Abstract
An enzyme-stabilized nucleophilic water molecule has been implicated at the transition state of Escherichia coli methylthioadenosine nucleosidase (EcMTAN) by transition state analysis and by crystallography. We analyzed the EcMTAN mass in complex with a femtomolar transition state analogue to determine if the inhibitor and nucleophilic water can be detected in the gas phase. Complexes of EcMTAN-inhibitor and EcMTAN-inhibitor-nucleophilic water were identified by high-resolution mass spectrometry under non-denaturing conditions. The stability of the enzyme-inhibitor-water complex is adequate to exist in the gas phase.
Compared with solution reactions, enzymes enhance the reaction rates by as much as twenty orders of magnitude.1 During the reaction coordinate of an enzyme-catalyzed reaction, a transition state is reached by a dynamic search of enzyme-reactant geometry.2,3 Molecules mimicking the structure of a transition state convert the rare enzymatic geometry of the transition state to a stable complex that binds with high affinity to the targeted enzymes.4–6 Transition state structures for enzymatic reactions have been established by the combination of kinetic isotope effects and quantum computational calculations.6–8 Using this approach, transition states of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidases (MTANs) from several bacterial species have been established.9,10 Transition state analogue inhibitors have been designed and synthesized with Ki values in the femtomolar range.11,12 This tight binding is proposed to support faithful mimicry of transition state structure. Transition state analysis and crystal structures of MTAN with transition state analogue inhibitors implicate a crystallographic water molecule in a position to act as the nucleophile. Tightly-bound inhibitors have slow release rates in solution and we wanted to see if a transition state analogue together with its nucleophilic water would be retained into the gas phase during mass spectrometry experiments.
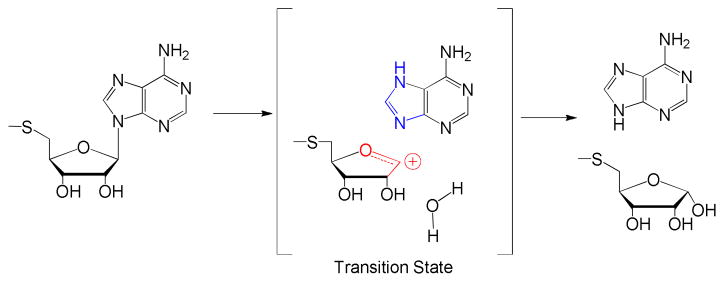
MTAN is a dimeric enzyme and catalyzes hydrolytic depurination of 5′-methylthioadenosine (MTA) and S-adenosylhomocysteine. The transition state for MTA hydrolysis by EcMTAN has been reported to be a 5-methylthioribocation and a neutral adenine, with a C1′ to N9 distance of 3 Å or more.13 A nucleophilic water molecule was proposed to be part of the transition state complex (Fig. 1). Water attacks C1′ of the ribocation to complete the reaction.13 Assuming a transition state lifetime of 10−14 s,14 the nucleophilic water must be pre-organized at the attacking position. The transition state lifetime is too short to permit significant water diffusion.15,16 Transition state analogues mimicking the geometry and molecular electrostatic potential of the transition state structure of MTANs have Ki values in the femtomolar range.11
Figure 1.
The transition state structure of MTA catalyzed by EcMTAN.13
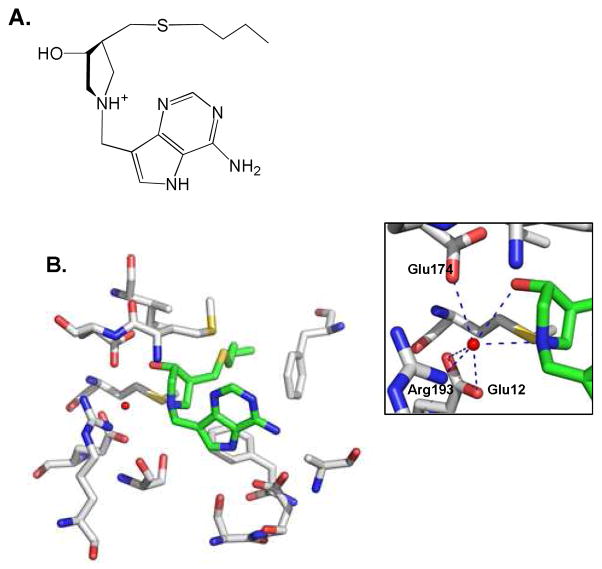
Crystal structures with transition state analogue complexes reveal an extensive bonding network with the enzyme (Fig. 2),17,18 consistent with the powerful binding interactions. BuT-DADMe-ImmA has a high affinity for EcMTAN (Ki = 0.3 pM) and its structure has been solved with Vibrio cholerae MTAN (VcMTAN), which shares 60% sequence identity with EcMTAN. Inhibitor is bound in the active site with 5 favorable hydrogen bonds to the protein and two hydrogen bonds to the nucleophilic water molecule and multiple hydrophobic interactions (Figs. 2B, S1). A highly-stabilized nucleophilic water molecule is positioned 2.7 Å from the cationic nitrogen (pKa ~9) that mimics the 1′–anomeric carbon of the ribocation transition state (Fig. 2).18 This complex contains all elements of the transition state complex, including the nucleophilic water. High-resolution mass spectrometry was applied to determine if the complex remains intact following extraction into the gas phase.
