Background: SMC differentiation is a complicated process involving many transcription factors.
Results: hnRNPA2/B1 regulates SMC differentiation gene expression transcriptionally and promotes neural crest cell migration and differentiation toward SMCs.
Conclusion: hnRNPA2/B1 promotes smooth muscle differentiation and development in vitro and in vivo.
Significance: This is the first report to demonstrate the functional involvements of hnRNPA2/B1 in SMC differentiation from stem cells and embryonic arteriogenesis.
Keywords: Cell Differentiation, Gene Regulation, Heterochromatin, Smooth Muscle, Stem Cells, Arteriogenesis, Chick Embryo, Heterogeneous Nuclear Ribonucleoprotein A2/B1
Abstract
Heterogeneous nuclear ribonucleoproteins (hnRNPs) play various roles in transcriptional and post-transcriptional modulation of gene expression. However, it remains unclear if hnRNPs are associated with smooth muscle cell (SMC) differentiation from stem cells and embryonic arteriogenesis. In this study, mouse embryonic stem (ES) cells were cultivated on collagen IV-coated plates and smooth muscle differentiation medium. We found that hnRNPA2/B1 gene and protein expression was significantly up-regulated following 3–7 days of cell differentiation. hnRNPA2/B1 knockdown resulted in down-regulation of specific smooth muscle markers and transcription factors, whereas enforced expression of hnRNPA2/B1 enhanced the expression of these genes. Moreover, we demonstrated by using luciferase and chromatin immunoprecipitation assays that hnRNPA2/B1 could transcriptionally regulate SMC gene expression through direct binding to promoters of Smαa and Sm22α genes. We further demonstrated that chromobox protein homolog gene 3, a previously identified SMC differentiation regulatory nuclear protein, is required for hnRNPA2/B1-mediated SMC differentiation gene expression. Importantly, specifically designed Hnrnpa2/b1 morpholinos for in vivo knockdown could inhibit the migration and differentiation of neural crest cells into SMCs in chick embryos. This resulted in the maldevelopment of branchial arch arteries and increased embryo lethality at a later developmental stage. Our findings demonstrated that hnRNPA2/B1 plays a functional role in SMC differentiation from stem cells in vitro and embryonic branchial arch artery development. This indicates that hnRNPA2/B1 is a potential modulating target for deriving SMCs from stem cells and cardiovascular regenerative medicine.
Introduction
Understanding the signaling pathways and molecular mechanisms in regulating stem/progenitor cell differentiation into cardiomyocytes and endothelial and smooth muscle cells (SMCs)5 is crucial. This is especially so for cardiovascular regenerative medicine that eventually benefits patients in clinical applications. Unlike endothelial cells and cardiomyocytes, which are terminally differentiated, adult SMCs retain significant plasticity to carry out different functions such as proliferation, migration, and synthesis of extracellular matrix protein in a series of cardiovascular pathogeneses (1). Therefore, studies focused on signaling pathways and molecular mechanisms in SMC differentiation are crucial for improving our understanding of the pathological process of cardiovascular diseases and developing novel therapeutic strategies for these diseases. Although the regulatory networks and transcriptional regulators of SMC differentiation have been extensively investigated in recent years by our group and other researchers (2–13), whether certain heterogeneous nuclear ribonucleoproteins (hnRNPs) are involved in SMC differentiation gene regulation remains obscure.
hnRNPs belong to the RNA-binding protein family and play important roles in regulating gene expression at both transcriptional and post-transcriptional levels (14). Each hnRNP includes at least one RNA-binding domain such as RNA recognition motif, hnRNP K homology domain, or arginine/glycine-rich box (14). The significance of these RNA-binding domains has been strongly linked to its importance in gene post-transcription, mature mRNA cytoplasm transportation, and protein translation through its binding to pre-RNAs (15, 16). However, accumulating evidence also suggests that RNA recognition motif and arginine/glycine-rich box of hnRNPs are involved in the binding of single-stranded DNA (17), telomere biogenesis (18, 19), and transcriptional regulation (20, 21). For example, it has been reported that hnRNPA2/B1 can bind to the promoters of c-myc, Apoe, vitamin D receptor, breast cancer 1, and gonadotropin-releasing hormone 1 genes and regulate the gene expressions (22–26). However, to date, there has been no report of the functional involvement of hnRNPA2/B1 in SMC differentiation and embryonic development. In this study, we first identified an essential role of hnRNPA2/B1 in SMC differentiation from embryonic stem (ES) cells. We demonstrated that hnRNPA2/B1 regulated the expression of SMC differentiation genes by directly binding to and activating its promoters of specific SMC genes such as SmαA and Sm22α. In addition, we further demonstrate for the first time that knockdown of hnRNPA2/B1 by using specifically designed Hnrnpa2/b1 morpholino could disrupt chick neural crest cell migration and differentiation into branchial arch arterial SMCs. Our findings highlight the importance of hnRNPA2/B1 for SMC differentiation from both stem cells and neural crest cells.
MATERIALS AND METHODS
Cell Culture and SMC Differentiation
Detailed protocols for mouse ES cell (ES-D3 cell line, CRL-1934; ATCC, Manassas, VA) culture and SMC differentiation were described in our previous studies (2–8). For SMC differentiation, undifferentiated ES cells were seeded on mouse collagen IV (5 μg/ml)-coated flasks or plates in differentiation medium (DM), α-minimal essential medium (Invitrogen) supplemented with 10% FBS, 0.05 mm β-mercaptoethanol, 100 units/ml penicillin, and 100 μg/ml streptomycin for 3–7 days prior to further treatments. The medium was refreshed every other day.
Generation of Hnrnpa2/b1 Gene Expression Plasmids
Mouse and chick full-length Hnrnpa2/b1 genes were amplified using RT-qPCR from day 3 differentiating ES cells and chick embryos with the primer set as shown in supplemental Table S1 and cloned into KpnI/PstI sites of the pCMV5-HA and pSPT18 expression vector, designated as pCMV5-HA-Hnrnpa2/b1 and pSPT18-Hnrnpa2/b1. All the vectors were verified by DNA sequencing.
Other Procedures
Experiments of nucleofection, siRNA experiments, indirect immunofluorescent staining for cells, and real time quantitative PCRs (RT-qPCR) were performed according to the standard procedures as described previously (2–8). Whole mount immunohistochemistry and in situ hybridization were carried out as described previously (7). For detailed “Materials and Methods,” see the supplemental material.
