A, pC-R/E, pN, and pX, as indicated. In refolding experiments, the samples were diluted to a final luciferase concentration of 0.04 μm in buffer B supplemented with ATP (5 mm), an ATP regeneration system, and chaperone proteins (1.5 μm DnaK, 0.1 μm DnaJ, 0.1 μm GrpE, 1.5 μm ClpB). Following a 1-h incubation at 25 °C, luciferase activity was determined in a Beckman scintillation counter using a luciferase assay system (Promega). The activity of renatured luciferase was compared with the activity of an equivalent amount of the native protein, diluted in buffer B supplemented with 0.2 mg/ml BSA, and kept on ice (the activity of the native enzyme was set as 100%).
MDH Denaturation and Refolding Experiments
MDH (2 μm) was heat-denatured for 30 min at 52 °C in buffer C (100 mm Tris-HCl, pH 7.4, 150 mm KCl, 10 mm dithiothreitol, 20 mm magnesium acetate) in the presence or absence of IbpA or its truncated variants (2 μm) and/or IbpB (7.8 μm), as indicated. After denaturation, MDH was diluted to a final concentration of 90 nm in buffer C supplemented with ATP (5 mm), an ATP regeneration system, and chaperone proteins (3 μm DnaK, 0.24 μm DnaJ, 0.3 μm GrpE, and 3 μm ClpB). The reactivation and disaggregation reaction was carried out for 1 h at 25 °C. MDH activity was measured at 25 °C in buffer D (150 mm potassium phosphate, pH 7.4, 2 mm dithiothreitol, 0.5 mm oxaloacetate, and 0.28 mm NADH). The time-dependent oxidation of NADH by MDH was monitored at 340 nm.
Electron Microscopy
To determine the morphology of sHsp or luciferase aggregates, IbpA, or its truncated variants (2 μm) was incubated with or without luciferase in buffer B for 10 min at 48 °C. To describe the effect of peptides on IbpA oligomerization and the morphology of luciferase aggregates formed in the presence of IbpA, the indicated components were incubated with the pC peptide or pC-AEA (1 mm), as above. Subsequently, the samples were applied on carbon coated copper grids. After allowing the proteins to adsorb for 3 min, excess solution was removed with filter paper. The grids were negatively stained with 1.5% uranyl acetate for 20 s. Electron micrographs were recorded at a nominal magnification of 15,500× using a Phillips CM 100 transmission electron microscope.
Sedimentation Experiments
To analyze sHsp oligomerization and sHsp-luciferase complex formation in the absence or presence of pC peptide or pC-AEA, the indicated components were incubated for 10 min at 48 °C in buffer B. After heat treatment, the samples were applied on a 3.5-ml 15- 45% (v/v) glycerol gradient in buffer B. The gradients were centrifuged at 4 °C in a Beckman SW 60 rotor at 40,000 rpm for 1.5 h, and following this, fractions were collected from the top. The protein pellet at the bottom of the tube was solubilized by incubation with Laemmli buffer at 95 °C. The proteins were separated by SDS-PAGE and visualized by Western blotting using anti-luciferase and anti-IbpAB antibodies.
Isolation of sHsp Complexes by Interaction with Ni-NTA-Agarose Resin
The experiments were performed essentially as described (5). His-IbpB (6 μm) was incubated with the indicated components: IbpA or its truncated variants (6 μm) and/or pC peptide (1 mm) in buffer E (50 mm Tris-HCl, pH 7.4, 150 mm KCl, 20 mm magnesium acetate) at different temperatures in a 30-μl reaction volume. After 20 min of incubation, Ni-NTA-agarose (20 μl) was added to the reaction mixture and incubated for an additional 10 min. Next, the supernatant was discarded, and the Ni-NTA-agarose was washed several times with buffer E supplemented with 20 mm imidazole. Proteins bound to the resin were eluted with the same buffer containing 300 mm imidazole and subjected to SDS-PAGE followed by staining with Coomassie Brilliant Blue.
RESULTS
N and C Termini of IbpA Are Essential for Its Oligomerization and Chaperone Function
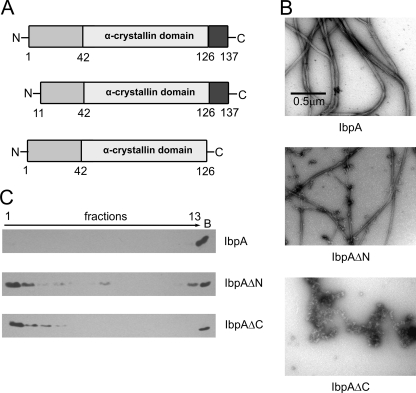
Jiao et al. (18) had shown that the 11 residues at both the N and the C termini of IbpB are highly flexible and important for association into oligomeric structures and are required for chaperone activity. Although IbpA and IbpB differently influence the substrate aggregation process (9), they are 48% identical at the amino acid level. Limited proteolysis experiments performed for IbpA compared with analogous results obtained for IbpB (Ref. 18 and results not shown) suggested that the N and C termini of both IbpA and IbpB have similar flexibility. With this information in hand, we prepared the two constructs, IbpAΔN and IbpAΔC (Fig. 1A), which result in proteins lacking the first or last 11 amino acid residues of IbpA, respectively. Both IbpA constructs still possess the intact α-crystallin domain. In the IbpAΔN mutant, only part of the N-terminal domain is deleted, whereas the change introduced in IbpAΔC removes the entire C-terminal domain (Fig. 1A).
FIGURE 1.
Deletion of the N or C terminus of IbpA impairs fibril formation. A, schematic presentation of IbpA and its two variants carrying a deletion in either the N or C terminus. B, representative electron micrographs of purified IbpA, IbpAΔN, and IbpAΔC. IbpA, IbpAΔN, or IbpAΔC (2 μm) was heat-treated at 48 °C for 10 min. Subsequently samples were stained with uranyl acetate and viewed by EM (nominal magnification, 15,500×). C, glycerol gradient sedimentation of IbpA, IbpAΔN, and IbpAΔC. IbpA, IbpAΔN, or IbpAΔC (2 μm) was heat-treated at 48 °C for 10 min, followed by sedimentation in a glycerol gradient. The fractions were collected from the top of the gradient, analyzed by SDS-PAGE, and visualized by Western blotting. Lane B shows proteins pelleted at the bottom of the tube.