Figure 2.
The complex of VcMTAN, BuT-DADMe-ImmA and a water molecule. A. The chemical structure of BuT-DADMe-ImmA is shown in the orientation corresponding to its position in the crystal structure. B. In the crystal structure (pdb 3DP9), BuT-DADMe-ImmA (green) and a water molecule (red dot) are surrounded by active site residues (gray). Inset: hydrogen bonding network to the water molecule. PYMOL was used to generate the graphs.
EcMTAN is a homodimer with a calculated monomer molecular weight of 26029.7 Da. The experimental mass for the apo enzyme was studied from a denatured sample containing 50% methanol and 0.1% formic acid using a nano electrospray quadrupole time of flight mass spectrometer (nanoESI-QTof). An experimental mass of 26029.6 Da was acquired, confirming the integrity of the expressed enzyme and the absence of bound water or ions (Fig. S2A).
Native dimeric EcMTAN in 5 mM ammonium acetate was analyzed and produced peaks at m/z 4339.3, 4005.6, 3719.5, 3471.7, and 3254.7. When deconvoluted for charge states, these correspond to the multiply protonated dimeric MTAN with the charge states of 12+, 13+, 14+, 15+ and 16+ respectively. The deconvoluted spectrum gave an experimental dimer molecular mass of 52059.6 Da, in good agreement with the calculated value of 52059.4 Da. Thus, denatured monomer and native dimer can be mobilized into the gas phase without additional water, ion or solute molecules (Fig. S2B).
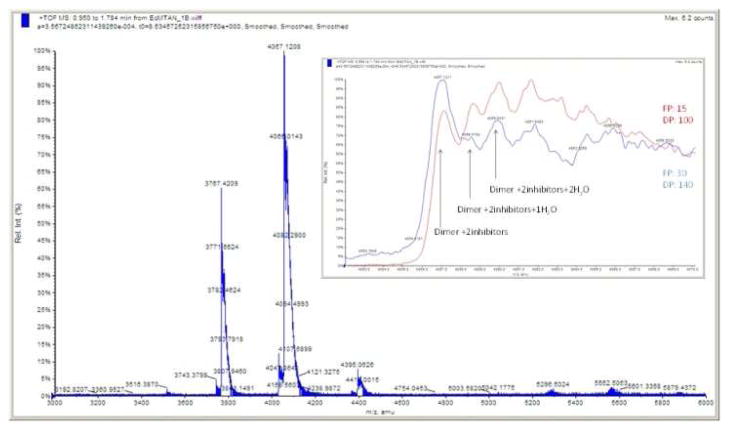
The calculated molecular weight of BuT-DADMe-ImmA of 336.18 was also confirmed by MS (Fig. S3). Molar equivalent amounts of BuT-DADMe-ImmA and EcMTAN were mixed and the mass spectrometer (MS) spectrum was acquired under conditions optimized for dimeric EcMTAN. Peaks generated from this complex shifted to higher m/z values than those of dimeric EcMTAN alone (Fig. 3). Spectral deconvolution revealed a species corresponding to two inhibitor molecules bound to dimeric EcMTAN, at 52730.8 Da. Another peak corresponds to dimer, two inhibitors and two water molecules to give the mass peak at 52766.4 Da. Other peaks are also observed and are consistent with addition of Na+ and/or K+ ions in the complex (Fig. 3).
Figure 3.
MS analysis of EcMTAN in complex with stoichiometric BuTDADMe-ImmA. Inset: with increased focusing potential and declustering potential, water molecules are shifting out of the enzyme-inhibitor complex.
Altered protein mass in MS analysis might possibly originate from oxidation of the protein or the inhibitor during high voltage electrospray. Oxidation produces mass shifts of 32 Da (two oxygen addition), compared to that of 36 Da generated from two water molecules. As indicated above and in the supplemental information, no oxidation was observed in either apo-MTAN or the inhibitor under our experimental conditions (Figs. S2, S3). Hence, oxidation of the dimeric enzyme and inhibitors complex was considered unlikely.
