Summary
The translation, localization, and degradation of cytoplasmic mRNAs are controlled by the formation and rearrangement of their mRNPs. The conserved Ded1/DDX3 DEAD-box protein functions in an unknown manner to affect both translation initiation and repression. We demonstrate that Ded1 first functions by directly interacting with eIF4G to assemble a Ded1-mRNA-eIF4F complex, which accumulates in stress granules. Following ATP hydrolysis by Ded1, the mRNP exits stress granules and completes translation initiation. Thus, Ded1 functions both as a repressor of translation, by assembling an mRNP stalled in translation initiation, and as an ATP-dependent activator of translation, by resolving the stalled mRNP. These results identify Ded1 as a translation initiation factor that assembles and remodels an intermediate complex in translation initiation.
Introduction
Eukaryotic mRNAs exist in different biochemical mRNP states, which affect the translation, decay, and localization of mRNAs. For example, a translating mRNA associates with translation factors and ribosomes, while translationally repressed mRNPs can accumulate in P-bodies complexed with mRNA decay and translation repression factors (Parker and Sheth, 2007). Non-translating mRNPs can also localize to stress granules (SGs) with a subset of translation initiation factors in the process of either entering or exiting translation (Buchan and Parker, 2009). Determining how mRNPs are assembled and remodeled is critical to understanding the control of translation, mRNA storage, and decay.
The highly conserved DEAD-box protein, Ded1, is a strong candidate for modulating the composition of mRNPs. In vitro, Ded1 acts as a RNA-dependent helicase or RNA chaperone and can remodel mRNP complexes (Bowers et al., 2006; Halls et al., 2007; Iost et al., 1999; Yang and Jankowsky, 2006). In vivo, Ded1 and its orthologs (DDX3, An3, PL10) have been implicated in translation initiation (Beckham et al., 2008; Chuang et al., 1997; de la Cruz et al., 1997; Lee et al., 2008), translation repression (Beckham et al., 2008; Lee et al., 2008; Shih et al., 2008), and RNA interference (Kanai et al., 2004; Raponi and Arndt, 2002; Ulvila et al., 2006). Ded1 orthologs localize to SGs, as well as neuronal and germinal mRNP granules that store repressed mRNAs (see below; Beckham et al., 2008; Goulet et al., 2008; Johnstone et al., 2005; Kanai et al., 2004; Lai et al., 2008). Ded1 also promotes the translation of brome mosaic virus RNA2 (Noueiry et al., 2000). Similarly, the mammalian ortholog, DDX3, promotes HCV replication (Ariumi et al., 2007; Randall et al., 2007) and the nuclear export of genomic HIV mRNAs (Yedavalli et al., 2004). Despite this broad biological importance, how Ded1 functions is unknown.
In this work we demonstrate that Ded1 functions by directly interacting with eIF4G to assemble a Ded1-mRNA-eIF4F complex, which accumulates in SGs. Following ATP hydrolysis by Ded1, the mRNP exits SGs and completes translation initiation. Thus, Ded1 can function both as a repressor of translation, by forming an mRNP stalled in translation initiation, and an activator of translation, via ATP-dependent activity. These results place Ded1 at an important regulatory step in translation following eIF4F assembly and suggest that control of Ded1's activities is critical in the regulation of mRNA storage and translation.
Results
General Strategy
To understand Ded1 function, our approach was to identify specific alleles of Ded1 that affected either its essential role in translation initiation, or its ability to repress translation. Such alleles could then be characterized for their effects on translation and mRNP granule assembly in vivo, translation in vitro, and interactions between Ded1 and other proteins.
Genetic approach to identify separation-of-function alleles of ded1
To identify functional domains of DED1, we tested how various mutations in DED1 (Table S4; Figure S1) affected its essential function in translation initiation (Chuang et al., 1997; de la Cruz et al., 1997) and the growth inhibition caused by DED1 over-expression, which reflects an inhibition of translation (Beckham et al., 2008). We observed two classes of mutants.
