Background: New antimicrobial targets and compounds against resistant pathogens are urgently needed.
Results: Exometabolomic and enzymologic studies identified the antimicrobial compound triphenylbismuthdichloride as an efficient inhibitor of the bacterial pyruvate dehydrogenase complex (PDHC).
Conclusion: The bacterial PDHC has attractive properties as an antimicrobial target.
Significance: We suggest that metabolomics can be very useful for studying the modes of action of antimicrobial compounds.
Keywords: Antibiotic Action, Bacterial Metabolism, Metabolomics, Pyruvate Dehydrogenase Complex, Staphylococcus aureus
Abstract
The desperate need for new therapeutics against notoriously antibiotic-resistant bacteria has led to a quest for novel antibacterial target structures and compounds. Moreover, defining targets and modes of action of new antimicrobial compounds remains a major challenge with standard technologies. Here we characterize the antibacterial properties of triphenylbismuthdichloride (TPBC), which has recently been successfully used against device-associated infections. We demonstrate that TPBC has potent antimicrobial activity against many bacterial pathogens. Using an exometabolome profiling approach, a unique TPBC-mediated change in the metabolites of Staphylococcus aureus was identified, indicating that TPBC blocks bacterial pyruvate catabolism. Enzymatic studies showed that TPBC is a highly efficient, uncompetitive inhibitor of the bacterial pyruvate dehydrogenase complex. Our study demonstrates that metabolomics approaches can offer new avenues for studying the modes of action of antimicrobial compounds, and it indicates that inhibition of the bacterial pyruvate dehydrogenase complex may represent a promising strategy for combating multidrug-resistant bacteria.
Introduction
Antibiotics have been extremely useful for combating bacterial infections, but the currently available compounds are becoming increasingly ineffective because of the rising numbers of bacterial strains resistant to most or even all clinically available antibiotics. Methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci (VRE),5 enterobacteria with extended-spectrum β-lactamases, multidrug-resistant Mycobacterium tuberculosis, and fluoroquinolone-resistant Clostridium difficile have dramatically increased over the last years with regular outbreaks that are hardly controllable even with strict hygiene regimes (1–3).
In recent years, it has become difficult to identify new inhibitors for the currently available targets, such as peptidoglycan biosynthetic enzymes, ribosome, gyrase, or folic acid biosynthesis. In addition, many genomics-based approaches for identifying novel targets for antibiotics have been disappointing. More sophisticated screening strategies have yielded some promising results recently (4–8), and new therapeutic concepts, such as the development of anti-virulence drugs (9, 10), the induction of bacterial programmed cell death (11), or the use of host-encoded defense peptides (12, 13), are increasingly the focus of research efforts. Nevertheless, of the small numbers of new antibiotics awaiting approval for clinical use in the near future, most are variants of ancient antibiotic classes (14, 15).
Although finding new antimicrobial compounds is a very challenging task, identifying the mode of antimicrobial action has been equally difficult. This holds particularly true for antibiotics with strong bactericidal activities that lead to an almost simultaneous halt of most vital processes in a bacterial cell, thereby limiting possibilities for elucidating what triggered the event. Genome-wide transcriptional or translational profiles are increasingly used to define signatures of gene or protein expression that are indicative of a particular type of target (16–18). However, such profiles often represent an indirect stress response rather than a direct consequence of the inhibitory event, which limits their use for identifying the mode of action of unknown compounds against new target structures (19). Metabolomics is a novel technology permitting a simultaneous qualitative and quantitative analysis of small metabolic intermediates and products by NMR- or MS-based techniques, which allows for a direct view of changes in vital metabolic pathways. Studying the total sets of extracellular metabolites has been defined as exometabolome profiling (quantitative) or footprinting (qualitative) (20). Although metabolomic analyses have so far mostly been used to study human diseases (21, 22) or to monitor metabolic changes in bacteria during different environmental conditions and in mutants (23–26), such strategies have only rarely been used to elucidate the modes of action of new antimicrobial compounds (27).
In this study, we analyzed antibacterial activities of the synthetic compound triphenylbismuthdichloride (TPBC), which has proven efficacy in preventing catheter-associated infections (28, 29). TPBC was found to have strong antimicrobial activity against major bacterial pathogens, among them many antibiotic-resistant strains, such as methicillin-resistant S. aureus and VRE. Using combined exometabolomic and enzymologic approaches, TPBC was shown to block the bacterial pyruvate dehydrogenase complex (PDHC), thereby abrogating central metabolic activities.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strains used in this work include S. aureus Sa113, S. aureus COL (methicillin-resistant S. aureus), S. aureus Newman, Staphylococcus epidermidis O-47, Bacillus subtilis DB2, Enterococcus faecalis VRE366, Enterococcus faecium VRE392, Pseudomonas aeruginosa MPA01, and Escherichia coli K12. Cultures prepared for metabolomic analyses were grown in modified RPMI 1640 medium (Sigma R7509) containing 2 mm l-glutamine and trace elements (69 μg/liter ZnCl2, 99 μg/liter MnCl3·4H2O, 6 μg/liter H3BO3, 350 μg/liter CoCl2, 2 μg/liter CuCl2, 24 μg/liter NiCl2·6H2O, and 36 μg/liter Na2MoO4·2H2O). For all other experiments, strains were grown in BM medium, containing 1% casein peptone, 0.5% yeast extract, 0.5% NaCl, 0.01% K2HPO4·3H2O, and 0.1% glucose.