Chromatin Immunoprecipitation (ChIP) Assays
As described previously (3, 5, 7, 8), differentiating ES cells transfected with pCMV5 or pCMV5-Hnrnpa2/b1 were treated with 1% (v/v) formaldehyde at room temperature for 10 min and then quenched with glycine at room temperature. After removing the medium, cells were harvested for sonication. The sheared samples were diluted into 1 ml of immunoprecipitation buffer containing 25 mm Tris-HCl, pH 7.2, 0.1% Nonidet P-40, 150 mm NaCl, and 1 mm EDTA. Immunoprecipitation was conducted with antibody raised against hnRNPA2/B1, together with single strand salmon sperm DNA saturated with protein-G-Sepharose beads. Normal IgG was used as a control. The immunoprecipitates were eluted from the beads using 100 μl of elution buffer (50 mm NaHCO3, 1% SDS). A total of 200 μl of proteinase K solution was added to a total elution volume of 300 μl and incubated at 60 °C overnight. Immunoprecipitated DNA was extracted, purified, and then used to amplify target DNA sequences by real time quantitative PCR (RT-qPCR). Relative DNA level was defined as the ratio of immunoprecipitated promoter DNA level to its input level with that of the control sample (pCMV5) set as 1.0. Target DNAs were almost undetectable in the normal IgG control samples. The data were obtained from three independent experiments.
Chick Embryo Cultivation and Staging
Fertilized chick eggs were obtained from Joyce and Hill (Farm) and incubated at 38.5 °C in a humidified incubator. Hamburger and Hamilton (HH) staging was applied throughout the study (27, 28).
In Vivo Electroporation on Chick Embryos
In vivo electroporation carried out on chick embryos was performed as described previously but with some modifications (7). Eggs were incubated in a humidified incubator at 38 °C for around 37 h. A window was opened at the top of the egg shell using a blade, and embryos at HH 10/10−/10+ were electroporated with Hnrnpa2/b1 morpholino (1 μm, 5′-TCTCTCTGTCGCCGCGAGGCATCTT-3′, Gene Tools, LLC) or control morpholino (1 μm, 5′-CCTCTTACCTCAGTTACAATTTATA-3′). Electroporator was set at 7.5 V, 4 pulses, length of 50 ms, and interval of 1000 ms. Oligonucleotides were mixed with 60% sucrose before electroporation. After electroporation, the eggs were sealed well and placed back into the incubator, window side up, and incubated until the required developmental stages.
Statistical Analysis
Data were expressed as means ± S.E. and analyzed using two-tailed Student's t test for two group comparisons or one-way analysis of variance for comparing different groups. χ2 analysis was performed using SPSS 17.0 software to compare the percentage of embryonic maldevelopment in two groups. A value of p < 0.05 was considered statistically significant.
RESULTS
hnRNPA2/B1 Is Involved in ES Cell Differentiation to SMC
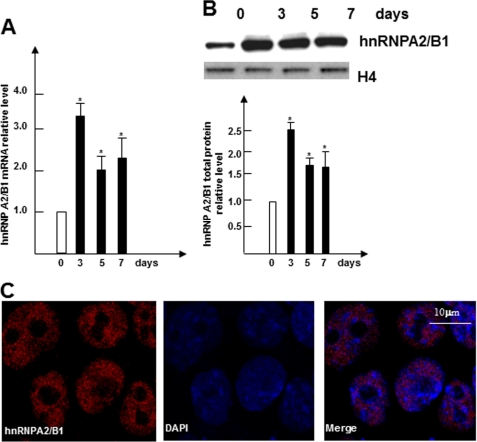
In parallel with SMC-specific gene inductions reported previously (2–8), the expression levels of hnRNPA2/B1, one of the several up-regulated nuclear proteins found during early SMC differentiation as demonstrated by our nuclear proteomics analysis (data not shown), were transiently elevated when ES cells were cultivated on collagen IV-coated plates in the absence of leukemia inhibitory factor to allow for SMC differentiation (Fig. 1, A and B). The levels of hnRNPA2/B1 expression peaked at day 3 and displayed a sustained signal until day 7, suggesting that hnRNPA2/B1 may play a role during ES cell differentiation to SMC (Fig. 1, A and B). As expected, immunofluorescent staining analyzed using confocal microscopy further confirmed the nuclear location of hnRNPA2/B1 proteins in differentiating cells (Fig. 1C).
FIGURE 1.
hnRNPA2/B1 is up-regulated during SMC differentiation. A, Hnrnpa2/b1 mRNA was up-regulated during SMC differentiation. Undifferentiated ES cells were plated onto dishes coated with 5 μg/ml collagen IV and cultured in DM. Total RNA from undifferentiated ES cells (day 0) or differentiating ES cells at days 3, 5, and 7 were harvested and subjected to RT-qPCR analysis with primers specific for Hnrnpa2/b1. B, nuclear protein from undifferentiated ESCs (day 0) or differentiating ES cells at the indicated time points were harvested and subjected to Western blot analysis. Upper panels show the representative images, and bottom panel show bar graphs representing means ± S.E. of densitometric analysis (n = 3) of the relative protein levels of hnRNPA2B1. C, nuclear localization of hnRNPA2/B1 in differentiating ES cells. Immunofluorescence staining was conducted on day 7 differentiating ES cells with antibodies against hnRNPA2/B1. The data presented here are representative or an average of three independent experiments. Significant difference from control, *, p < 0.05.
Essential Roles of hnRNPA2/B1 in SMC Differentiation from ES Cells
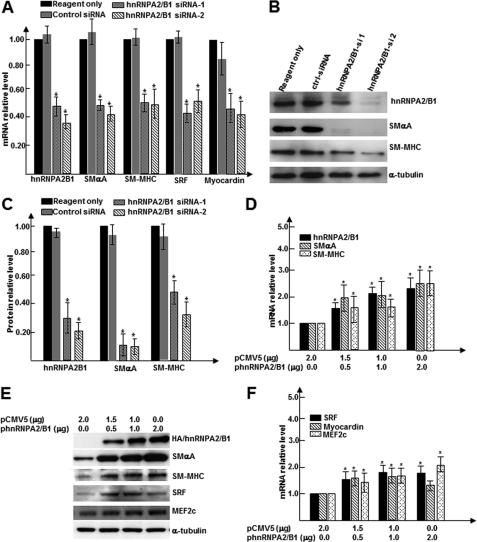
To determine the role of hnRNPA2/B1 in SMC differentiation, the effects of endogenous hnRNPA2/B1 knockdown on the expression of SMC-specific genes were examined. It showed that the knockdown of endogenous Hnrnpa2/b1 gene expression significantly decreased mRNA expression of multiple SMC-specific differentiation genes (Smαa and Smmhc) and transcription factors (Srf and Myocardin) (Fig. 2A). Similarly, Western blot analysis also revealed decreased protein levels of these genes in hnRNPA2/B1 knockdown cells (Fig. 2, B and C). To further investigate if hnRNPA2/B1 expression is sufficient for specific SMC gene expression, we generated Hnrnpa2/b1 expression plasmids. It is noteworthy that only the A2 isoform of hnRNPA2/B1 was successfully generated in our study as demonstrated by DNA sequencing of the expression plasmids (data not shown). Overexpression carried out using different amounts of Hnrnpa2/b1 plasmids was conducted in differentiating ES cells. RT-qPCR and Western blot analyses revealed that overexpressing hnRNPA2/B1 significantly increased SMC gene expressions at both mRNA (Fig. 2D) and protein (Fig. 2E) levels in a dose-dependent manner. Importantly, gene expression levels of SMC transcription factors were also dramatically increased in overexpressing cells (Fig. 2, E and F). Taken together, the above findings strongly suggest that Hnrnpa2/b1 gene expression is essential for SMC differentiation from stem cells.