Following purification, we examined the ability of these two truncated proteins to form fibrillar structures characteristic of IbpA. EM studies showed that IbpAΔC does not form fibrils. IbpAΔN forms fibril structures, yet the amount of observed structures was much less, and the fibrils were thinner than those observed for IbpA (Fig. 1B). To further evaluate the influence of the IbpA termini on the oligomeric state of IbpA, we analyzed the sedimentation properties of the truncated versions of IbpA. We used this technique instead of standard sizing chromatography because the IbpA fibrils interact with the chromatography resin. For both IbpAΔN and IbpAΔC, the majority of protein was found in the upper fractions, as opposed to the wild type IbpA, which, because of its fibrillar state, sedimented to the bottom of the tube (Fig. 1C). Under our experimental conditions, bacterial RNA polymerase, a multisubunit protein (β, β′, 2α; molecular mass, ∼400 kDa), sedimented to fraction 3 (25).
Next, we analyzed the chaperone activity of IbpAΔN and IbpAΔC. For this purpose, we thermally denatured the substrate luciferase alone or in the presence of IbpB and either IbpA, IbpAΔN, or IbpAΔC. Subsequently, the denatured luciferase was disaggregated and refolded by the ClpB-DnaK, DnaJ, GrpE bichaperone system. As previously reported (5), the presence of IbpA and IbpB during the luciferase denaturation step efficiently stabilized it in a disaggregation-competent state and enabled its subsequent reactivation (Fig. 2A). In contrast, neither IbpAΔN nor IbpAΔC accompanied by IbpB was able to stabilize luciferase in a disaggregation-competent state at any of the concentrations tested (Fig. 2A). The efficiency of the reactivation reaction was similar to that obtained for luciferase denatured in the absence of any sHsps (Fig. 2A). A similar result was obtained when malate dehydrogenase was used as a substrate instead of luciferase. Neither IbpAΔN nor IbpAΔC, in cooperation with IbpB, was able to stabilize malate dehydrogenase in a disaggregation-competent state (Fig. 2B).
FIGURE 2.
IbpAΔN and IbpAΔC do not promote reactivation of heat-inactivated substrates by the ClpB-KJE bichaperone system. A, luciferase (1.5 μm) was inactivated at 48 °C for 10 min in the presence of IbpAΔN or IbpAΔC (2 or 4 μm) and IbpB (7.8 μm), as indicated. Refolding of aggregated luciferase was started by shifting the samples to 25 °C and adding chaperones (DnaK, DnaJ, GrpE, and ClpB). After 1 h of incubation, luciferase activity was measured and normalized to the activity of the native protein (set as 100%). For comparison, luciferase was aggregated alone or in the presence of either IbpA (2 μm), IbpB (7.8 μm), or both IbpA (2 μm) and IbpB (7.8 μm), followed by subsequent reactivation, as stated above. The data are the means (± S.D.) of three independent experiments. B, MDH (2 μm) was heat-treated at 52 °C for 30 min alone, in the presence of either IbpA (2 μm), IbpB (7.8 μm), or both IbpB (7.8 μm) and wild type or mutated variants of IbpA (2 μm), as indicated. For disaggregation, the samples were supplemented with chaperones (DnaK, DnaJ, GrpE, and ClpB) and incubated for 1 h at 25 °C. Next, MDH activity was measured and compared with the activity of the equivalent amount of native enzyme (set as 100%). The data are the means (±S.D.) of three independent experiments.
Functional Analysis of N and C Termini Truncated IbpA Mutants
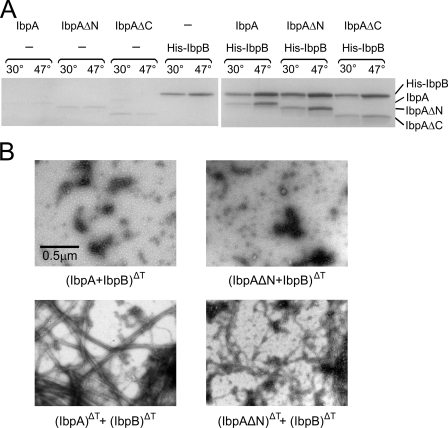
Our previous work (9) suggested that IbpA possesses at least two distinct activities: (i) the ability to interact with IbpB and (ii) the ability to associate with aggregating polypeptides. Thus, we were interested to know which of these two activities is impaired in IbpAΔN and IbpAΔC. First, the interaction of the two truncated IbpA mutants with IbpB was examined. We took advantage of the availability of hexahistidine-tagged IbpB (His-IbpB) and performed an affinity chromatography experiment. His-IbpB was incubated alone or in the presence of IbpA, IbpAΔN, or IbpAΔC at a low (30 °C) or high temperature (47 °C), followed by the addition of Ni-NTA-agarose. Proteins associated with the resin were analyzed by SDS-PAGE. Both IbpAΔC and IbpAΔN associated with Ni-NTA-agarose, similarly to IbpA, if incubated in the presence of His-IbpB (Fig. 3A). For all three proteins, the higher incubation temperature increased their association with IbpB. This experiment suggests that IbpAΔC and IbpAΔN interact with IbpB similarly to full-length IbpA.
FIGURE 3.
The N- and C-terminal regions of IbpA are not essential for interaction with IbpB. A, IbpA, IbpAΔN, or IbpAΔC was incubated in the absence or presence of His-IbpB at indicated temperatures, followed by the addition of Ni-NTA-agarose. After extensive washing, the proteins associated with the Ni-NTA-agarose were eluted with a high concentration of imidazole, subjected to SDS-PAGE, and stained with Coomassie Brilliant Blue. B, interaction between IbpAΔN and IbpB blocks the fibril formation. Representative electron micrographs of negatively stained complexes between IbpA or IbpAΔN with IbpB. IbpA or IbpAΔN (2 μm) were heat-treated in presence of IbpB (7.8 μm) at 48 °C for 10 min (upper panels). In the control experiments (lower panels), the indicated proteins were incubated separately as above. Afterward, the samples were cooled and combined together at room temperature. Nominal magnification, 15,500×.