Determination of Antimicrobial Activities and Toxicity
Antibiotic stock solutions were prepared with appropriate solvents (e.g. TPBC was dissolved in DMSO based on its relatively high octanol/water coefficient log P value of 6.94 (see the TDR Targets Database Web site). Bacterial control cultures were incubated with equivalent amounts of corresponding solvent. Minimal inhibitory concentrations (MICs) for bacterial strains were determined using a standard protocol (30). Briefly, ∼106 bacteria were added to 5 ml of BM containing serially diluted inhibitory molecules. Cultures were incubated at 37 °C and shaken for 24 h, and A578 was measured to determine antibiotic concentrations leading to 50% reduced growth. Human monocytic MM6 cells were precultured in VLE RPMI 1640 medium (Biochrom AG) containing 1× nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mm oxaloacetate, 0.45 mm pyruvate, and 0.2 units/ml insulin. After 48 h of incubation, cells were harvested by centrifugation (250 × g for 5 min at room temperature) and resuspended in an appropriate volume of fresh medium. ∼1–2 × 107 cells/ml were incubated for 3 h with increasing concentrations of inhibitor, and the percentage of dead cells was calculated after trypan blue staining and subsequent counting of stained and non-stained cells. IC50 values were defined as antibiotic concentrations that lead to 50% reduction in viability.
Quantification of Extracellular Metabolites by 1H NMR
Culture supernatants of untreated and treated S. aureus cultures were prepared as described above. Antibiotics were added when A578 = 0.5 was reached, and this time point was defined as the starting point (0 h). Supernatants were filter-sterilized (0.22-μm pore size). The samples were then prepared for 1H NMR analysis as described previously (31). Briefly, 200 μl of phosphate buffer (pH 7.0, 0.2 m) were mixed with 400 μl of sample in a 5-mm NMR tube, and spectra were recorded using a Bruker®Avance II 600-MHz spectrometer operating with TOPSPIN 2.0 (Bruker®Biospin). Metabolites were identified using pure reference compounds and quantified using signal integration in relation to the internal standard trimethylsilylpropionic acid-d4 (1 mm). The identity of the key metabolite pyruvate was confirmed by GC/MS analysis as described previously (32).
Preparation of Cell-free Extracts
Cell lysates from E. coli were prepared by resuspending fresh cells in 2.5 ml of MOPS buffer (150 mm, pH 7.4) containing 10 μg/ml DNase I (Fermentas)/g cell wet weight. The resuspended cells were subjected to three subsequent rounds of disruption using a French press (Aminco) at 20,000 p.s.i. Insoluble components were removed from the extracts by centrifugation (15,000 × g for 20 min at 4 °C), and the clear culture supernatants were filter-sterilized and used immediately for enzymatic assays. S. aureus Newman cells were resuspended in 2.5 ml/g cell wet weight MOPS buffer (50 mm, pH 7.4), and 2,250 μl of the suspension were mixed with 750 μl of lysostaphin (0.025 mg/ml; Genmedics) and 30 μl of DNase I (10 mg/ml; Fermentas). Enzyme-containing suspensions were incubated for 60 min at 37 °C, and 3 × 1 ml of lysate were transferred into separate screw top polystyrene tubes each containing 400 mg of glass beads (G1145; Sigma). Cells were mechanically disrupted using a FastPrep-24 instrument (MP Biomedicals Europe) in four subsequent cycles of 20 s at lowest speed (4.0). Each cycle was interrupted for at least 30 s to avoid overheating of the sample. Insoluble components were finally removed from the extracts by centrifugation (14,000 × g for 10 min at 4 °C). 4 × 500 μl of the resulting filter-sterilized lysate were subjected to ultracentrifugation for 1 h using a Beckman TLA-55 rotor at 50,000 rpm and 4 °C. The obtained pellets were stored on ice for 1.5 h and were carefully resuspended in a total volume of 800 μl of MOPS buffer for analysis of PDHC activity.
Purification of E. coli PDHC
PDHC was purified from E. coli cells as described previously (33). Briefly, cell lysates were subjected to ultracentrifugation (4 h at 150,000 × g). Pellets were then washed with a small amount of potassium phosphate buffer (50 mm, pH 7.5) and centrifuged at 12,000 × g for 10 min. Subsequently, pellets were redissolved again, and the process of concentration and washing was repeated twice. The PDHC-containing fraction obtained after the third round of ultracentrifugation was loaded onto a calcium phosphate column, and PDHC was eluted using a linear gradient of potassium phosphate buffer (0.05-0.5 m). Active fractions were pooled and concentrated by ultracentrifugation as above.
Determination of Enzymatic Activities
PDHC activity was assayed essentially according to the method described previously (34) and modified (35). Reaction mixtures contained 50 mm MOPS (pH 7.4), 0.2 mm MgCl2, 0.01 mm CaCl2, 0.3 mm thiamine diphosphate, 0.12 mm CoA, 2.0 mm β-NAD+, 5.1 mm pyruvate, 0.1 mm 1-methoxy-5-methylphenazinium methyl sulfate, and 0.4 mm iodonitrotetrazolium formazan in an assay volume of 1.5 ml. Enzymatic activity was measured spectrophotometrically at 500 nm and 20 °C. Units of activity were calculated using an absorption coefficient of 12.5 mm−1 cm−1 for iodonitrotetrazolium formazan.
E1 subunit activity was determined by measuring the reduction of potassium ferricyanide (K3(Fe(CN)6)), similar to the procedure described previously (36). A single reaction contained 45 mm Tris/HCl, 1 mm MgCl2, 0.1 mm thiamine diphosphate, 5 mm pyruvate, 1.8 mm K3(Fe(CN)6), and an appropriate amount of purified enzyme. E3 subunit activity was determined by monitoring the oxidation of dihydrolipoic acid (37). The reaction mix for a single reaction contained 100 mm potassium phosphate buffer (pH 7.5), 4.8 mm dl-thioctic acid (Sigma), 2.0% (v/v) acetone, 0.2 mm NAD+, and purified enzyme from E. coli.