FIGURE 2.
hnRNPA2/B1 is critical for SMC differentiation from ES cells. A–C, hnRNPA2/B1 knockdown down-regulated SMC-specific transcription factors and SMC markers. Two sets of hnRNPA2/B1-specific small interfering RNAs (hnRNPA2B1si-1 and -2) and random siRNA controls were transfected into day 3 differentiating ES cells; after an additional 2–3 days of culture, total RNA and protein were harvested and subjected to RT-qPCR (A) and Western blot analyses (B and C). B shows the representative images, and C shows bar graphs representing means ± S.E. of densitometric analysis (n = 3) of the relative protein levels of target proteins. D–F, hnRNPA2/B1 overexpression enhanced collagen IV-induced SMC differentiation. Undifferentiated ES cells were nucleofected by nucleofector II with different amounts of hnRNPA2/B1 expression plasmids pCMV5-Hnrnpa2/b1. Nucleofected cells were plated on dishes coated with 5 μg/ml of collagen IV and cultured for 3–4 days in DM. Total RNA and protein were harvested and subjected to RT-qPCR analysis for SMC gene expression (D), transcription factors (F), and Western blot analysis (E). An appropriate amount of empty vector pCMV5 was included as plasmid amount compensation. Same amount of protein was loaded into each well, blotted, and probed with specific antibodies for HA (represents exogenous hnRNPA2/B1), SMαA, SRF, MEF2c, and SM-MHC. α-Tubulin was included as internal control (B and E). The data presented here were representative or an average of three independent experiments. *, p < 0.05 versus control.
hnRNPA2/B1-mediated SMC Differentiation through Transcription Activation
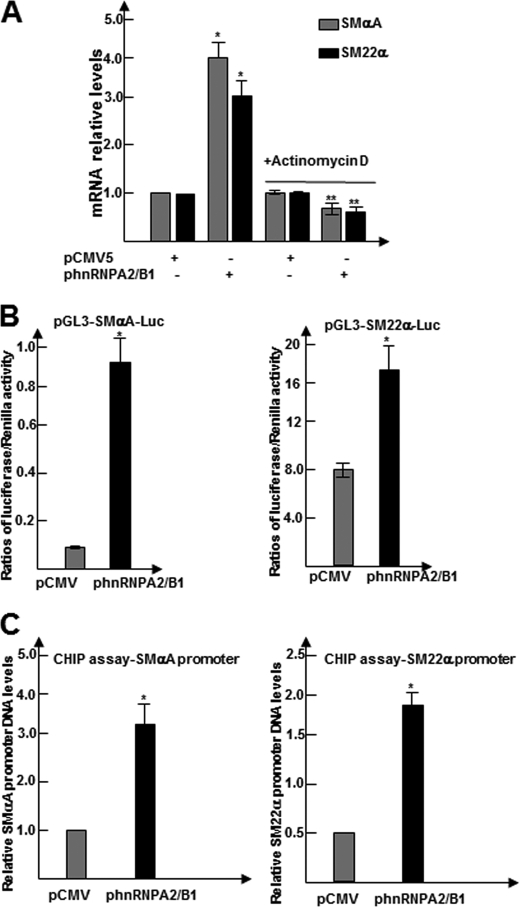
As demonstrated above, hnRNPA2/B1 influences the levels of SMC marker expression in differentiating ES cells. To understand how hnRNPA2/B1 regulates SMC gene expression, day 3 differentiating ES cells were transfected with pCMV5 or pCMV5-Hnrnpa2/b1 and treated on day 5 with the RNA synthase inhibitor actinomycin D (1 μg/ml) for 6 h. Total RNA was harvested and subjected to RT-qPCR analysis to examine Smαa and Sm22α gene expression. Treatment with actinomycin D ablated the effect of Hnrnpa2/b1 overexpression on Smαa and Sm22α gene expression (Fig. 3A), which suggested that hnRNPA2/B1 may activate Smαa and Sm22α gene expression at a transcriptional level.
FIGURE 3.
hnRNPA2/B1 transcriptionally activates SMC gene expression through direct binding to the promoters of SMC differentiation genes. A, actinomycin D ablated hnRNPA2/B1 overexpression-mediated SMC gene expression. Day 3 differentiating ES cells transfected with pCMV5-Hnrnpa2/b1 or pCMV5 (2.0 μg/1 × 106 cells) were treated with or without actinomycin D (1 μg/ml) for 6 h and harvested for RT-qPCR analysis of Smαa and Sm22α gene expressions. DMSO was included as a control. The data presented here are the average of three independent experiments. *, p < 0.05 (2nd column versus 1st column); **, p < 0.05 (4th column versus 2nd column). B, hnRNPA2/B1 regulated the promoter activities of SMC differentiation genes. ES cells were cultured on collagen IV and transfected at day 3 with luciferase reporter plasmids pGL3-SMαA-Luc (0.3 μg/5 × 104 cells) together with pCMV5-Hnrnpa2/b1 or pCMV5 (0.4 μg/5 × 104 cells). pGL3-Renilla (0.05 μg/5 × 104cells) was included as luciferase plasmid control. Luciferase and Renilla activity assays were detected 48 h after transfection. Luciferase activity was normalized or defined as the ratio of luciferase activity to Renilla activity. The data presented here are an average of three independent experiments. *, p < 0.05 (versus control). C, hnRNPA2/B1 bound directly to the promoter regions within SMC differentiation genes. ChIP assays were performed using antibodies against hnRNPA2/B1 and analyzed as described before. The data presented here are an average of three independent experiments. *, p < 0.05 (versus control).
To further test our hypothesis that hnRNPA2/B1 activates the specific SMC gene transcription, luciferase gene reporter and ChIP assays were performed. The plasmids used in the luciferase assay, pGL3-Smαa-Luc and pGL3-Sm22α-Luc, were previously designed and cloned in our laboratory (3). ES cells were cultured on collagen IV-coated 12-well plates and co-transfected at day 3 of differentiation. The cells were either transfected with Hnrnpa2/b1 overexpressing vector (400 ng/5 × 104cells) or the control vector, alongside with the luciferase reporter gene under the control of Smαa or Sm22α promoters. pGL3-Renilla vector was included in all samples as an internal control to assess transfection efficiency. The luciferase activity and Renilla signal were detected 48 h after transfection. Data shown in Fig. 3B revealed that the overexpression of hnRNPA2/B1 in differentiating ES cells significantly increased Smαa or Sm22α promoter activities. This indicates that hnRNPA2/B1 directly activates specific SMC gene promoters. Furthermore, ChIP assays were conducted using an hnRNPA2/B1 antibody in the differentiating ES cells to further verify if hnRNPA2/B1 activates specific SMC gene transcription through direct binding to their promoters. It revealed that hnRNPA2/B1 directly bound to the promoters of Smαa and Sm22α (Fig. 3C), and such binding was dramatically enhanced by hnRNPA2/B1 overexpression. These findings demonstrate for the first time that hnRNPA2/B1 regulates Smαa and Sm22α gene expressions during SMC differentiation through direct binding to the promoter region of specific SMC genes.