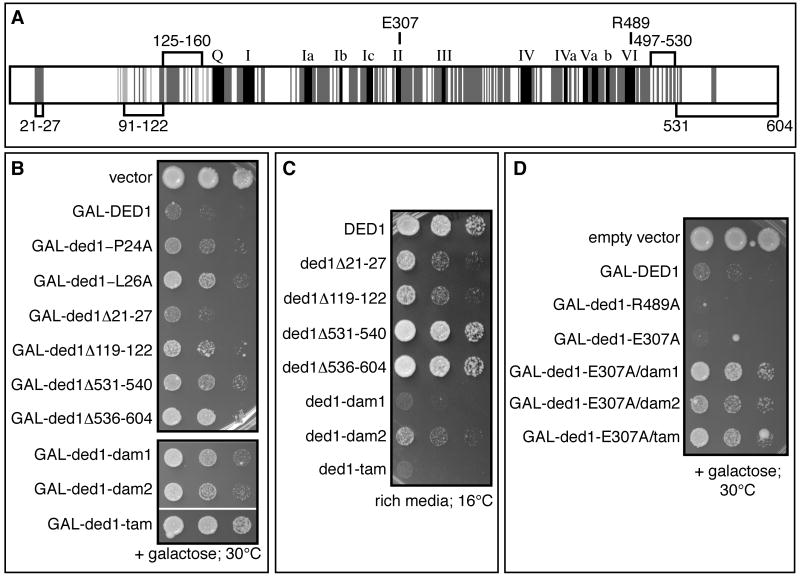
In the first class, we identified three regions of Ded1, referred to as assembly domains (Figure 1A; see below) required for Ded1's function in translation repression as assessed by growth inhibition upon over-expression. Specifically, point mutations in amino acids 21-27, small deletions in amino acids 91-122 or deletion of amino acids 531-540 or 536-604, partially relieve the over-expression lethality (Figure 1B), but still complement ded1Δ for viability (see Table S4 for all mutations and phenotypes). Moreover, some of the alleles are slightly cold-sensitive (Figure 1C). Combining mutations in these three regions gave stronger phenotypes. Specifically, strains with the ded-dam1 (double assembly mutant; Δ21-27+Δ119-122), ded1-dam2 (Δ21-27+Δ531-540), or ded1-tam alleles (triple assembly mutant; Δ21-27+Δ119-122+Δ531-540) showed less inhibition of growth when over-expressed compared to the single mutants (Figure 1B) and were more cold-sensitive for growth than any of the single deletions (Figure 1C & Table S4).
Figure 1. Genetic approach to identify separation-of-function alleles of DED1.
A. The conserved motifs of the DEAD-box protein family are shown in black (Fairman-Williams et al., 2010). Conservation among Ded1 orthologs is shown in dark grey (from yeast to humans) or light grey (among fungi). The numbers mark amino acid domains of Ded1 that are implicated in promoting translation (amino acids labeled above schematic) or in promoting assembly of a Ded1-eIF4F-mRNA complex (amino acids labeled below the schematic). B. Effects of over-expression of ded1 alleles from a galactose inducible promoter in the presence of wild-type, endogenous Ded1. C. Growth of yeast expressing single copies of wild type or mutant ded1 at low temperature. Strains containing a plasmid with wild type or mutant DED1 under the control of its endogenous promoter as the sole copy of DED1 (yRP2799) were spotted at the same concentration on rich media and grown at 16°C for 7 days. D. Cells were treated as in B. See also Table S4 and Figure S2.
A second class of mutants failed to complement a ded1Δ strain (Figure S2A, Table S4) but still inhibited growth when ded1 was over-expressed (Figure 1D), suggesting they are defective in translation initiation. These alleles included point mutations in the ATPase domain, ded1-E307A (helicase motif II) or R489A (helicase motif VI), and small deletions near the ATPase domain of Ded1 (e.g. Δ141-150; Table S4). These observations are consistent with the essential function of Ded1 being provided by its ATPase/RNA helicase activities (Iost et al., 1999) and indicate that the growth defect conferred by DED1 over-expression is independent of the ATPase activity. Compared to wild-type, the ded1-E307A or R489A mutants, which are defective in ATP binding and hydrolysis (Iost et al., 1999; data not shown), showed a stronger growth defect upon over-expression (Figure 1D). This suggests that ATP binding or turnover relieves translation inhibition by Ded1, and functions downstream of translation repression as mediated by the assembly domains defined above. Consistent with that possibility, we observed that the ded1-dam1, dam2, or tam alleles partially suppressed the over-expression growth defect of the ded1-E307A allele (Figure 1D).
Taken together, our genetic analysis has identified three domains that contribute to translation repression by Ded1, and suggested these domains function in steps upstream of the ATP-dependent step, which appears to relieve repression and promote translation.