RESULTS
TPBC Has Potent Antimicrobial Activity against Multidrug-resistant Bacterial Pathogens
The synthetic compound TPBC (1) (Fig. 1) was shown in the 1960s (38) to have antimicrobial activity and has hardly been further investigated since then. Only recently has TPBC been used as an additive of catheter coatings to prevent device-associated infections, which yielded very promising results in clinical studies (28, 29) and prompted us to analyze the antimicrobial properties of TPBC against clinically relevant bacterial pathogens. TPBC had antibacterial activity against Gram-positive pathogens, such as staphylococci and enterococci, including methicillin-resistant S. aureus and VRE strains (Table 1). The MICs of TPBC were found to be 0.15 and 0.28 μg/ml for S. aureus and E. faecalis, respectively, whereas S. epidermidis and E. faecium were slightly less susceptible. The Gram-negative bacteria P. aeruginosa and Escherichia coli were also inhibited albeit at 84- and 32-fold higher concentrations compared with S. aureus COL (Table 1). Gram-negative bacteria are known to be less susceptible to certain antibiotics than Gram-positive bacteria because of the outer membrane, which represents an efficient diffusion barrier. When E. coli was incubated with subinhibitory concentrations of TPBC plus the outer membrane-permeabilizing substance polymyxin B, TPBC exhibited potent antimicrobial activity against E. coli (Fig. 2), indicating that TPBC can inhibit Gram-negative bacteria once the outer membrane is disrupted.
FIGURE 1.
Chemical structures of TPBC (1), triphenylbismuth (2), bismuthsalicylate (3), and fluoropyruvate (4).
TABLE 1.
Activity spectrum of TPBC
| Organism | Inhibitory concentrationsa |
|---|---|
| μg/ml | |
| Antimicrobial activity (MIC) | |
| S. aureus SA113 | 0.15 ± 0.01 |
| S. aureus Col (MRSA) | 0.28 ± 0.00 |
| S. epidermidis O-47 | 1.92 ± 0.07 |
| B. subtilis DB2 | 0.11 ± 0.05 |
| E. faecalis VRE 366 (VRE) | 0.16 ± 0.01 |
| E. faecium VRE 392 (VRE) | 1.34 ± 0.08 |
| P. aeruginosa MPA01 | 23.4 ± 1.4 |
| E. coli K12 | 8.84 ± 0.53 |
| Toxicity (IC50) | |
| MonoMac 6 (MM6) | >100 |
a Means ± S.E. of three independent experiments are shown.
FIGURE 2.

TPBC inhibits E. coli growth when the outer membrane is destabilized by polymyxin. Growth of E. coli is shown in the presence of TPBC (8 μg/ml) (○), TPBC and polymyxin B (8 and 0.3 μg/ml, respectively) (□), polymyxin B only (0.3 μg/ml) (▴), or in the absence of antibiotics (■). Data represent means and S.E. of three independent experiments.
The antimicrobial activities of TPBC and of the related compounds triphenylbismuth (2) and the anti-Helicobacter pylori drug bismuth-subsalicylate (3) (Fig. 1) were compared. TPBC exhibited by far the most potent antimicrobial activity against S. aureus, with bismuth-salicylate having no anti-staphylococcal activity at all (Table 2). The toxicity of TPBC toward human MM6 macrophages was above 100 μg/ml (Table 1), suggesting that TPBC may not be harmful to human cells.
TABLE 2.
Antimicrobial activities of TPBC and chemically or functionally related compounds against S. aureus SA113
| Compound | MICa |
|---|---|
| μg/ml | |
| TPBC | 0.15 ± 0,01 |
| Triphenylbismuth | 0.88 ± 0.25 |
| Bismuth(III) salicylate | >362 |
| Fluoropyruvate | 16.4 ± 0.2 |
a Means ± S.E. of three independent experiments are shown.
Exometabolome Analysis Indicates That TPBC Blocks Bacterial Pyruvate Catabolism
The hydrophobic properties of TPBC suggested that the substance might reach the bacterial cytoplasm and may interfere with essential cellular processes. To investigate the impact of TPBC on the primary metabolism of S. aureus, we chose a 1H NMR-based exometabolome profiling approach, which permits the simultaneous quantification of key metabolites in culture supernatants (31). Overall changes in the exometabolome of S. aureus Sa113 cultures grown in synthetic minimal medium in the presence or absence of TPBC are given in Fig. 3 and supplemental Fig. S1. In the absence of inhibitory substances, culture supernatants rapidly lost glucose and amino acids and accumulated acetate, the primary catabolic product of S. aureus (Figs. 3 and 4, b and c). During exponential, aerobic growth, S. aureus is known to oxidize glucose largely to acetate and to generate additional ATP from acetyl coenzyme A (Fig. 5). Only at late growth stages, when reduced carbon sources are limited, is acetate further oxidized to CO2 (39, 40). The central glycolysis intermediate pyruvate was detectable at low concentrations with a small, transient peak at 2 h of growth (Fig. 4d). Fermentation products, such as lactate, acetoine, or butandiole, were produced only at very low levels as expected for S. aureus growth under aerobic conditions (Fig. 4, e–g).
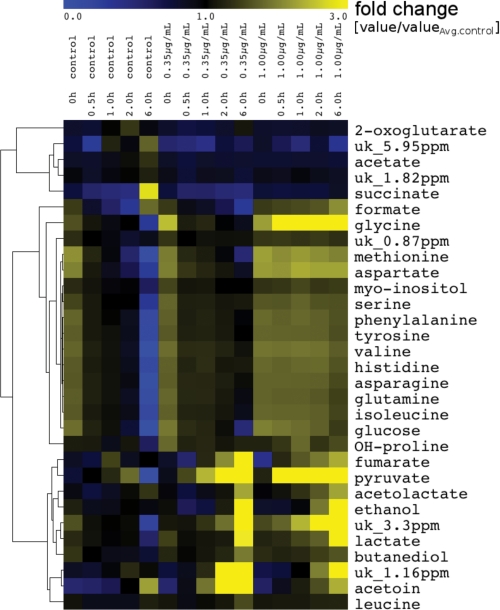
FIGURE 3.