Additional tests for the nature of the EcMTAN complexes in MS analysis involved different experimental conditions. During MS analysis, ions are generated, desolvated and accelerated by potential voltage differences. This was achieved by applying different voltages to components between the ESI tip and the detector. Higher potential accelerates ions faster and results in more energetic collision with nitrogen along the ion path.19,20 More energetic collisions are expected to result in the dissociation of non-covalently bound molecules such as water or inhibitor in the complex.
Combinations of focusing potential (FP) and declustering potential (DP) generated by differential voltage application were used to produce different potential MS energies. First, mass spectra of the inhibitor bound complex were acquired with FP 100 and DP 15 (arbitrary potential voltage units) (Fig. 3 inset, red). Then, a higher potential was applied (FP 140 and DP 30) to investigate possible changes (Fig. 3 inset, blue). Mass differences between the three peaks arising from oxidation would not change the peak intensity ratios. However, the high potential combination increased the peak intensity at m/z 4057.1 compared to the original conditions (Fig. 3, inset). Increased intensity of the peak at m/z 4057.1 implies that more energetic collisions under higher potential conditions caused the loss of water molecules. The apoenzyme produced a single peak at each charge, regardless of change to the potential. These results support the presence of bound water molecules only in the complex of EcMTAN together with transition state analogues in the gas phase, and not to the apoenzyme.
Detection of the water molecule agrees with the chemical mechanism of EcMTAN (Fig. 1), in which a nucleophilic water attacks C1′ at the transition state. Because of the short life-time of enzymatic transition states (< 10−14 s),15 the water molecule must be pre-organized near C1′ at the transition state. During the transition state lifetime, there is insufficient time for water diffusion. This preorganization explains the affinity of bound water in complex with the transition state analogue. The crystal structure shows the presumptive water molecule hydrogen bonded with the 3′-OH and N1′ of BuT-DADMe-ImmA, as well as the side chains of Arg193, Glu12 and Glu174 of MTAN (Fig. 2). Consequently, we propose that the water molecules identified by MS are the nucleophilic waters.
We did not observe the EcMTAN dimer bound to only one inhibitor unless we intentionally formed this complex by sub-stoichiometric inhibitor titration (Fig. S4). Thus, the inhibitor binds to both subunits with sufficient affinity to be retained in the gas phase. The stoichiometric binding of the inhibitor with EcMTAN results from the slow dissociation rate constant (koff). In solution, the association rate constant (kon) and slow-onset conformational change for tight binding inhibitors to MTAN is in the range of 103 s−1. To achieve a Ki value in the femtomolar range, koff should be in the range of 10−9 s−1. Consistent with this approximation of koff, incubation of the enzyme-inhibitor complex in solution with a near-saturation concentration of MTA (2 mM) did not restore significant enzymatic activity in a three hour incubation.
BuT-DADMe-ImmA has a Ki value of 0.3 pM with EcMTAN, and binds to the enzyme with geometry similar to the substrate.21 Both inhibitor and enzyme contacts are needed to bind the nucleophilic water molecule in the enzyme. The number of favorable hydrogen bonds and hydrophobic interactions increase as a substrate analogue of MTA is replaced with BuT-DADMe-ImmA at the catalytic site.21,22 In addition, a new ionic interaction is formed between the inhibitor cation and the immobilized nucleophilic water.18,22 Thus, BuT-DADMe-ImmA is an excellent transition state analogue with high binding affinity to EcMTAN and an ability to stabilize the nucleophilic water molecule. A second crystallographic water in contact with the nucleophilic water is weakly bound with only one favorable hydrogen bond (Fig. S1).
Although BuT-DADMe-ImmA and EcMTAN function to stabilize a catalytic site water, the water molecule has weaker affinity to the protein than the inhibitor. Complexes were detected by MS with sub-stoichiometric amounts of water, reflecting the dynamic hydrogen-bond nature of water and its weaker hydrogen-bond network. Supersaturation with inhibitor produced two additional smaller peaks with m/z ratios of 4085.7 and 4111.5 (Fig. S5), corresponding to an enzyme complex with one and two additional inhibitors bound per dimer, but with no additional water molecules. These peaks are attributed to non-specific binding of the inhibitor (acting as a cation) and the enzyme.
An attempt to measure the exchange rate of 16O water from the enzyme-inhibitor-water complex used dilution experiments into 18O water followed by MS analysis. However, expected mass change could not be reliably distinguished from the enzyme dimer + potassium ion mass under our experimental conditions.