hnRNPA2/B1 Regulates Cbx3 Gene Expression
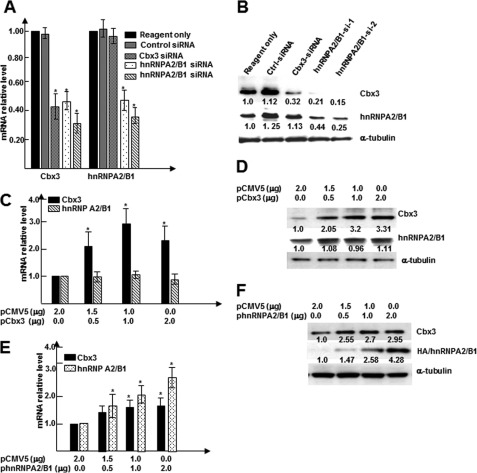
We have previously reported that nuclear protein Cbx3 plays a crucial role in SMC differentiation from stem cells and chick embryonic arteriogenesis (7). Interestingly, by examining the genomic locations of Hnrnpa2/b1 and Cbx3, we noticed that both mouse Cbx3 and Hnrnpa2/b1 genes localize in chromosome 6 and are closely linked to each other (29). In particular, the 5′-upstream region of the Cbx3 gene exhibits a high degree of similarity to a portion of the Hnrnpa2/b1 gene. Therefore, we wondered whether there is a relationship between these two genes. To test our hypothesis, knockdown experiments were conducted with Cbx3 and Hnrnpa2/b1 siRNAs in differentiating ES cells. The levels of gene and protein expressions of both genes were then analyzed using RT-qPCR (Fig. 4A) and Western blot analysis (Fig. 4B). Data shown in Fig. 4, A and B, revealed that both Cbx3 gene and protein levels were significantly down-regulated by the knockdown of Cbx3 gene itself as well as by the knockdown of the Hnrnpa2/b1 gene. Neither gene nor protein expression of hnRNPA2/B1 was down-regulated by the knockdown of the Cbx3 gene, suggesting that Cbx3 acts downstream of hnRNPA2/B1. Importantly, overexpression of hnRNPA2/B1 alone significantly up-regulated Cbx3 gene expression at both mRNA (Fig. 4C) and protein levels (Fig. 4D), whereas the gene (Fig. 4E) and protein (Fig. 4F) expression of hnRNPA2/B1 were not affected by Cbx3 overexpression. These data strongly suggest that Cbx3 is regulated by hnRNPA2/B1.
FIGURE 4.
hnRNPA2/B1 regulates Cbx3 gene expression. A and B, knockdown of hnRNPA2/B1 down-regulated Cbx3 gene expression. Cbx3-/Hnrnpa2/b1-specific siRNAs and random siRNA control were transfected into day 3 differentiating cells; after an additional 48 or 72 h of culture, total RNA and protein were harvested and subjected to RT-qPCR (A) and Western blot analysis (B). α-Tubulin was included as internal control. C and D, Hnrnpa2/b1 gene expression was not affected by Cbx3 overexpression. E and F, hnRNPA2/B1 overexpression significantly increased Cbx3 gene and protein expression levels. Undifferentiated ES cells were nucleofected using nucleofector II with different amounts of hnRNPA2/B1 expression plasmids pCMV5-Hnrnpa2/b1. Nucleofected cells were plated on dishes coated with 5 μg/ml collagen IV and cultured for 3–4 days in DM. Total RNA and protein were harvested and subjected to RT-qPCR (E) and Western blot analyses (F). Appropriate amounts of empty vector pCMV5 (control) were included to ensure total DNA concentrations for all experiments were equal. The data presented here are representative or mean ± S.E. of three independent experiments, *, p < 0.05. Numbers below the bands in B, D, and F indicate the average relative expression levels of target proteins from three independent experiments.
hnRNPA2/B1 Induces SMC Differentiation, at Least in Part, through Regulation of Cbx3 Gene Expression
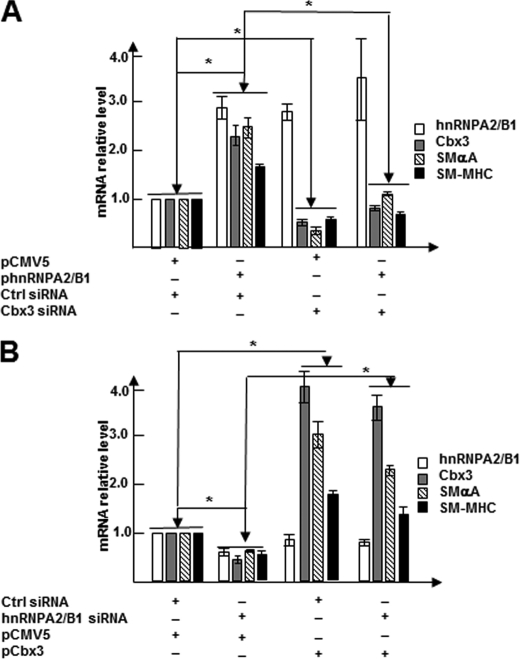
Next, we wondered if Cbx3 plays a role in hnRNPA2/B1-mediated SMC differentiation. Knockdown of Cbx3 with specific siRNA significantly abrogated the expression of SMC differentiation genes induced by hnRNPA2/B1 overexpression (Fig. 5A), suggesting that Cbx3 is crucial for hnRNPA2/B1-mediated SMC differentiation. To confirm that Cbx3 is essential in hnRNPA2/B1-mediated SMC gene expression, day 3 differentiating ES cells were co-transfected with control (pCMV5) or Cbx3 expression plasmids pCMV5-Cbx3 (2 μg/106 cells) and control random siRNA or Hnrnpa2/b1 siRNAs (100 nm). Data shown in Fig. 5B demonstrated that knockdown of hnRNPA2/B1 significantly down-regulated Smαa and Sm-mhc gene expression, although overexpression of Cbx3 dramatically up-regulated the expression of these genes. Importantly, compared with hnRNPA2/B1 siRNA transfection alone, co-transfection of hnRNPA2/B1 siRNAs and Cbx3 expression plasmid significantly increased Smαa and Sm-mhc gene expression. This suggests that Cbx3 overexpression can rescue the inhibitory effects of SMC differentiation gene expression mediated by Hnrnpa2/b1 siRNA. Taken together, these data strongly indicate that apart from the transcriptional regulation of SMC gene expression, hnRNPA2/B1 can also regulate Cbx3 to mediate SMC differentiation.