Exometabolome changes in S. aureus cultures upon TPBC treatment. Hierarchical clustering analysis groups the 1H NMR-detected extracellular metabolite profiles over the three different growth conditions (0.0 μg/ml (control), 0.30 μg/ml, and 1.0 μg/ml TPBC) into different profile classes. Blue and yellow indicate decreases and increases, respectively, in concentration. Concentrations were set in relation to measured A578 values at a given time point. uk, unknown metabolite.
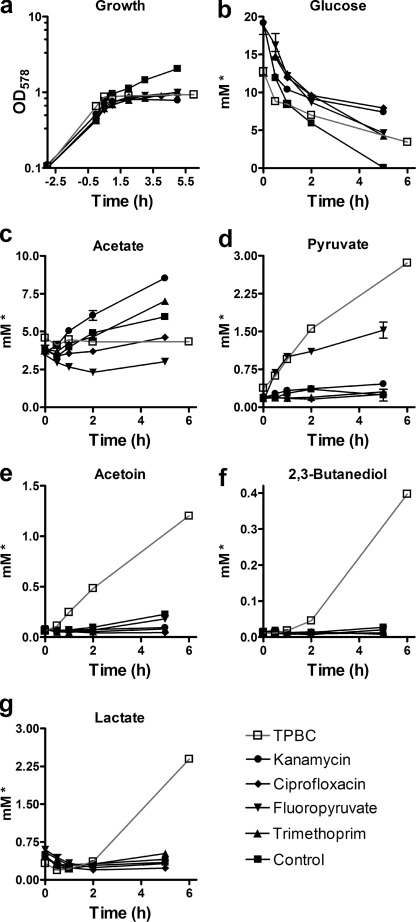
FIGURE 4.
S. aureus treatment with TPBC leads to accumulation of pyruvate and fermentation products. Exposure of S. aureus Sa113 to TPBC leads to immediate growth inhibition (a), to halt of glucose consumption (b) and acetate accumulation (c), and to accumulation of pyruvate (d) plus fermentation products, such as acetoine (e), 2,3-butanediole (f), and lactate (g), in culture supernatants as determined by 1H NMR-based exometabolome analysis. *, concentrations relative to measured A578 values at a given time point. They represent means and S.E. (error bars) of three biological replicates.
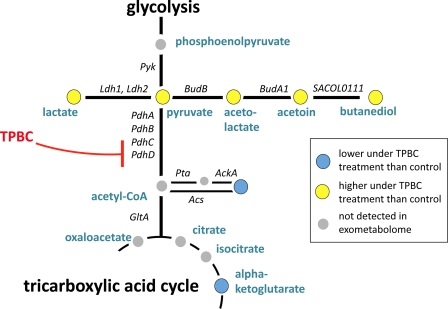
FIGURE 5.
Schematic representation of the pyruvate-metabolic pathways in S. aureus. Pyruvate is metabolized to acetyl-CoA via PDHC, which is formed by four protein subunits (PdhA, -B, -C, and -D) or reduced to lactate or to butandiole via acetolactate and acetoine. Acetyl-CoA is either funnelled into the tricarboxylic acid cycle, used for various biosynthetic pathways, or hydrolyzed to form free acetate.
After the addition of TPBC, growth, glucose and amino acid consumption, and acetate accumulation were suppressed (Figs. 3 and 4, a–c). Unexpectedly, pyruvate concentrations strongly increased in cultures containing inhibitory concentrations of TPBC over the entire duration of the experiment (Fig. 4d), indicating that glucose oxidation could only proceed to the level of pyruvate, whereas later steps were blocked. In parallel, the pyruvate-derived fermentation products acetolactate, acetoine, butanediole, lactate, formate, and ethanol accumulated over time (Figs. 3 and 4, e–g), suggesting that pyruvate was in part directed into alternative pathways. Together, these data suggested that TPBC interferes with the bacterial pyruvate catabolism.
Inhibition of Bacterial Pyruvate Catabolism Is Specific Consequence of TPBC Treatment
In order to investigate whether the accumulation of pyruvate is a specific response of S. aureus to TPBC or a general consequence of antibiotic treatment, we extended the exometabolomic analyses with a set of antibiotics of known modes of action. Again, the antibiotics were added to the bacterial cultures during aerobic, exponential growth at inhibitory but sublethal concentrations (Fig. 6a). The ribosomal inhibitor kanamycin, the DNA gyrase inhibitor ciprofloxacin, and the tetrahydrofolic acid biosynthesis inhibitor trimethoprim all caused a reduction of glucose consumption as expected (Fig. 5B). However, these antibiotics did not induce the accumulation of pyruvate or the production of pyruvate-derived fermentation products (Fig. 6, d–g), indicating that the TPBC-mediated change in the exometabolite profile is characteristic for this compound and does not represent a general response of staphylococci to antibiotic stress.
FIGURE 6.
Accumulation of pyruvate is a specific response of S. aureus to treatment with TPBC and the PDHC inhibitor fluoropyruvate. Treatment of S. aureus with TPBC (0.35 μg/ml; red line), kanamycin (12.5 μg/ml), ciprofloxacin (150 μg/ml), trimethoprim (25 μg/ml), or fluoropyruvate (FPA; 200 μg/ml) results in inhibition of growth (a), glucose consumption (b), and, with most of these antibiotics, altered acetate release (c), as determinded by 1H NMR-based exometabolome analysis. However, only TPBC and fluoropyruvate led to accumulation of pyruvate (d), and only TPBC induced accumulation of acetoine (e), 2,3-butanediole (f), and lactate (g). *, concentrations relative to measured A578 values at a given time point. They represent means and S.E. (error bars) of three biological replicates.