We additionally analyzed the interaction between IbpAΔN and IbpB by EM. Because IbpB blocks the formation of IbpA fibrils (Fig. 3B and Ref. 20) and IbpAΔN is, to some extent, able to form fibrils, we assessed the morphology of IbpAΔN in the presence of IbpB by EM. When IbpA or IbpAΔN were incubated with IbpB at 48 °C, the fibrils were not observed (Fig. 3B). In control, the fibrils were detected when IbpAΔN and IbpB were heat-activated separately, cooled, and mixed at 25 °C. This result is in agreement with His-tagged affinity experiment (Fig. 3A) and confirms that IbpAΔN interacts with IbpB.
Because the deletion of either the N- or C-terminal part of IbpA does not influence its interaction with IbpB, we examined the ability of the two truncated proteins to change the properties of substrate aggregates during the denaturation process. For this purpose, the influence of IbpAΔN and IbpAΔC on the size and morphology of luciferase aggregates was analyzed under EM. When luciferase was heat-denatured alone at 48 °C, this resulted in the formation of large aggregates, positively stained with uranyl acetate (Fig. 4A). Heat denaturation of luciferase in the presence of IbpA resulted in the formation of a different type of aggregate. Not only were the aggregates much smaller, but they also resembled oligomeric structures and showed negative staining with uranyl acetate typical of proteins (Fig. 4A and Ref. 9). When IbpAΔN or IbpAΔC was used instead of IbpA, the size and morphology of luciferase aggregates did not change, and positively stained large aggregates were observed (Fig. 4A).
FIGURE 4.
The N- and C-terminal regions of IbpA are required for interaction with luciferase during its thermal denaturation. A, the presence of IbpAΔN or IbpAΔC during thermal denaturation of luciferase does not change the morphology of aggregates. Representative electron micrographs of negatively stained luciferase heat-treated in the presence of IbpA, IbpAΔN, or IbpAΔC. Luciferase (1.5 μm) was thermally inactivated (48 °C, 10 min) in the absence or presence of IbpA, IbpAΔN, or IbpAΔC (2 μm), as indicated. The samples were stained with uranyl acetate and viewed by EM. Nominal magnification, 15,500-. B, the presence of IbpAΔN or IbpAΔC during thermal denaturation does not change the size of luciferase aggregates. Luciferase (1.5 μm) was heat-treated ( 48 °C, 10 min) in the presence of IbpA, IbpAΔN, or IbpAΔC (2 μm) followed by sedimentation in a glycerol gradient. The fractions were collected from the top of the gradient. Every second fraction was analyzed by SDS-PAGE, followed by Western blotting to visualize the individual proteins.
To further verify the results obtained by EM, we analyzed the ability of IbpAΔN or IbpAΔC to change the sedimentation properties of luciferase aggregates. The conditions of the sedimentation analysis (15–45% glycerol gradient, 40,000 rpm, Beckman SW60 rotor, 1.5 h) were optimized to allow the differentiation of the size of aggregates (25). When luciferase was heat-denatured in the presence of IbpAΔN or IbpAΔC, it sedimented to the bottom of the tube (Fig. 4B). When wild type IbpA was used, a substantial portion of the luciferase aggregates sedimented more slowly and was found in the middle of the gradient. This was accompanied by the cosedimentation of IbpA to the same fractions (Fig. 4B). Sizing chromatography performed with the IbpA-luciferase complexes suggested that they cover a wide spectrum of masses with no apparent peak. Under the same experimental conditions, luciferase denatured in the absence of IbpA did not enter the chromatographic resin (result not shown).
All of the above experiments show that IbpAΔN and IbpAΔC, in contrast to wild type IbpA, are not able to change the size of luciferase aggregates. We conclude that the deletion of either the N or C terminus of IbpA results in the inability of the protein to stably interact with the substrate during the denaturation process.
Another approach taken to determine the importance of the N and C termini in IbpA function was based on competition experiments involving wild type IbpA and peptides corresponding to its N and C termini. Several peptides were synthesized: the pN peptide corresponding to the first 12 amino acids of IbpA (MRNFDLSPLYRS); the pC peptide corresponding to the last 12 amino acids (PEAKKPRRIEIN); pC-AEA, which is the pC peptide with two isoleucines replaced by alanines (PEAKKPRRAEAN); and the pX peptide (SSPGKPPRLVGGPMDA) corresponding to the 1–15-amino acid fragment of processed human cystatin C, which was used as an additional control. The pC-AEA peptide was synthesized because the IEI motif present in the C terminus of IbpA was suggested by studies of other sHsps (26–28) to be involved in the intermolecular interaction with the α-crystallin domain of another IbpA molecule in the oligomerization process.