FIGURE 5.
hnRNPA2/B1 mediates SMC differentiation partially through regulation of Cbx3 gene expression. A, Cbx3 gene activation was essential for hnRNPA2/B1-mediated SMC gene expression. Day 3 differentiating ES cells were co-transfected with control (pCMV5) or hnRNPA2/B1 expression plasmids pCMV5-Hnrnpa2/b1 (2 μg/106 cells) and control random siRNA or Cbx3 siRNAs (100 nm). Total RNA were harvested 72 h after transfection and subjected to RT-qPCR analysis. B, impairment of SMC gene expression mediated by hnRNPA2/B1 knockdown was rescued by Cbx3 overexpression. Day 3 differentiating ES cells were co-transfected with control random siRNA or Hnrnpa2/b1 siRNAs (100 nm) and control (pCMV5) or Cbx3 expression plasmids pCMV5-Cbx3 (2 μg/106 cells). Total RNA was harvested 72 h after transfection and subjected to RT-qPCR analysis. The data presented here are a representative or mean ± S.E. of three independent experiments, *, p < 0.05.
hnRNPA2/B1 Knockdown in Neural Crest Cells Causes Maldevelopment of Chick Embryos
To explore the in vivo relevance of hnRNPA2/B1 in SMC differentiation and cardiovascular development, whole mount immunohistochemical staining with hnRNPA2/B1 antibody and whole mount in situ hybridization staining with digoxin-labeled antisense RNA probe specific for Hnrnpa2/b1 were performed on chick embryos as described previously (7). We observed a consistent positive staining on the developing heart and the first four branchial arch arteries (data not shown). Importantly, hnRNPA2/B1 expression co-localized with SMαA on the floor of dorsal aorta as demonstrated by triple immunofluorescent staining (supplemental Fig. S1), suggesting that hnRNPA2/B1 might be involved in vascular smooth muscle development in the chick embryo.
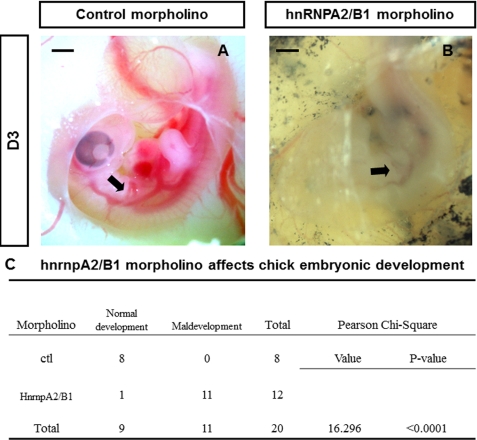
To examine the involvement of hnRNPA2/B1 in the differentiation of neural crest cells into SMCs, specifically designed Hnrnpa2/b1 or control morpholinos (Gene Tools, LLC) were introduced into the right-hand side of the neural crest using electroporation. The electroporator was set at 7.5 V, 4 pulses, length of 50 ms, and interval of 1000 ms as described previously (7). Twenty four hours after transfection, embryos were checked, and only those with the correct fluorescence labeling (Hnrnpa2/b1 morpholino versus control morpholino, n = 12 versus 8, respectively) were included for further evaluation. Whole mount immunohistochemistry showed significant inhibition of hnRNPA2/B1 protein expression in neural tubes that had been transfected with Hnrnpa2/b1 morpholino, whereas the expression of hnRNPA2/B1 in neural tubes electroporated with control morpholino was not affected (supplemental Fig. S2). Importantly, we observed that chick embryos electroporated with Hnrnpa2/b1 morpholino grew slowly, and most of them were smaller than embryos in the control group. Particularly, 48 h after electroporation, branchial arch arteries in the Hnrnpa2/b1 morpholino chick embryos did not seem to function well, with either nil or reduced blood circulation. In comparison, no observed embryos in the control morpholino group developed abnormally, even at a later stage (Fig. 6, A and B). χ2 tests showed that the percentage of embryos with maldevelopment in Hnrnpa2/b1 morpholino group was dramatically higher than that of the control morpholino group (p < 0.0001, Fig. 6C).
FIGURE 6.
Inhibition of hnRNPA2/B1 causes maldevelopment of chick embryos. A and B, representative images of embryos transfected with hnRNPA2/B1 and control morpholino-oligonucleotides. Most embryos in the Hnrnpa2/b1 morpholino group have reduced blood circulation in branchial arch arteries (B) in comparison with embryos electroporated with control morpholino (A). Black arrow indicates branchial arch arteries. C, maldevelopment of chick embryos was caused by inhibition of hnRNPA2/B1 expression in NCCs. χ2 analysis was performed using SPSS 17.0 software. It was that found Hnrnpa2/b1 morpholino significantly affected the chick embryonic development (p < 0.0001). Scale bar, 1 mm.
hnRNPA2/B1 Knockdown Inhibits Neural Crest Cell Migration and Differentiation
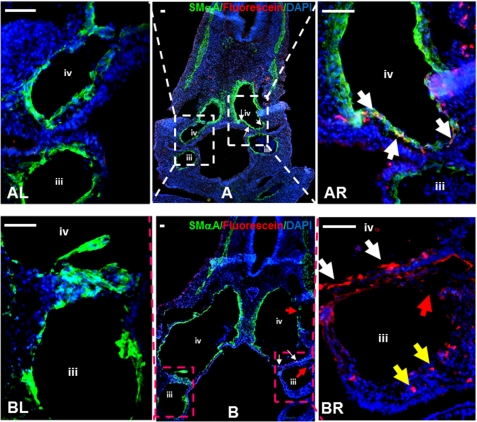
To investigate the underlying mechanisms in the maldevelopment or death of embryos, five embryos from each group were selected randomly to perform double immunofluorescent staining for SMαA and fluorescein expression to trace SMC differentiation and migration of neural crest cells in the developing chick embryos. Our data revealed that the smooth muscle of branchial arch arteries, particularly the 3rd and 4th arteries on the right-hand side of the embryos in the Hnrnpa2/b1 morpholino group did not develop completely (Fig. 7BR, red arrow; supplemental Fig. S3). However, branchial arch arteries at the left side of the Hnrnpa2/b1 morpholino group and both sides of embryos in control morpholino group (Fig. 7, AL, AR, and BL; supplemental Fig. S3) developed normally. Such developmental defects could result from either the delayed migration of neural crest cells transfected with Hnrnpa2/b1 morpholino (indicated by yellow arrows in Fig. 7BR; supplemental Fig. S3) or the differentiation of neural crest cells into SMCs was abolished if migration is not affected (indicated by white arrows in Fig. 7BR; supplemental Fig. S3). In comparison, neural crest cells transfected with control morpholino not only migrate into the vessel wall but also differentiate into SMαA-positive cells (white arrows in Fig. 7AR; supplemental Fig. S3). These data strongly suggest that the knockdown of hnRNPA2/B1 in neural crest cells delays the cell migration and inhibits SMC differentiation, resulting in developmental defects of branchial arch arteries of chick embryos. Subsequently, these lead to the breakage of arteries (red arrows in Fig. 7, B and BR; supplemental Fig. S3) and lethality of embryos.
FIGURE 7.