To further verify that TPBC blocks the oxidation of pyruvate, we compared the activity of TPBC with that of fluoropyruvate (4) (Fig. 1), a known competitive inhibitor of PDHC (41). We first noted that fluoropyruvate also has inhibitory properties against S. aureus (Table 2), albeit at much higher concentrations than TPBC. When the impact of fluoropyruvate on metabolite composition in staphylococcal cultures was analyzed, we observed accumulation of pyruvate in a very similar fashion as found for TPBC (Fig. 6d). However, consistent with its likely role as a competitive inhibitor of other pyruvate-utilizing enzymes, such as lactate dehydrogenase (42), fluoropyruvate did not stimulate the release of the pyruvate-derived fermentation products lactate, acetoine, or butandiole. These results provided further evidence for an interference of TPBC with the bacterial pyruvate metabolism and indicated that TPBC uses an inhibitory mechanism similar but not identical to that of fluoropyruvate.
TPBC Is Potent Inhibitor of the Bacterial PDHC
Most strictly or facultatively aerobic bacteria convert the catabolic intermediate pyruvate into CO2 and acetyl CoA via the PDHC, and PDHC-generated acetyl CoA is necessary for many biosynthetic pathways. Accordingly, the enzyme has been shown to be essential for viability of S. aureus (43–45). Conversely, direct inhibition of PDHC by TPBC would explain the observed accumulation of pyruvate in the exometabolome analyses and the antimicrobial properties of this compound. To explore this possibility, we prepared cell lysates from S. aureus, enriched PDHC by ultracentrifugation, and monitored PDHC activity according to an established protocol that relies on the pyruvate-, CoA-, and NAD+-dependent reduction of the chromogenic electron acceptor iodonitrotetrazolium (35) in the absence or presence of TPBC. Notably, TPBC blocked PDHC activity very efficiently in a dose-dependent manner (Fig. 7a). The same inhibition was observed with E. coli lysates or with E. coli PDHC, which had been isolated by chromatographic purification (Fig. 7, b–f). PDHC was also efficiently inhibited by fluoropyruvate, as expected (Fig. 6e), whereas triphenylbismuth and bismuth-salicylate did not affect PDHC activity at all. Taken together, these data confirm that TPBC inhibits very efficiently the PDHC of different bacterial kingdoms, ranging from the firmicute S. aureus to the γ-proteobacterium E. coli. Of note, inhibition of PDHC activity and of S. aureus growth followed similar dose-response curves (Fig. 7a).
FIGURE 7.
TPBC is an efficient, uncompetitive inhibitor of the bacterial PDHC. a, TPBC-mediated inhibition of growth and of activity of enriched S. aureus PDHC correlate closely. b, inhibition of purified enzyme (open bars) or of cell lysates (gray bars) obtained from E. coli is dose-dependent and occurs at very low concentrations. c, kinetics of PDHC inhibition by TPBC. Experiments were performed with purified enzyme from E. coli and 0 (□), 0.13 (▴), 0.27 (▾), 0.53 (♦), 1.07 (●), and 2.13 μg/ml TPBC (■). d, enzymatic analyses of subunit activities in purified PDHC from E. coli show that TPBC has no effect on the activities of the E1 and E3 subunits even when administered at high concentrations. e and f, Michaelis-Menten kinetics of PDHC from E. coli incubated with TPBC reveals a decrease of Km and Vmax, which is typical for uncompetitive enzyme inhibitors. Fluoropyruvate (FPA), by contrast, affects PDHC kinetics in a way that is typical for competitive enzyme inhibition. Data obtained with purified enzyme are given for one representative experiment. All other data represent means and S.E. (error bars) of three independent experiments.
TPBC Is Uncompetitive Inhibitor of PDHC
PDHC is a multicomponent enzyme complex consisting of three functional subunits that mediate (i) decarboxylation of pyruvate and formation of hydroxyethyl thiamine pyrophosphate (E1 subunit), (ii) oxidation of the hydroxyethyl group by the lipoic acid cofactor and transfer of resulting acetyl group to free CoA (E2 subunit), and (iii) oxidation of dihydrolipoic acid by NAD+ via the flavine adenine dinucleotide cofactor (E3 subunit). Although fluoropyruvate has been described as being a competitive inhibitor of the E1 subunit of E. coli PDHC (41), we aimed at elucidating how TPBC may inhibit the PDHC. We first performed Michaelis-Menten kinetics using enriched PDHC from E. coli and inhibitory concentrations of TPBC or fluoropyruvate. Consistent with published data (41), fluoropyruvate caused a strong increase of Km but did not alter the Vmax of PDHC (Fig. 7e), which is typically observed with competitive inhibitors (46). In contrast, TPBC led to a decrease of both Km and Vmax (Fig. 7f), which is typical for uncompetitive enzyme inhibitors that depend on an active enzyme-substrate complex to exert their inhibitory function (46).
In order to elucidate which step of the PDHC pathway may be inhibited by TPBC, we monitored the pyruvate-dependent reduction of ferricyanide (E1 reaction) (36) or the dihydrolipoic acid-dependent reduction of NAD+ (E3 reaction) (37) in the presence of increasing concentrations of TPBC. TPBC had no influence on the E1 and E3 subreactions even at very high concentrations, whereas it clearly blocked the entire PDHC reaction (Fig. 7d), suggesting that TPBC may interfere with the E2 subunit, whose activity cannot be measured in a physiological manner or that it can exert its activity only in the context of the complete enzymatic reaction of the PDHC complex.