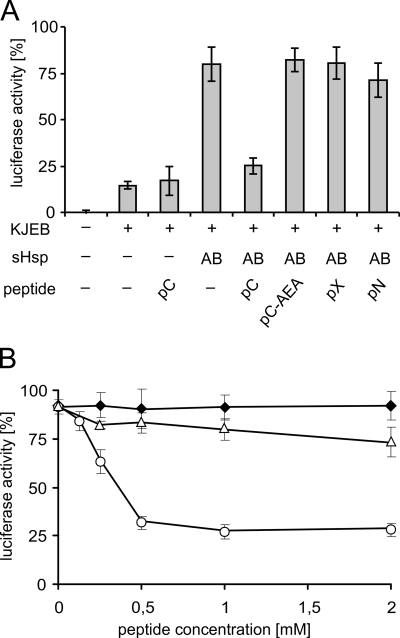
We first analyzed the influence of the peptides on the chaperone activity of IbpA and IbpB. For this purpose, we thermally denatured the luciferase substrate either alone or in the presence of IbpA, IbpB, and the peptides. Subsequently, the denatured luciferase was disaggregated and refolded by the ClpB-DnaK/DnaJ/GrpE bichaperone system. A substantial decrease in the amount of reactivated luciferase was observed for the reaction in which the pC peptide was present during the luciferase thermal denaturation step. No effect was observed with the other peptides at any concentration tested (Fig. 5A and results not shown). The inhibition of luciferase reactivation by the pC peptide may potentially result from (i) direct inhibition of the ClpB-DnaK/DnaJ/GrpE bichaperone system or (ii) the influence of the pC peptide on the quality of the luciferase aggregates formed in the presence of IbpA and IbpB. We ruled out the first possibility by using the luciferase assay, in which luciferase was denatured only in the presence of the pC peptide. In this case, the pC peptide does not inhibit the disaggregation activity of the ClpB-DnaK/DnaJ/GrpE chaperone system (Fig. 5A). To further verify that it is indeed the pC peptide that influences the quality of the luciferase aggregates formed in the presence of IbpA and IbpB, we performed an experiment in which we titrated the amount of peptide. Efficient inhibition of the luciferase reactivation reaction was observed with the pC peptide at a concentration of 0.5 mm (Fig. 5B), which is 250-fold higher than the concentration of IbpA protein in the assay. It must be remembered that the pC peptide competes with the C-terminal tails of interacting IbpA dimers. It was shown that the interaction between sHsp dimers also depends on the contact between α-crystallin domains (26). This may position the IbpA C-terminal tail in the neighborhood of its interaction site in the α-crystallin domain, and therefore the competing peptide must be present in a large excess to observe an effect. The inhibition was observed only when the pC peptide was present together with the IbpA and IbpB proteins during the luciferase heat denaturation step. The addition of the pC peptide following heat denaturation did not result in a decrease in the amount of reactivated luciferase (Fig. 5B), which also demonstrates that the pC peptide does not inhibit the disaggregation activity of the ClpB-DnaK/DnaJ/GrpE chaperone system acting on substrate-sHsps complexes. In the same experiment, we titrated the pC-AEA peptide and showed that its presence during the luciferase denaturation step inhibited the reactivation of luciferase very weakly (Fig. 5B). It is also worth noting that the presence of the pC peptide during the heat denaturation step decreases the amount of reactivated luciferase to a level similar to that observed when IbpA and IbpB are absent during the denaturation step (Fig. 5A). All of these results suggest that the pC peptide inhibits the activity of IbpA or possibly IbpB during the luciferase denaturation step, resulting in the formation of luciferase aggregates possessing limited competence for chaperone-dependent disaggregation.
FIGURE 5.
pC peptide present during heat inactivation of luciferase with IbpA/B inhibits subsequent luciferase reactivation by the ClpB-KJE bichaperone system. A, luciferase (1.5 μm) was inactivated at 48 °C for 10 min in the absence or presence of IbpA (2 μm) and IbpB (7.8 μm) and peptides (1 mm), as indicated. Refolding of aggregated luciferase was started by shifting the samples to 25 °C and adding chaperones (DnaK, DnaJ, GrpE, and ClpB), as indicated. After 1 h of incubation, luciferase activity was measured and normalized to the activity of the native protein (set as 100%). B, luciferase (1.5 μm) was heat-denatured in the presence of a fixed concentration of IbpA (2 μm), IbpB (7.8 μm), and increasing concentrations of pC (○) or pC-AEA (△). The reactivation was performed in the presence of the ClpB-KJE bichaperone system, as above. In the control experiment, pC peptide was added to the reaction following the heat denaturation procedure (♦). The data are the means (± S.D.) of three independent experiments.
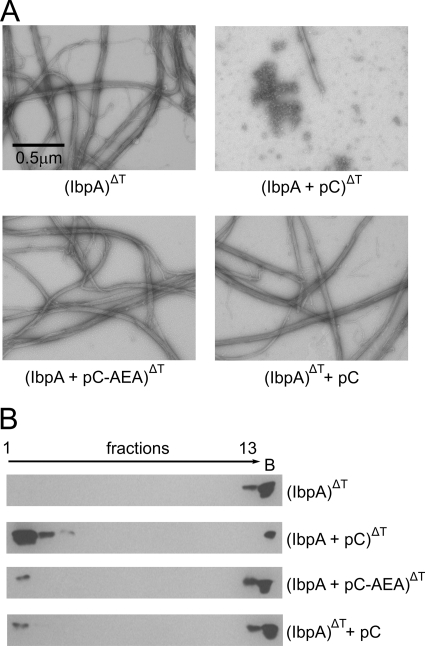
Next, we evaluated the influence of the pC peptide on the ability of IbpA to form fibrils. IbpA was incubated at 48 °C with or without pC or pC-AEA peptide and analyzed by EM. The incubation of pC peptide and IbpA at a high temperature strongly inhibited the formation of fibrils (Fig. 6A). The inhibition effect required the presence of both the peptide and IbpA during incubation of the sample at a high temperature, because IbpA fibrils were easily detected when IbpA and the pC peptide were heated separately and then mixed in at the later step. The observed inhibition of IbpA fibril formation is specific for the pC peptide; when the pC-AEA peptide was used instead, IbpA fibrils were indistinguishable from those observed for the wild type protein. Further analysis of the influence of the pC peptide on the oligomeric state of IbpA was performed by sedimentation. Incubation of the pC peptide with IbpA at a high temperature resulted in a major change in the sedimentation properties of IbpA. The majority of IbpA was found in the upper fractions of the gradient as opposed to the situation when IbpA was incubated at a high temperature alone, or with the pC-AEA peptide, or when the pC peptide and IbpA were incubated at a high temperature separately before mixing and centrifugation (Fig. 6B). All of these experiments show that the pC peptide specifically inhibits the formation of fibrillar structures by IbpA. Additionally these experiments point out the importance of the universally conserved IEI motif (26–28) for IbpA oligomerization.
FIGURE 6.
The pC peptide blocks the formation of IbpA fibrils. A, representative electron micrographs of IbpA heat-treated in the presence of the pC peptide or pC-AEA peptide. IbpA (2 μm) was heat-treated alone or in the presence of the pC or pC-AEA peptide (1 mm). In the control experiment (bottom right panel), following heat treatment of IbpA, the sample was cooled to room temperature and supplemented with pC peptide. The samples were stained with uranyl acetate and viewed by EM (nominal magnification, 15,500×) B, glycerol gradient sedimentation of IbpA heat-treated in the presence of pC or pC-AEA peptide. IbpA (2 μm) was heat-treated in the presence of pC or pC-AEA peptide (1 mm). In the control experiment, pC peptide was added after the sample, containing heat-treated IbpA, was cooled to room temperature. Following this, the samples were applied on glycerol gradients and sedimented. The fractions were collected from the top of the gradient, analyzed by SDS-PAGE, and then visualized by Western blotting. Lane B shows proteins pelleted at the bottom of the tube.