Inhibition of hnRNPA2/B1 in NCCs results in developmental defects of 3rd and 4th branchial arch arteries. A and B, triple stainings for DAPI (blue), SMαA (green), and morpholino-oligonucleotides (red fluorescein) with sections from embryos electroporated with control (A) or Hnrnpa2/b1 (B) morpholino-oligonucleotides. In Hnrnpa2/b1 morpholino group, fluorescein-labeled NCCs could not migrate into branchial arch arteries at the right-hand side (BR, yellow arrows) and differentiate into SMCs (BR, white arrows), leading to the developmental defect of smooth muscle in the 3rd and 4th branchial arches (BR, indicated by red arrows). In contrast, the left-hand side of branchial arch arteries (BL) developed normally. In control morpholino group, both sides of branchial arch arteries showed complete smooth muscle formation (AR and AL). The fluorescein-labeled NCCs could not only migrate into branchial arch arteries wall at the right-hand side but also differentiate into SMCs in the 4th branchial arch artery. White arrows indicate those NCCs that are differentiating to SMCs. Representative images from five embryos in each group are presented here. Scale bar, 50 μm. Isotype IgG substituted primary antibody as negative control during staining process. iii, 3rd branchial arch artery; iv, 4th branchial arch artery. Original unmerged images of AR and BR were provided in supplemental Fig. S3.
DISCUSSION
Adult SMCs retain significant plasticity to carry out different functions. After vascular injury and in response to local inflammation factors or cytokine stimulations, vascular contractile SMCs undergo phenotype switching to acquire either synthetic or dedifferentiated phenotypes. These SMCs then participate in the formation of neointima by decreasing the expression of SMC differentiation markers (1). Similarly, during the progression of atherosclerosis, recruited SMCs also acquired a transition to a synthetic phenotype in lesion formation (30). Studies on the mechanism of SMC de/differentiation are not only crucial for exploring human pathologies but are also useful for isolating and differentiating stem cells or vascular progenitors to SMCs, which is required for the tissue-engineered vessel. In this study, we uncovered important roles of nuclear protein hnRNPA2/B1 in SMC differentiation from ES cell and chick embryo arteriogenesis. Our current data strongly suggest that hnRNPA2/B1 is a potential modulating target to efficiently derive SMCs from stem cells for cardiovascular regenerative medicine.
hnRNPA2/B1 and SMC Differentiation
The Hnrnpa2/b1 gene gives rise to two different isoforms, A2 and B1. B1 mRNA is alternatively spliced from the primary Hnrnpa2/b1 gene transcript by including a 36-bp exon, which adds 12 amino acids near the amino terminus in B1 isoform (15). Kamma et al. (31) showed that hnRNPA2/B1 is localized in the nucleus with considerable tissue-specific variation in rat. This is consistent with the immunofluorescent staining that we performed in differentiating SMCs, where hnRNPA2/B1 localized in the nucleus.
hnRNPA2/B1 is involved in various different nuclear activities, such as post-transcriptional regulation of RNA splicing and transportation or transcriptional regulation of c-Myc, apolipoprotein E (apo E), vitamin receptor gene, breast cancer 1 (BRCA1), and gonadotropin-releasing-hormone 1 (GnRH1) (22–26). Most hnRNPA2/B1 studies were carried out to study cancer cell metabolism and phenotype switch, which are related to alternative splicing (32–37). However, little is known about hnRNPA2/B1 in stem cell differentiation, especially in SMC lineage and embryonic vascular development. At the start of the study, hnRNPA2/B1 was revealed using nuclear proteomics as one of several nuclear proteins whose protein levels were significantly up-regulated during early SMC differentiation from ES cells (data not shown), which strongly implies that hnRNPA2/B1 may play a role in SMC differentiation.
Subsequently, the importance of hnRNPA2/B1 in SMC differentiation from ES cell was further demonstrated using gain and loss of Hnrnpa2/b1 gene functions in differentiating ES cells. We have demonstrated that enhanced Hnrnpa2/b1 gene expression significantly promotes SMC differentiation, although knockdown of endogenous Hnrnpa2/b1 gene expression dramatically inhibits SMC differentiation. No such effects were observed in differentiating ES cells on other cell lineage gene expression such as markers from endothelial cell (Flt-1 and CD144), hematopoietic progenitor cells (CD133 and Alcam), cardiac fibroblast (CD90 and Ddr2), cardiomyocyte (Actc-1 and Tnnc-1), trophoblast (Dlx3 and Tpbg), mesenchymal stem cell (CD29 and CD44), and neural progenitors (Nestin and Gap43) (data not shown). Taken together, our findings indicate that hnRNPA2/B1 plays a critical role in mediating SMC differentiation from ES cells.
hnRNPA2/B1 Regulates SMCs Differentiation through a Transcriptional Mechanism
Although evidence showed that hnRNPA2/B1 regulates gene expression mainly via post-transcriptional modification and transport of pre-RNAs (15, 16), recent research also strongly suggests that hnRNPA2/B1 can regulate gene expression via a transcriptional mechanism. For instance, studies showed that hnRNPA2/B1 regulates c-myc, apoE, vitamin D receptor, Brca1, and Gnrh1 gene expression by binding to related gene promoters (22–26). hnRNPA2/B1 has been reported as a gene transcriptional activator or repressor in different gene regulations. For example, hnRNPA2 transcriptionally represses expression of vitamin D receptor, but it works more like an activator for Brca1 gene transcription (14, 38). In this study, our current findings also revealed that hnRNPA2/B1 is an activator that regulates SMC Smαa and Sm22 gene expression during SMC differentiation from ES cells. Although G-quadruplex formation in the promoter region and protein-protein interaction was proposed in Hnrnpa2/b1 gene regulation (39, 40), the mechanism of hnRNPA2/B1 in transcriptional regulation remains unclear. Moreover, whether other functions of hnRNPA2/B1 such as alternative splicing and nuclear-cytosome transportation of mRNA are involved in SMC differentiation remains to be elucidated.
hnRNPA2/B1 and Cbx3 Interaction in SMC Differentiation
We have identified nuclear protein hnRNPA2/B1 as a critical SMC differentiation regulator in this study. Also, we previously reported that nuclear protein Cbx3 plays a crucial role in SMC differentiation from stem cells and in chick embryonic arteriogenesis (7). Interestingly, evidence has revealed a close link between Hnrnpa2/b1 and Cbx3 gene (29). All this evidence has prompted us to investigate the functional interactions of hnRNPA2/B1 and Cbx3 and its significance in SMC differentiation from stem cells. Our data revealed that hnRNPA2/B1 is an upstream regulator of Cbx3. By applying expression plasmids and siRNA co-transfection strategies, we further demonstrated that Cbx3 is required for hnRNPA2/B1-mediated SMC differentiation. These data strongly indicate that apart from direct activation of SMC gene transcription, hnRNPA2/B1 also regulates Cbx3 to promote SMC differentiation.
hnRNPA2/B1 Is Involved in Embryonic Arteriogenesis
hnRNPA2/B1 is expressed in multiple organogenesis such as neural, gastrointestinal, skeletal, and cardiovascular systems during early embryonic development. We found that hnRNPA2/B1 was strongly expressed in the dorsal aorta and branchial arches arteries and co-localized with the SMC-specific marker SMαA on the floor of dorsal aorta in chick embryos. SMαA-positive cells in HH 16 chick embryos were restricted to the aortic floor, where the first smooth muscle starts to form during embryonic aorta angiogenesis (41). Therefore, we speculated that hnRNPA2/B1 may play a role in the earlier induction of SMC differentiation during embryonic arteriogenesis. To address this possibility, instead of using siRNAs, electroporation of morpholino-oligonucleotides on the neural crest in chick embryos was applied.