DISCUSSION
Our study demonstrates the power of metabolomics approaches for elucidating the mode of action of new antimicrobial compounds that interfere with metabolic processes and suggests that the bacterial PDHC may become useful as a potential target for antibiotics. Most of the currently employed antibiotic targets are confined to cell wall biosynthetic enzymes and precursor molecules, such as lipid II, ribosome subunits, gyrase, RNA polymerase, and folic acid biosynthetic enzymes (47). Nevertheless, recent studies have yielded previously unrecognized potential drug targets, such as the bacterial protein degradation pathway (5), the bacterial ATP synthase (4), the cell division protein FtsZ (7), the Gram-negative lipoprotein pathway (8), or the biosynthesis of siderophores (6).
The bacterial PDHC has been investigated for more than 4 decades, and a number of studies have assigned vital and essential roles to the PDHC in staphylococci (43–45) and other bacteria (48, 49). However, to the best of our knowledge, there are no reports on PDHC-specific antibiotic compounds in the available literature. The PDHC inhibitor fluoropyruvate was first described in 1954 (50) but has not been regarded as a potential antibiotic compound. This is presumably due to the relatively high concentrations required to achieve antimicrobial effects. Using a combined exometabolomic and biochemical approach, we demonstrate that PDHC can be efficiently inhibited by TPBC, whose mode of action has previously remained unknown. In contrast to fluoropyruvate, TPBC inhibited PDHC in an uncompetitive fashion and at very low concentrations (>0.2 μg/ml). Because TPBC did not affect the activity of the E1 and E3 reaction, TPBC may only be able to exert its inhibitory action in the full course of the enzymatic cascade, or it may involve the E2 subunit. The inhibition of PDHC by TPBC may relate to the previously described capacity of TPBC to phenylate ketones and enols or enolate anions (51). Detailed structural analyses of PDHC, which represent a huge multicomponent protein complex (52), will be necessary to unravel the exact mode of inhibition of PDHC by TPBC. The substantially higher antimicrobial activity of TPBC compared with fluoropyruvate may be due to a more efficient inhibitory mode (uncompetitive versus competitive) and its much more pronounced hydrophobicity, which favors the penetration across the cytoplasmic membrane. PDHC is related to other α-ketoacid dehydrogenases. However, the amounts of α-ketoglutarate, the substrate of α-ketoglutarate dehydrogenase, were hardly affected by TPBC (Fig. 3), indicating that this enzyme is probably not inhibited by TPBC.
TPBC has interesting antimicrobial properties against bacterial PDHCs from unrelated organisms, such as firmicutes and proteobacteria, and it has been successfully used to coat catheters as a preventive strategy against device-related infections (28, 29). Thorough toxicological studies will be necessary in the future to explore if TPBC or related compounds may become candidates for further antiinfective applications. The frequent and successful use of bismuth(III) salicylate or related compounds against H. pylori infections demonstrates the potential of bismuth-containing organic compounds as antimicrobial drugs and indicates that toxicity is not necessarily a problem. In fact, many bismuth-containing drugs are considered safe due to their non-toxic and non-carcinogenic properties (53).
Although numerous attempts have been undertaken to elucidate the antimicrobial mechanisms of new antimicrobial substances, it remains extremely difficult to identify the major target molecule(s). We suggest that the application of metabolomic techniques may represent a powerful approach to identify novel targets of compounds with known antimicrobial properties but unknown modes of action. This approach may be particularly promising for inhibitors of enzymes that metabolize small compounds, such as pyruvate. Although we did not see characteristic metabolite changes in bacterial culture supernatants upon the addition of ribosome, DNA gyrase, or tetrahydrofolic acid biosynthetic inhibitors, detailed analyses of intracellular metabolomes may yield characteristic signatures for all classes of antibiotics in the future. Our ongoing efforts will help to further assess the power of metabolomics in antimicrobial drug discovery.
The antimicrobial properties of TPBC along with its proven efficacy in preventing catheter-associated infections put TPBC on the list of promising antibiotic lead substances. Our study represents a basis for using TPBC-derived drugs or novel compounds to target the bacterial PDHC as a new therapeutic option against highly antibiotic-resistant bacteria, such as methicillin-resistant S. aureus, VRE, and others.
Supplementary Material
Acknowledgments
We thank Anne Sedlag and Nadja Heinz for technical support and A. Schnell, R. Deppisch, and Kornelius Zeth for helpful discussion.
This work was supported by grants from the German Ministry of Education and Research with Gambro Inc. (ABBA-Tech), the German Science Foundation (TRR34, SFB766), and the IZKF Program of the Medical Faculty, University of Tübingen (to A. P.).

This article contains supplemental Fig. S1.
- VRE
- vancomycin-resistant enterococci
- TPBC
- triphenylbismuthdichloride
- PDHC
- pyruvate dehydrogenase complex
- MIC
- minimal inhibitory concentration.