Because the pC peptide specifically impairs the ability of IbpA and IbpB to mediate the formation of luciferase aggregates competent for Hsp100-Hsp70-dependent disaggregation, we decided to examine which step of the reaction is influenced by the pC peptide. As suggested previously, two steps of the reaction can be distinguished: (i) interaction of IbpA with IbpB and (ii) association of IbpA with aggregating polypeptides. First, we analyzed the possible influence of the pC peptide on the interaction between IbpA and IbpB. We used hexahistidine-tagged IbpB and performed an affinity chromatography experiment. His-IbpB was incubated in the presence of IbpA and the pC peptide at a high temperature (47 °C), followed by the addition of Ni-NTA-agarose. The proteins associated with the resin were analyzed by SDS-PAGE. IbpA interacted with IbpB both in the absence and presence of the pC peptide (Fig. 7A), suggesting that the pC peptide does not influence this step of the reaction.
FIGURE 7.
The pC peptide does not disrupt interaction between IbpA and the IbpB cochaperone but inhibits the ability of IbpA to change the morphology of luciferase aggregates. A, the pC peptide does not inhibit complex formation between IbpA and IbpB. IbpA was incubated at 48 °C in the absence or presence of His-IbpB and the pC peptide, followed by the addition of Ni-NTA-agarose. After extensive washing, proteins bound to the resin were eluted with a high concentration of imidazole and subjected to SDS-PAGE, followed by staining with Coomassie Brilliant Blue. B, IbpA in the presence of the pC peptide is unable to change the morphology of luciferase aggregates. Representative electron micrographs of luciferase heat-treated in the presence of IbpA and the pC peptide. Luciferase (1.5 μm) was inactivated at 48 °C for 10 min alone or in the presence of IbpA (2 μm) in the absence or presence of the pC peptide (1 mm). In the control experiment (bottom right panel), following the denaturation of luciferase in the presence of IbpA, the sample was cooled to room temperature and supplemented with the pC peptide. The samples were stained with uranyl acetate and viewed by EM (nominal magnification, 15,500×).
Next, we performed EM studies to analyze the influence of the pC peptide on the ability of IbpA to change the size and morphology of luciferase aggregates. As shown previously (9), heat-induced luciferase aggregates obtained in the absence of IbpA are large and stain positively with uranyl acetate (Fig. 7B). Heat denaturation of luciferase in the presence of IbpA leads to the formation of much smaller, oligomeric structures, which showed negative staining with uranyl acetate, typical of proteins (Fig. 7B and Ref. 9). The addition of the pC peptide to the reaction containing luciferase and IbpA at a high temperature did not allow IbpA to change the size or morphology of luciferase aggregates. The aggregates viewed by EM appeared the same as those obtained in the absence of IbpA (Fig. 7B). When the pC peptide was added to the reaction at room temperature after the heat denaturation of luciferase in the presence of IbpA, it did not influence the size and morphology of aggregates. Thus, small aggregates negatively stained with uranyl acetate were still observed (Fig. 7B). This experiment shows that the pC peptide, when present during the heat denaturation step, efficiently blocks the ability of IbpA to change the size and morphology of aggregating substrate protein.
The Arg Residue Neighboring the IEI Motif in the C Terminus of IbpA Is Important for Chaperone Function and Higher Order Structure Formation
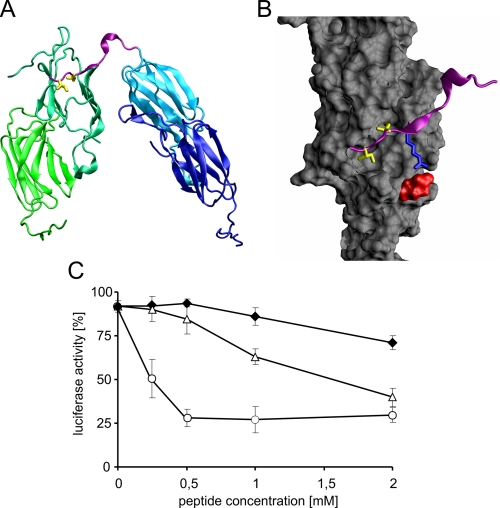
We analyzed the importance of the C terminus and the IEI motif in IbpA by in silico studies. Homology modeling followed by molecular dynamics studies enabled us to build a model of two interacting IbpA dimers (Fig. 8A). In this model, the IEI motif in IbpA was found to be involved in interactions with the hydrophobic pocket (phenylalanine 56, isoleucines 63 and 119, alanine 65, and leucines 70, 112, and 117) at the edge of the α-crystallin domain in the other IbpA molecule (Fig. 8B). We noticed the possible involvement of arginine (position 133 in the IbpA amino acid sequence), found at position −1 with respect to the IEI motif, which in our model makes close contact with glutamic acid at position 62 in the α-crystallin domain (Fig. 8B). Such an interaction might be important for the proper positioning of the C-terminal region of IbpA within the interacting surface of the α-crystallin domain. To verify the importance of arginine 133 for the chaperone activity of IbpA, we synthesized two additional peptides, pC-R/A (PEAKKPRAIEIN) and pC-R/E (PEAKKPREIEIN), in which the arginine was replaced by alanine or glutamic acid, respectively. Next, we analyzed the ability of these peptides to inhibit the chaperone activity of IbpA and IbpB during the luciferase denaturation step and compared the results with those obtained for the pC peptide. For both the pC-R/A and pC-R/E peptides, the observed inhibition of chaperone activity of IbpA and IbpB was minor compared with that of the pC peptide (Fig. 8C). The pC peptide at 0.5 mm concentration efficiently inhibited the ability of IbpA and IbpB to stabilize the luciferase in a disaggregation-competent state, preventing subsequent reactivation, whereas the pC-R/A and pC-R/E peptides at the same concentration did not inhibit this reaction (Fig. 8C). Only at higher concentrations of pC-R/A but not pC-R/E was the inhibitory effect observed. These experiments suggest the importance of arginine 133 for the proper positioning of the C terminus of IbpA during the intermolecular interaction with the α-crystallin domain of another IbpA molecule in the oligomerization process.