First, although specific knockdown strategies using morpholino-oligonucleotides or siRNA have been successfully proven to be powerful tools to study gene functions in different organisms in vivo, the off-target effects independent to siRNA sequence occasionally cause embryo developmental defects or even death (42, 43). Whereas no such side effects was reported with morpholino, the use of morpholino-oligonucleotides to inhibit protein translation of certain genes was therefore a preferable approach for knockdown experiments during chick development (44).
Second, neural crest cells residing at the levels of rhombomere 2, 4, , and 7 of chick embryos at stages HH10−/HH10/HH10+ were chosen to perform in vivo electroporation using Hnrnpa2/b1 morpholino-oligonucleotides. It is well known that neural crest cells are the only cell lineage contributing to smooth muscle during branchial arch angiogenesis (45, 46). In particular, neural crest cells residing at the level of rhombomere 2, 4, 6, and 7 will migrate into branchial arch 1–4 and differentiate toward SMCs at day 3 and 4 in chick embryonic development (47–49). Therefore, these neural crest cells serve as a specific target for studying the differentiation of embryonic progenitors into SMCs and the functional importance of any genes in branchial arch artery development. As expected, stainings on cryosections clearly showed that the smooth muscle of the 3rd and 4th branchial arch arteries at the right-hand side of embryos transfected with Hnrnpa2/b1 morpholino did not form completely. However, branchial arch arteries at the left-hand side and both sides of the embryos in the control morpholino group developed normally with at least the formation of a single layer of smooth muscle cells. Thus, the inhibition of hnRNPA2/B1 is responsible for the developmental defects of these branchial arteries in chick embryos, eventually leading to embryo death.
Summary
The functional role of hnRNPA2/B1 in SMC differentiation from stem cells and embryonic branchial arteriogenesis has been demonstrated in this study. We showed that hnRNPA2/B1 mediates SMC differentiation by direct binding and activates the promoters of SMC differentiation genes (Smαa and Sm22α). We further identified a regulatory role of hnRNPA2/B1 on Cbx3 gene expression during SMC differentiation from stem cells. Our studies highlight the importance of hnRNPA2/B1 in SMC differentiation from both stem cells and chick embryonic arteriogenesis. These findings will significantly contribute to the understanding of the mechanisms in embryonic arteriogenesis and benefit future application in engineering tissue vessels.
Supplementary Material
This work was supported in part by the British Heart Foundation and Oak Foundation. This work forms part of the research themes contributing to the translational research portfolio of Barts and the London Cardiovascular Biomedical Research Unit supported and funded by the National Institute of Health Research.

This article contains supplemental “Materials and Methods,” Figs. S1–S3, Table 1, and additional references.
- SMC
- smooth muscle cell
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- qPCR
- quantitative PCR
- DM
- differentiation medium
- HH
- Hamburger and Hamilton
- NCC
- neural crest cell.
REFERENCES
- 1. Yoshida T., Kaestner K. H., Owens G. K. (2008) Conditional deletion of Krüppel-like factor 4 delays down-regulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ. Res. 102, 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xiao Q., Luo Z., Pepe A. E., Margariti A., Zeng L., Xu Q. (2009) Embryonic stem cell differentiation into smooth muscle cells is mediated by Nox4-produced H2O2. Am. J. Physiol. Cell Physiol. 296, C711–C723 [DOI] [PubMed] [Google Scholar]
- 3. Margariti A., Xiao Q., Zampetaki A., Zhang Z., Li H., Martin D., Hu Y., Zeng L., Xu Q. (2009) Splicing of HDAC7 modulates the SRF-myocardin complex during stem cell differentiation toward smooth muscle cells. J. Cell Sci. 122, 460–470 [DOI] [PubMed] [Google Scholar]
- 4. Xiao Q., Zeng L., Zhang Z., Hu Y., Xu Q. (2007) Stem cell-derived Sca-1+ progenitors differentiate into smooth muscle cells, which is mediated by collagen IV-integrin α1/β1/αv and PDGF receptor pathways. Am. J. Physiol. Cell Physiol. 292, C342–C352 [DOI] [PubMed] [Google Scholar]
- 5. Pepe A. E., Xiao Q., Zampetaki A., Zhang Z., Kobayashi A., Hu Y., Xu Q. (2010) Crucial role of nrf3 in smooth muscle cell differentiation from stem cells. Circ. Res. 106, 870–879 [DOI] [PubMed] [Google Scholar]
- 6. Xiao Q., Wang G., Luo Z., Xu Q. (2010) The mechanism of stem cell differentiation into smooth muscle cells. Thromb. Haemost. 104, 440–448 [DOI] [PubMed] [Google Scholar]
- 7. Xiao Q., Wang G., Yin X., Luo Z., Margariti A., Zeng L., Mayr M., Ye S., Xu Q. (2011) Chromobox protein homolog 3 is essential for stem cell differentiation to smooth muscles in vitro and in embryonic arteriogenesis. Arterioscler. Thromb. Vasc. Biol. 31, 1842–1852 [DOI] [PubMed] [Google Scholar]
- 8. Zhang L., Jin M., Margariti A., Wang G., Luo Z., Zampetaki A., Zeng L., Ye S., Zhu J., Xiao Q. (2010) Sp1-dependent activation of HDAC7 is required for platelet-derived growth factor-BB-induced smooth muscle cell differentiation from stem cells. J. Biol. Chem. 285, 38463–38472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mack C. P. (2011) Signaling mechanisms that regulate smooth muscle cell differentiation. Arterioscler. Thromb. Vasc. Biol. 31, 1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 11. Lindahl P., Johansson B. R., Levéen P., Betsholtz C. (1997) Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242–245 [DOI] [PubMed] [Google Scholar]
- 12. Yamashita J., Itoh H., Hirashima M., Ogawa M., Nishikawa S., Yurugi T., Naito M., Nakao K., Nishikawa S. (2000) Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature 408, 92–96 [DOI] [PubMed] [Google Scholar]
- 13. Hautmann M. B., Madsen C. S., Owens G. K. (1997) A transforming growth factor β (TGFβ) control element drives TGFβ-induced stimulation of smooth muscle α-actin gene expression in concert with two CArG elements. J. Biol. Chem. 272, 10948–10956 [DOI] [PubMed] [Google Scholar]
- 14. He Y., Smith R. (2009) Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell. Mol. Life Sci. 66, 1239–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ball E. E., Rehm E. J., Goodman C. S. (1991) Cloning of a grasshopper cDNA coding for a protein homologous to the A1, A2/B1 proteins of mammalian hnRNP. Nucleic Acids Res. 19, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dreyfuss G., Matunis M. J., Piñol-Roma S., Burd C. G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62, 289–321 [DOI] [PubMed] [Google Scholar]