During the reaction coordinate of EcMTAN, a fully dissociated ribocation transition state is formed. Sugar cations are highly reactive and susceptible to attack by any nearby nucleophile.23 Thus, enzymes forming sugar cation transition states must stabilize the attacking group within electron-reorganization distance of the reaction center. The related enzyme purine nucleoside phosphorylase immobilizes a phosphate nucleophile for reaction of the phosphate with the ribocation and motion at the reaction coordinate involves altered ribose sugar pucker that occurs on the time scale of 70 fs, the reaction coordinate lifetime, of which 10 fs is the lifetime of the ribocation transition state.15 In order for a transition-state analogue to mimic the transition state, it is required to mimic both the ribocationic feature of the transition state and also to stabilize the attacking nucleophile. Using MS under non-denaturing conditions at neutral pH, we were able to identify the presumptive nucleophilic water molecule in complex with the transition state analogue BuT-DADMe-ImmA and EcMTAN. The results support the proposed chemical mechanism and demonstrate the utility of high-resolution MS to detect interesting non-covalent complexes formed along the reaction coordinate.
Supplementary Material
Acknowledgments
We thank Dr. Richard Furneaux and Dr. Peter Tyler of Industrial Research Laboratory, Inc. for providing BuT-DADMe-ImmA. This work was financially supported by NIH research grant GM41916.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available. Experimental procedures, Figures S1 – S5. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Radzicka A, Wolfenden R. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 2.Pauling L. Am Sci. 1948;36:51–58. [PubMed] [Google Scholar]
- 3.Wolfenden R. Nature. 1969;223:704–705. doi: 10.1038/223704a0. [DOI] [PubMed] [Google Scholar]
- 4.Wolfenden R. Biophys Chem. 2003;105:559–572. doi: 10.1016/s0301-4622(03)00066-8. [DOI] [PubMed] [Google Scholar]
- 5.Wolfenden R, Snider MJ. Acc Chem Res. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 6.Schramm VL. J Biol Chem. 2007;282:28297–28300. doi: 10.1074/jbc.R700018200. [DOI] [PubMed] [Google Scholar]
- 7.Schramm VL, Horenstein BA, Kline PC. J Biol Chem. 1994;269:18259–18262. [PubMed] [Google Scholar]
- 8.Cleland WW. Arch Biochem Biophys. 2005;433:2–12. doi: 10.1016/j.abb.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Schramm VL. Arch Biochem Biophys. 2005;433:13–26. doi: 10.1016/j.abb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 10.Schramm VL. Annu Rev Biochem. 2011;80:703–732. doi: 10.1146/annurev-biochem-061809-100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh V, Evans GB, Lenz DH, Mason JM, Clinch K, Mee S, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 12.Schramm VL. Biochim Biophys Acta. 2002;1587:107–117. doi: 10.1016/s0925-4439(02)00073-x. [DOI] [PubMed] [Google Scholar]
- 13.Singh V, Lee JE, Nunez S, Howell PL, Schramm VL. Biochemistry. 2005;44:11647–11659. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- 14.Albery WJ. Adv Phys Org Chem. 1993;28:139–171. [Google Scholar]
- 15.Saen-Oon S, Quaytman-Machleder S, Schramm VL, Schwartz SD. Proc Natl Acad Sci U S A. 2008;105:16543–16548. doi: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghanem M, Murkin AS, Schramm VL. Chem Biol. 2009;16:971–979. doi: 10.1016/j.chembiol.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh V, Shi W, Evans GB, Tyler PC, Furneaux RH, Almo SC, Schramm VL. Biochemistry. 2004;43:9–18. doi: 10.1021/bi0358420. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho MC, Almo SC, Schramm VL. Nat Chem Biol. 2009;5:251–257. doi: 10.1038/nchembio.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 20.Gabelica V, De Pauw E. Mass spectrometry reviews. 2005;24:566–587. doi: 10.1002/mas.20027. [DOI] [PubMed] [Google Scholar]
- 21.Lee JE, Smith GD, Horvatin C, Huang DJ, Cornell KA, Riscoe MK, Howell PL. J Mol Biol. 2005;352:559–574. doi: 10.1016/j.jmb.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 22.Lee JE, Singh V, Evans GB, Tyler PC, Furneaux RH, Cornell KA, Riscoe MK, Schramm VL, Howell PL. J Biol Chem. 2005;280:18274–18282. doi: 10.1074/jbc.M414471200. [DOI] [PubMed] [Google Scholar]
- 23.Ghanem M, Murkin AS, Schramm VL. Chem Biol. 2009;116:971–979. doi: 10.1016/j.chembiol.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.