FIGURE 8.
Arginine 133 in the pC peptide is critical for the ability of the peptide to inhibit luciferase reactivation by the ClpB-KJE bichaperone system. A, arrangement of dimers interacting through the C-terminal IEI motif. The C-terminal fragment marked in pink corresponds to the length of peptides used in this study. Isoleucines 134 and 136 in the IEI motif are marked yellow. B, details of C-terminal fragment binding to the hydrophobic pocket in the α-crystallin domain (colored gray). Arg-133, shown in blue, forms a salt bridge with Glu-62 (red surface); isoleucines 134 and 136 (yellow) are buried in the hydrophobic pocket. Homology model of the IbpA protein was obtained based on the Xanthomonas axonopodis HspA crystal structure (Protein Data Bank entry 3GLA) using the Swissmodel server (34) and assembled as interacting dimers by fitting the subunits onto the oligomeric structure of M. jannaschii Hsp16.5 (Protein Data Bank entry 1SHS) using Spdbv software. The oligomeric model was then subjected to energy minimization and 10 cycles of simulated annealing in Gromacs (35). The structures were drawn with the VMD program (36). C, luciferase (1.5 μm) was heat-denatured in the presence of fixed concentrations of IbpA (2 μm) and IbpB (7.8 μm) and increasing concentrations of pC (○), pC-R/A (△), or pC-R/E (♦). The refolding of aggregated luciferase was started by shifting the samples to 25 °C and adding chaperones (DnaK, DnaJ, GrpE, and ClpB). After 1 h of incubation, luciferase activity was measured and normalized to the activity of the native protein (set as 100%). The data are the means (± S.D.) of three independent experiments.
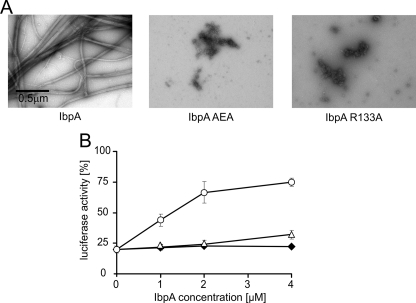
To directly analyze the importance of arginine 133 for IbpA activity, we used site-directed mutagenesis to change this arginine into alanine. We also mutagenized the neighboring IEI motif by changing both isoleucines into alanines. Both mutated proteins were purified and then analyzed. Neither IbpA R133A nor IbpA AEA formed the characteristic IbpA fibrils (Fig. 9A). When either IbpA R133A or IbpA AEA was present with IbpB during heat denaturation of luciferase, no increase in ClpB-DnaK/DnaJ/GrpE chaperone disaggregation activity was observed. Neither IbpA R133A nor IbpA AEA accompanied by IbpB was able to stabilize luciferase in a disaggregation-competent state at any of the concentrations tested (Fig. 9B). The efficiency of the reactivation reaction was similar to that obtained for luciferase denatured in the absence of any sHsps (Fig. 9B). These results show that, in addition to the IEI motif, the neighboring arginine 133 is important for the chaperone function of IbpA.
FIGURE 9.
Both arginine 133 and the IEI motif are important for IbpA chaperone activity and fibril formation. A, representative electron micrographs of purified IbpA, IbpA R133A, and IbpA AEA. IbpA, IbpA R133A, or IbpA AEA (2 μm) was heat-treated at 48 °C for 10 min. Subsequently the samples were stained with uranyl acetate and viewed by EM (nominal magnification, 15,500×). B, IbpA R133A and IbpA AEA do not promote reactivation of heat-inactivated luciferase by the ClpB-KJE bichaperone system. Luciferase (1.5 μm) was inactivated at 48 °C for 10 min in the presence of different concentrations of IbpA (○) or IbpA R133A (△), or IbpA AEA (♦) and a constant concentration of IbpB (7.8 μm). Refolding of aggregated luciferase was started by shifting the samples to 25 °C and adding chaperones (DnaK, DnaJ, GrpE, and ClpB). After 1 h of incubation, luciferase activity was measured and normalized to the activity of the native protein (set as 100%). The data are the means (± S.D.) of three independent experiments.
DISCUSSION
Here we have investigated the role of the N- and C-terminal regions of IbpA, the E. coli small Hsp, for self-oligomerization and chaperone function. The chaperone activity of IbpA and its truncated versions was analyzed in a system requiring six chaperone proteins for the reactivation of the heat-aggregated substrate. Two of these chaperones, namely IbpA and IbpB, cooperate at the substrate denaturation step, and their presence is required for the formation of disaggregation-competent substrate aggregates. Four others, namely ClpB, DnaK, DnaJ, and GrpE, are necessary at the disaggregation and reactivation step. The deletion of either the N- or C-terminal region of IbpA resulted not only in the defect in fibril formation specific for IbpA but also impaired the chaperone activity of IbpA. Experiments performed in the absence of the Hsp100-Hsp70 chaperone system show that the observed defect in the chaperone function of the N- or C-terminally truncated versions of IbpA is due to the inability of these proteins to change the properties of aggregates. At the same time, these versions of IbpA interact with IbpB in a manner similar to the wild type IbpA.
The importance of the N terminus for oligomerization and substrate binding was previously reported for other sHsps (18, 28–32). Recently, Jaya et al. (28), using a cross-linking approach, showed that several hydrophobic amino acids in the N-terminal arm of sHsp18.1 from pea form strong contacts with two substrates, firefly luciferase and malate dehydrogenase. These authors also showed that defined regions of the α-crystallin domain and C-terminal extension are able to form substrate specific cross-links, but at substantially lower intensity compared with those of the N-terminal region (28). It was additionally reported that switching the N-terminal arms between two closely related plant sHsps effectively switches the relative chaperone activity of these two proteins with the substrates luciferase and citrate synthase (29). The N-terminal domain of the yeast Saccharomyces cerevisiae Hsp26 chaperone was also shown to be essential for both the association with non-native proteins and formation of oligomeric complexes (32). Thus, our result, showing that IbpA with a partial deletion of the N-terminal region is not able to change the properties of luciferase aggregates, is in agreement with those reported for other sHsps. It is also worth mentioning that the peptide pN, corresponding to the first 12 amino acids of IbpA, does not inhibit the chaperone activity of IbpA, which suggests that (i) the pN peptide itself is not able to stably interact with the substrate and titrate the possible interactions sites on it or (ii) the peptide does not stably interact with IbpA, which would, in turn, block its activity. This suggests that it is rather the integrity of the N-terminal part of IbpA that is important for its functions.