REFERENCES
- 1. Arias C. A., Murray B. E. (2009) Antibiotic-resistant bugs in the 21st century. A clinical super-challenge. N. Engl. J. Med. 360, 439–443 [DOI] [PubMed] [Google Scholar]
- 2. Lowy F. D. (2003) Antimicrobial resistance. The example of Staphylococcus aureus. J. Clin. Invest. 111, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Proctor R. A., von Eiff C., Kahl B. C., Becker K., McNamara P., Herrmann M., Peters G. (2006) Small colony variants. A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305 [DOI] [PubMed] [Google Scholar]
- 4. Andries K., Verhasselt P., Guillemont J., Göhlmann H. W., Neefs J. M., Winkler H., Van Gestel J., Timmerman P., Zhu M., Lee E., Williams P., de Chaffoy D., Huitric E., Hoffner S., Cambau E., Truffot-Pernot C., Lounis N., Jarlier V. (2005) A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227 [DOI] [PubMed] [Google Scholar]
- 5. Brötz-Oesterhelt H., Beyer D., Kroll H. P., Endermann R., Ladel C., Schroeder W., Hinzen B., Raddatz S., Paulsen H., Henninger K., Bandow J. E., Sahl H. G., Labischinski H. (2005) Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 6. Ferreras J. A., Ryu J. S., Di Lello F., Tan D. S., Quadri L. E. (2005) Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 1, 29–32 [DOI] [PubMed] [Google Scholar]
- 7. Haydon D. J., Stokes N. R., Ure R., Galbraith G., Bennett J. M., Brown D. R., Baker P. J., Barynin V. V., Rice D. W., Sedelnikova S. E., Heal J. R., Sheridan J. M., Aiwale S. T., Chauhan P. K., Srivastava A., Taneja A., Collins I., Errington J., Czaplewski L. G. (2008) An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321, 1673–1675 [DOI] [PubMed] [Google Scholar]
- 8. Pathania R., Zlitni S., Barker C., Das R., Gerritsma D. A., Lebert J., Awuah E., Melacini G., Capretta F. A., Brown E. D. (2009) Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat. Chem. Biol. 5, 849–856 [DOI] [PubMed] [Google Scholar]
- 9. Smith P. A., Romesberg F. E. (2007) Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 3, 549–556 [DOI] [PubMed] [Google Scholar]
- 10. Weidenmaier C., Kristian S. A., Peschel A. (2003) Bacterial resistance to antimicrobial host defenses. An emerging target for novel antiinfective strategies? Curr. Drug Targets 4, 643–649 [DOI] [PubMed] [Google Scholar]
- 11. Rice K. C., Bayles K. W. (2008) Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 72, 85–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hancock R. E., Sahl H. G. (2006) Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557 [DOI] [PubMed] [Google Scholar]
- 13. Schneider T., Kruse T., Wimmer R., Wiedemann I., Sass V., Pag U., Jansen A., Nielsen A. K., Mygind P. H., Raventós D. S., Neve S., Ravn B., Bonvin A. M., De Maria L., Andersen A. S., Gammelgaard L. K., Sahl H. G., Kristensen H. H. (2010) Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328, 1168–1172 [DOI] [PubMed] [Google Scholar]
- 14. Lentino J. R., Narita M., Yu V. L. (2008) New antimicrobial agents as therapy for resistant gram-positive cocci. Eur. J. Clin. Microbiol. Infect. Dis. 27, 3–15 [DOI] [PubMed] [Google Scholar]
- 15. Projan S. J., Bradford P. A. (2007) Late stage antibacterial drugs in the clinical pipeline. Curr. Opin. Microbiol. 10, 441–446 [DOI] [PubMed] [Google Scholar]
- 16. Bandow J. E., Brötz H., Leichert L. I., Labischinski H., Hecker M. (2003) Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47, 948–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jang H. J., Nde C., Toghrol F., Bentley W. E. (2008) Microarray analysis of toxicogenomic effects of ortho-phenylphenol in Staphylococcus aureus. BMC Genomics 9, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sass V., Pag U., Tossi A., Bierbaum G., Sahl H. G. (2008) Mode of action of human β-defensin 3 against Staphylococcus aureus and transcriptional analysis of responses to defensin challenge. Int. J. Med. Microbiol. 298, 619–633 [DOI] [PubMed] [Google Scholar]
- 19. Hutter B., Schaab C., Albrecht S., Borgmann M., Brunner N. A., Freiberg C., Ziegelbauer K., Rock C. O., Ivanov I., Loferer H. (2004) Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48, 2838–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mashego M. R., Rumbold K., De Mey M., Vandamme E., Soetaert W., Heijnen J. J. (2007) Microbial metabolomics. Past, present, and future methodologies. Biotechnol. Lett. 29, 1–16 [DOI] [PubMed] [Google Scholar]
- 21. Vinayavekhin N., Homan E. A., Saghatelian A. (2010) Exploring disease through metabolomics. ACS Chem. Biol. 5, 91–103 [DOI] [PubMed] [Google Scholar]
- 22. Bathe O. F., Shaykhutdinov R., Kopciuk K., Weljie A. M., McKay A., Sutherland F. R., Dixon E., Dunse N., Sotiropoulos D., Vogel H. J. (2011) Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol. Biomarkers Prev. 20, 140–147 [DOI] [PubMed] [Google Scholar]
- 23. Bundy J. G., Willey T. L., Castell R. S., Ellar D. J., Brindle K. M. (2005) Discrimination of pathogenic clinical isolates and laboratory strains of Bacillus cereus by NMR-based metabolomic profiling. FEMS Microbiol. Lett. 242, 127–136 [DOI] [PubMed] [Google Scholar]
- 24. Liebeke M., Meyer H., Donat S., Ohlsen K., Lalk M. (2010) A metabolomic view of Staphylococcus aureus and its Ser/Thr kinase and phosphatase deletion mutants. Involvement in cell wall biosynthesis. Chem. Biol. 17, 820–830 [DOI] [PubMed] [Google Scholar]
- 25. White A. P., Weljie A. M., Apel D., Zhang P., Shaykhutdinov R., Vogel H. J., Surette M. G. (2010) A global metabolic shift is linked to Salmonella multicellular development. PLoS One 5, e11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sadykov M. R., Zhang B., Halouska S., Nelson J. L., Kreimer L. W., Zhu Y., Powers R., Somerville G. A. (2010) Using NMR metabolomics to investigate tricarboxylic acid cycle-dependent signal transduction in Staphylococcus epidermidis. J. Biol. Chem. 285, 36616–36624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Halouska S., Chacon O., Fenton R. J., Zinniel D. K., Barletta R. G., Powers R. (2007) Use of NMR metabolomics to analyze the targets of d-cycloserine in mycobacteria. Role of d-alanine racemase. J. Proteome Res. 6, 4608–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schnell A., Dietrich R., Hagg S., Haug U., Ruhl U., Deppisch R. (2007) Surface degradation of Pur based vascular access catheters in aqueous lock solutions leading to enhanced coagulation and biofilm deposition can be prevented by a specific reactive coating. Int. J. Artif. Organs 30, 746–746 [Google Scholar]