- 17. Cobianchi F., Karpel R. L., Williams K. R., Notario V., Wilson S. H. (1988) Mammalian heterogeneous nuclear ribonucleoprotein complex protein A1. Large scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J. Biol. Chem. 263, 1063–1071 [PubMed] [Google Scholar]
- 18. Moran-Jones K., Wayman L., Kennedy D. D., Reddel R. R., Sara S., Snee M. J., Smith R. (2005) hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res. 33, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishikawa F., Matunis M. J., Dreyfuss G., Cech T. R. (1993) Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 13, 4301–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xia H. (2005) Regulation of γ-fibrinogen chain expression by heterogeneous nuclear ribonucleoprotein A1. J. Biol. Chem. 280, 13171–13178 [DOI] [PubMed] [Google Scholar]
- 21. Leverrier S., Cinato E., Paul C., Derancourt J., Bemark M., Leanderson T., Legraverend C. (2000) Purification and cloning of type A/B hnRNP proteins involved in transcriptional activation from the Rat spi 2 gene GAGA box. Biol. Chem. 381, 1031–1040 [DOI] [PubMed] [Google Scholar]
- 22. Takimoto M., Tomonaga T., Matunis M., Avigan M., Krutzsch H., Dreyfuss G., Levens D. (1993) Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem. 268, 18249–18258 [PubMed] [Google Scholar]
- 23. Campillos M., Lamas J. R., García M. A., Bullido M. J., Valdivieso F., Vázquez J. (2003) Specific interaction of heterogeneous nuclear ribonucleoprotein A1 with the −219T allelic form modulates APOE promoter activity. Nucleic Acids Res. 31, 3063–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen H., Hewison M., Hu B., Adams J. S. (2003) Heterogeneous nuclear ribonucleoprotein (hnRNP) binding to hormone response elements. A cause of vitamin D resistance. Proc. Natl. Acad. Sci. U.S.A. 100, 6109–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thakur S., Nakamura T., Calin G., Russo A., Tamburrino J. F., Shimizu M., Baldassarre G., Battista S., Fusco A., Wassell R. P., Dubois G., Alder H., Croce C. M. (2003) Regulation of BRCA1 transcription by specific single-stranded DNA binding factors. Mol. Cell. Biol. 23, 3774–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao S., Korzan W. J., Chen C. C., Fernald R. D. (2008) Heterogeneous nuclear ribonucleoprotein A/B and G inhibits the transcription of gonadotropin-releasing hormone 1. Mol. Cell. Neurosci. 37, 69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamburger V. (1992) The stage series of the chick embryo. Dev. Dyn. 195, 273–275 [DOI] [PubMed] [Google Scholar]
- 28. Hamburger V., Hamilton H. L. (1992) A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 195, 231–272 [DOI] [PubMed] [Google Scholar]
- 29. Sato M., Miyado K., Kimura M. (2001) Cloning and characterization of 5′-upstream sequence of the M32 gene for a mouse homologue of Drosophila heterochromatin protein 1 (HP1). DNA Seq. 12, 97–106 [DOI] [PubMed] [Google Scholar]
- 30. Xu Q. (2008) Stem cells and transplant arteriosclerosis. Circ. Res. 102, 1011–1024 [DOI] [PubMed] [Google Scholar]
- 31. Kamma H., Horiguchi H., Wan L., Matsui M., Fujiwara M., Fujimoto M., Yazawa T., Dreyfuss G. (1999) Molecular characterization of the hnRNPA2/B1 proteins. Tissue-specific expression and novel isoforms. Exp. Cell Res. 246, 399–411 [DOI] [PubMed] [Google Scholar]
- 32. Clower C. V., Chatterjee D., Wang Z., Cantley L. C., Vander Heiden M. G., Krainer A. R. (2010) The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 1894–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. David C. J., Chen M., Assanah M., Canoll P., Manley J. L. (2010) hnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Golan-Gerstl R., Cohen M., Shilo A., Suh S. S., Bakàcs A., Coppola L., Karni R. (2011) Splicing factor hnRNPA2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 71, 4464–4472 [DOI] [PubMed] [Google Scholar]
- 35. Liang Y., Shi S. L., Li Q. F., Chen L. Y., Jing G. J., Tan G. W., Wang S. Y., Wu F. Y. (2011) The localization of hnRNPA2/B1 in nuclear matrix and the aberrant expression during the RA-induced differentiation of human neuroblastoma SK-N-SH cells. J. Cell. Biochem. 112, 1722–1729 [DOI] [PubMed] [Google Scholar]
- 36. McGlincy N. J., Tan L. Y., Paul N., Zavolan M., Lilley K. S., Smith C. W. (2010) Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNPA2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics 11, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tauler J., Zudaire E., Liu H., Shih J., Mulshine J. L. (2010) hnRNPA2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 70, 7137–7147 [DOI] [PubMed] [Google Scholar]
- 38. He Y., Brown M. A., Rothnagel J. A., Saunders N. A., Smith R. (2005) Roles of heterogeneous nuclear ribonucleoproteins A and B in cell proliferation. J. Cell Sci. 118, 3173–3183 [DOI] [PubMed] [Google Scholar]
- 39. Kostadinov R., Malhotra N., Viotti M., Shine R., D'Antonio L., Bagga P. (2006) GRSDB. A database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 34, D119–D124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vera J., Jaumot M., Estanyol J. M., Brun S., Agell N., Bachs O. (2006) Heterogeneous nuclear ribonucleoprotein A2 is a SET-binding protein and a PP2A inhibitor. Oncogene 25, 260–270 [DOI] [PubMed] [Google Scholar]
- 41. Wiegreffe C., Christ B., Huang R., Scaal M. (2007) Sclerotomal origin of smooth muscle cells in the wall of the avian dorsal aorta. Dev. Dyn. 236, 2578–2585 [DOI] [PubMed] [Google Scholar]
- 42. Scacheri P. C., Rozenblatt-Rosen O., Caplen N. J., Wolfsberg T. G., Umayam L., Lee J. C., Hughes C. M., Shanmugam K. S., Bhattacharjee A., Meyerson M., Collins F. S. (2004) Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007) p53 activation by knockdown technologies. PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mende M., Christophorou N. A., Streit A. (2008) Specific and effective gene knockdown in early chick embryos using morpholinos but not pRFPRNAi vectors. Mech. Dev. 125, 947–962 [DOI] [PubMed] [Google Scholar]
- 45. Jiang X., Rowitch D. H., Soriano P., McMahon A. P., Sucov H. M. (2000) Fate of the mammalian cardiac neural crest. Development 127, 1607–1616 [DOI] [PubMed] [Google Scholar]
- 46. Nakamura T., Colbert M. C., Robbins J. (2006) Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ. Res. 98, 1547–1554 [DOI] [PubMed] [Google Scholar]
- 47. Trainor P. A., Sobieszczuk D., Wilkinson D., Krumlauf R. (2002) Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Development 129, 433–442 [DOI] [PubMed] [Google Scholar]
- 48. Lumsden A., Sprawson N., Graham A. (1991) Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development 113, 1281–1291 [DOI] [PubMed] [Google Scholar]
- 49. Trainor P. A., Krumlauf R. (2000) Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat. Rev. Neurosci. 1, 116–124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.