The importance of the C-terminal part of sHsps for their chaperone activity has been less studied. Structural studies showed that the C terminus contains the consensus sequence (I/V)X(I/V), which plays a crucial role in the formation of sHsp oligomers by interacting with the hydrophobic pocket formed at the edge of the α-crystallin domain in the sHsp molecule being bound (26, 27). Our experimental data are in good agreement with these results. Not only was IbpAΔC unable to form fibrillar structures characteristic of IbpA, but also the competition experiments performed with the pC peptide containing the IEI motif showed the efficient inhibition of IbpA fibril formation. The latter process requires the presence of the pC peptide during incubation of IbpA at a high temperature. Thus, we can speculate that an increase in temperature triggers the IbpA defibrillation process, disrupting the interaction between the IEI motif of one IbpA molecule and the α-crystallin domain of the other. This enables the interaction of the competing pC peptide with the α-crystallin domain. Binding of the pC peptide to the hydrophobic pocket in the α-crystallin domain blocks the interaction between the neighboring IbpA molecule and the hydrophobic pocket, resulting in inhibition of the IbpA fibrillation process. Replacing the IEI motif in peptide pC with the AEA motif reverses the inhibition of fibril formation, pointing out the importance of the IEI motif for IbpA oligomerization.
Our results show that, in addition to the universal (I/V)X(I/V) motif, the positively charged amino acids neighboring the motif are involved in the higher order structure formation mediated by the C termini of sHsps. Arginine 133 neighboring the IEI motif of IbpA appears to be important for the chaperone activity of IbpA, as shown by our experimental and in silico studies. Our model showed close contact of this arginine with glutamic acid at position 62 in the α-crystallin domain (Fig. 8B), which suggested its importance for the proper positioning of the C-terminal region of IbpA within the α-crystallin domain of the interacting molecule. In agreement, the mutated peptides pC-R/A and pC-R/E in which arginine 133 was replaced by either alanine or glutamic acid, respectively, were severely impaired in the ability to inhibit the IbpA/IbpB chaperone activity as compared with the pC peptide when allowed to compete with the C-terminal tail of IbpA. Thus, our results suggest that, in addition to the IEI motif, the neighboring arginine plays an important role in the interaction of the IbpA C terminus with the α-crystallin domain. Note that peptides pC-R/A and pC-R/E were nearly similar to pC-AEA, lacking the IEI motif, in their inability to inhibit the IbpA/IbpB chaperone activity. These observations were further confirmed by analysis of the IbpA mutants in which either arginine 133 or the IEI motif were changed into alanines. Neither IbpA R133A nor IbpA AEA forms the characteristic IbpA fibrils, and neither possesses chaperone activity. These results point out the importance of both structural elements for the interaction of the C-terminal tail with the α-crystallin domain. Because the contribution of a charged amino acid for the proper positioning of the sHSP C terminus has not been previously addressed experimentally, we analyzed two published crystal structures of oligomeric sHsps (26, 33). A similar contact between lysine 141 in position −3 with respect to the IXI motif, and glutamic acid (position 78) in the α-crystallin domain was observed in Hsp16.5 from Methanococcus jannaschii (26). There is also a contact between lysine 145, −2 with respect to the IXI motif, and glutamic acid (position 119) in the α-crystallin domain of sHsp 16.9 from wheat (33). Additionally, the analysis of amino acid sequences of sHsps revealed a high degree of conservation of positively charged amino acids in position −1 with respect to the IXI motif in the C termini of sHsps. In 618 analyzed sequences of sHsps from proteobacteria, arginine was present in 45% and lysine was present in 30% at this position.
The fact that the pC peptide also specifically inhibits the process in which IbpA changes the morphology of luciferase aggregates at a high temperature suggests the involvement of the C termini of IbpA in this process. However, it does not necessarily mean that the analyzed C-terminal region of IbpA or the hydrophobic pocket at the edge of the α-crystallin domain is directly involved in the interaction with substrate. The defect might be due to the fact that the presence of the competing pC peptide during substrate aggregation at high temperature interferes with the formation of higher order structures, which depends on the interaction between dimers of IbpA, which in turn involves the C termini of IbpA. Such interactions might be required for the formation of substrate aggregates competent for disaggregation.
Interestingly, when the pN peptide was used instead of pC in similar competition experiments, it was not able to inhibit either the oligomerization or chaperone activity of IbpA, even though IbpAΔN was impaired in fibril formation and chaperone activity similarly to IbpAΔC. These observations suggest that the N- and C-terminal regions of IbpA play unique roles with respect to its biochemical properties and chaperone functions.
In total, the results presented here demonstrate that the mechanism by which IbpA associates with the substrate in the presence of IbpB, its cochaperone, and prevents the irreversible aggregation of the substrate during heat denaturation is complex. Both the N- and C-terminal ends are required for its proper functioning. We hypothesize that the N terminus is involved in the interaction with the substrate, whereas the C terminus is important for the temperature-dependent changes in the oligomeric status of IbpA. In addition to the universally conserved IEI motif present in the C terminus of IbpA, arginine 133 at position −1 to this motif was shown to be important for chaperone activity and oligomerization of IbpA.
Acknowledgments
We thank Dr. Debbie Ang for discussions and critical reading of the manuscript. We thank Szymon Żwirowski for analysis of amino acid conservation and discussions.
This work was supported by Polish Ministry of Science and Higher Education Grant N N301 197239.