- 29. Schindler R., Heemann U., Haug U., Stoelck B., Karatas A., Pohle C., Deppisch R., Beck W., Hollenbeck M. (2010) Bismuth coating of non-tunneled haemodialysis catheters reduces bacterial colonization. A randomized controlled trial. Nephrol. Dial. Transplant. 25, 2651–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peschel A., Vuong C., Otto M., Götz F. (2000) The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44, 2845–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liebeke M., Brözel V. S., Hecker M., Lalk M. (2009) Chemical characterization of soil extract as growth media for the ecophysiological study of bacteria. Appl. Microbiol. Biotechnol. 83, 161–173 [DOI] [PubMed] [Google Scholar]
- 32. Liebeke M., Wunder A., Lalk M. (2010) A rapid microwave-assisted derivatization of bacterial metabolome samples for gas chromatography/mass spectrometry analysis. Anal. Biochem. 40, 312–314 [DOI] [PubMed] [Google Scholar]
- 33. Bisswanger H. (1981) Substrate specificity of the pyruvate dehydrogenase complex from Escherichia coli. J. Biol. Chem. 256, 815–822 [PubMed] [Google Scholar]
- 34. Brown J. P., Perham R. N. (1976) Selective inactivation of the transacylase components of the 2-oxo acid dehydrogenase multienzyme complexes of Escherichia coli. Biochem. J. 155, 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hinman L. M., Blass J. P. (1981) An NADH-linked spectrophotometric assay for pyruvate dehydrogenase complex in crude tissue homogenates. J. Biol. Chem. 256, 6583–6586 [PubMed] [Google Scholar]
- 36. Schwartz E. R., Old L. O., Reed L. J. (1968) Regulatory properties of pyruvate dehydrogenase from Escherichia coli. Biochem. Biophys. Res. Commun. 31, 495–500 [DOI] [PubMed] [Google Scholar]
- 37. Danson M. J., Perham R. N. (1976) Evidence for two lipoic acid residues per lipoate acetyltransferase chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem. J. 159, 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leebrick J. R. (October 11, 1966) Organo bismuth biocide. United States Patent 3,239,411, 1–8
- 39. Somerville G. A., Chaussee M. S., Morgan C. I., Fitzgerald J. R., Dorward D. W., Reitzer L. J., Musser J. M. (2002) Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70, 6373–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Somerville G. A., Saïd-Salim B., Wickman J. M., Raffel S. J., Kreiswirth B. N., Musser J. M. (2003) Correlation of acetate catabolism and growth yield in Staphylococcus aureus. Implications for host-pathogen interactions. Infect. Immun. 71, 4724–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Flournoy D. S., Frey P. A. (1989) Inactivation of the pyruvate dehydrogenase complex of Escherichia coli by fluoropyruvate. Biochemistry 28, 9594–9602 [DOI] [PubMed] [Google Scholar]
- 42. Urban P., Lederer F. (1988) Inactivation of flavocytochrome b2 with fluoropyruvate. Reaction at the active-site histidine. Eur. J. Biochem. 173, 155–162 [DOI] [PubMed] [Google Scholar]
- 43. Chaudhuri R. R., Allen A. G., Owen P. J., Shalom G., Stone K., Harrison M., Burgis T. A., Lockyer M., Garcia-Lara J., Foster S. J., Pleasance S. J., Peters S. E., Maskell D. J., Charles I. G. (2009) Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridization (TMDH). BMC Genomics 10, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adler L. A., Arvidson S. (1988) Cloning and expression in Escherichia coli of genes encoding a multiprotein complex involved in secretion of proteins from Staphylococcus aureus. J. Bacteriol. 170, 5337–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Forsyth R. A., Haselbeck R. J., Ohlsen K. L., Yamamoto R. T., Xu H., Trawick J. D., Wall D., Wang L., Brown-Driver V., Froelich J. M., Kg C., King P., McCarthy M., Malone C., Misiner B., Robbins D., Tan Z., Zhu Zy Z. Y., Carr G., Mosca D. A., Zamudio C., Foulkes J. G., Zyskind J. W. (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43, 1387–1400 [DOI] [PubMed] [Google Scholar]
- 46. Cook P. F., Cleland W. W. (2007) Enzyme kinetics and mechanisms, Garland Press, London [Google Scholar]
- 47. Fischbach M. A., Walsh C. T. (2009) Antibiotics for emerging pathogens. Science 325, 1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Herbert M., Kraiss A., Hilpert A. K., Schlör S., Reidl J. (2003) Aerobic growth-deficient Haemophilus influenzae mutants are non-virulent. Implications on metabolism. Int. J. Med. Microbiol. 293, 145–152 [DOI] [PubMed] [Google Scholar]
- 49. Nakano M. M., Dailly Y. P., Zuber P., Clark D. P. (1997) Characterization of anaerobic fermentative growth of Bacillus subtilis. Identification of fermentation end products and genes required for growth. J. Bacteriol. 179, 6749–6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mager J., Blank I. (1954) Synthesis of fluoropyruvic acid and some of its biological properties. Nature 173, 126–127 [DOI] [PubMed] [Google Scholar]
- 51. Barton D. H., Blazejewski J. C., Charpiot B., Finet J. P., Motherwell W. B., Papoula M. T., Stanforth S. P. (1985) Pentavalent organobismuth reagents. Part 3 phenylation of enols and of enolate and other anions. J. Chem. Soc. Perkin Trans. 1, 2667–2675 [Google Scholar]
- 52. de Kok A., Hengeveld A. F., Martin A., Westphal A. H. (1998) The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385, 353–366 [DOI] [PubMed] [Google Scholar]
- 53. Sun H., Zhang L., Szeto K. Y. (2004) Bismuth in medicine. Met. Ions Biol. Syst. 41, 333–378 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.