- sHsp
- small heat shock protein
- MDH
- malate dehydrogenase
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Mogk A., Schlieker C., Friedrich K. L., Schönfeld H. J., Vierling E., Bukau B. (2003) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278, 31033–31042 [DOI] [PubMed] [Google Scholar]
- 2. Haslbeck M., Miess A., Stromer T., Walter S., Buchner J. (2005) Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J. Biol. Chem. 280, 23861–23868 [DOI] [PubMed] [Google Scholar]
- 3. Lee G. J., Vierling E. (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 122, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mogk A., Deuerling E., Vorderwülbecke S., Vierling E., Bukau B. (2003) Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50, 585–595 [DOI] [PubMed] [Google Scholar]
- 5. Matuszewska M., Kuczyńska-Winik D., Laskowska E., Liberek K. (2005) The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J. Biol. Chem. 280, 12292–12298 [DOI] [PubMed] [Google Scholar]
- 6. Veinger L., Diamant S., Buchner J., Goloubinoff P. (1998) The small heat shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J. Biol. Chem. 273, 11032–11037 [DOI] [PubMed] [Google Scholar]
- 7. Lee G. J., Roseman A. M., Saibil H. R., Vierling E. (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ehrnsperger M., Gräber S., Gaestel M., Buchner J. (1997) Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 16, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ratajczak E., Zietkiewicz S., Liberek K. (2009) Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J. Mol. Biol. 386, 178–189 [DOI] [PubMed] [Google Scholar]
- 10. Narberhaus F. (2002) α-Crystallin-type heat shock proteins. Socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 66, 64–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Montfort R., Slingsby C., Vierling E. (2001) Structure and function of the small heat shock protein/α-crystallin family of molecular chaperones. Adv. Protein Chem. 59, 105–156 [DOI] [PubMed] [Google Scholar]
- 12. Haslbeck M. (2002) sHsps and their role in the chaperone network. Cell Mol. Life Sci. 59, 1649–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005) Some like it hot. The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12, 842–846 [DOI] [PubMed] [Google Scholar]
- 14. Allen S. P., Polazzi J. O., Gierse J. K., Easton A. M. (1992) Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J. Bacteriol. 174, 6938–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laskowska E., Wawrzynów A., Taylor A. (1996) IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie 78, 117–122 [DOI] [PubMed] [Google Scholar]
- 16. Kuczyńska-Winik D., Kedzierska S., Matuszewska E., Lund P., Taylor A., Lipińska B., Laskowska E. (2002) The Escherichia coli small heat-shock proteins IbpA and IbpB prevent the aggregation of endogenous proteins denatured in vivo during extreme heat shock. Microbiology 148, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 17. Shearstone J. R., Baneyx F. (1999) Biochemical characterization of the small heat shock protein IbpB from Escherichia coli. J. Biol. Chem. 274, 9937–9945 [DOI] [PubMed] [Google Scholar]
- 18. Jiao W., Qian M., Li P., Zhao L., Chang Z. (2005) The essential role of the flexible termini in the temperature-responsiveness of the oligomeric state and chaperone-like activity for the polydisperse small heat shock protein IbpB from Escherichia coli. J. Mol. Biol. 347, 871–884 [DOI] [PubMed] [Google Scholar]
- 19. Kitagawa M., Miyakawa M., Matsumura Y., Tsuchido T. (2002) Escherichia coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. Eur. J. Biochem. 269, 2907–2917 [DOI] [PubMed] [Google Scholar]
- 20. Ratajczak E., Strózecka J., Matuszewska M., Zietkiewicz S., Kuczyńska-Winik D., Laskowska E., Liberek K. (2010) IbpA the small heat shock protein from Escherichia coli forms fibrils in the absence of its cochaperone IbpB. FEBS Lett. 584, 2253–2257 [DOI] [PubMed] [Google Scholar]
- 21. Chuang S. E., Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. (1993) Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene 134, 1–6 [DOI] [PubMed] [Google Scholar]
- 22. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 23. Zylicz M., Ang D., Liberek K., Georgopoulos C. (1989) Initiation of λ DNA replication with purified host- and bacteriophage-encoded proteins. The role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 8, 1601–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woo K. M., Kim K. I., Goldberg A. L., Ha D. B., Chung C. H. (1992) The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J. Biol. Chem. 267, 20429–20434 [PubMed] [Google Scholar]
- 25. Zietkiewicz S., Lewandowska A., Stocki P., Liberek K. (2006) Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J. Biol. Chem. 281, 7022–7029 [DOI] [PubMed] [Google Scholar]
- 26. Kim K. K., Kim R., Kim S. H. (1998) Crystal structure of a small heat-shock protein. Nature 394, 595–599 [DOI] [PubMed] [Google Scholar]
- 27. Giese K. C., Vierling E. (2004) Mutants in a small heat shock protein that affect the oligomeric state. Analysis and allele-specific suppression. J. Biol. Chem. 279, 32674–32683 [DOI] [PubMed] [Google Scholar]
- 28. Jaya N., Garcia V., Vierling E. (2009) Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc. Natl. Acad. Sci. U.S.A. 106, 15604–15609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Basha E., Friedrich K. L., Vierling E. (2006) The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 281, 39943–39952 [DOI] [PubMed] [Google Scholar]
- 30. Fu X., Zhang H., Zhang X., Cao Y., Jiao W., Liu C., Song Y., Abulimiti A., Chang Z. (2005) A dual role for the N-terminal region of Mycobacterium tuberculosis Hsp16.3 in self-oligomerization and binding denaturing substrate proteins. J. Biol. Chem. 280, 6337–6348 [DOI] [PubMed] [Google Scholar]
- 31. Leroux M. R., Melki R., Gordon B., Batelier G., Candido E. P. (1997) Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J. Biol. Chem. 272, 24646–24656 [DOI] [PubMed] [Google Scholar]
- 32. Stromer T., Fischer E., Richter K., Haslbeck M., Buchner J. (2004) Analysis of the regulation of the molecular chaperone Hsp26 by temperature-induced dissociation. The N-terminal domail is important for oligomer assembly and the binding of unfolding proteins. J. Biol. Chem. 279, 11222–11228 [DOI] [PubMed] [Google Scholar]
- 33. van Montfort R. L., Basha E., Friedrich K. L., Slingsby C., Vierling E. (2001) Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 8, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 34. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace. A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 35. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 36. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]