Ded1 accumulates in and affect stress granules
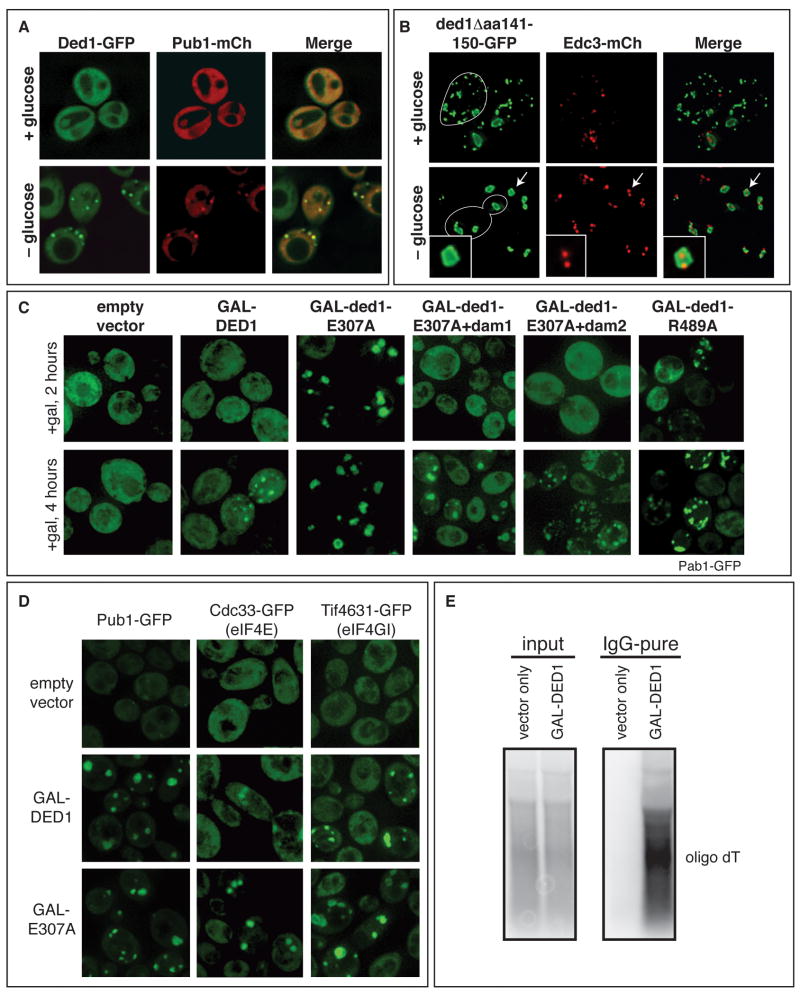
If Ded1's inhibition of translation when over-expressed leads to the accumulation of translationally inhibited mRNPs, these mRNPs may accumulate in P-bodies or stress granules with defined compositions. To understand how mutations in Ded1 affected the formation of different mRNP granules, we examined the location of Ded1 and various components of SGs and P-bodies in wild-type and various Ded1 mutants. Ded1-GFP accumulates in cytoplasmic foci during stress that show greater than 99% co-localization with the SG marker Pub1-mCherry (mCh; Figure 2A), indicating that Ded1 is a component of yeast SGs, like its mammalian ortholog (Goulet et al., 2008; Johnstone et al., 2005; Kanai et al., 2004; Lai et al., 2008). Some Ded1 foci overlap with P-bodies (Beckham et al., 2008), presumably due to the overlap of P-bodies and SGs in yeast (Buchan et al., 2008). Examination of various Ded1 mutants yielded the following observations.
Figure 2. Ded1 affects the accumulation of stress granules that contain a subset of translation initiation factors.
A. Plasmids containing Ded1-GFP (pRP1556) or Pub1-mCh (pRP1661) were transformed into wild type yeast (yRP2065) and tested for co-localization with or without glucose. B. Strain yRP2065 was transformed with plasmids containing ded1Δ141-150-GFP (pRP2071) and Edc3-mCh (pRP1574). Foci formation was analyzed as in A. White traces show the outline of a cell. The arrows point to the foci in the inset. C. Strain yRP2065 was transformed with plasmids containing Pab1-GFP (pRP1657) and either an empty vector (pRP245) or wild type (pRP2086) or mutant DED1 (i.e. pRP2118) under a galactose-inducible promoter. Foci formation was assessed after 2 or 4 hours of induction in galactose. The same strains, when grown in sucrose where DED1 is not over-expressed, all resemble the empty vector control (data not shown). D. Yeast strains with GFP integrations in the chromosome (see Table S1) were transformed with either an empty vector (pRP1827) or wild type (pRP1559) or mutant DED1 (pRP1564) under a galactose-inducible promoter. Foci formation was assessed after 4 hours in galactose. E. Wild type DED1 (pRP1559) was over-expressed in yeast and immunoprecipitated under native conditions. RNA associated with Ded1 was extracted and analyzed by Northern blot with an oligo dT(36) probe. See also Figure S3.
First, we observed that defects in Ded1's ATPase domain or over-expression of Ded1 led to the accumulation of SGs. Specifically, we observed that the lethal deletion mutants (i.e. ded1Δ141-150 and ded1Δ497-504), when conjugated to GFP, dominantly drive the assembly of constitutive SGs (Figure 2B, S3C). These SGs often have P-bodies inside or docked next to them, as shown by the localization of P-body marker Edc3-mCh (Figure 2B). During glucose deprivation, these ded1 foci are enlarged and their co-localization with P-bodies increased (Figure 2B). We interpret these constitutive ded1 foci to be SGs since they co-localize with SG factors (Pub1, Pab1, eIF4E, and eIF4G; Figure S3A,C) and associate with P-bodies (Figure 2B). We also found that, over a time course of induction, both wild type DED1 and ded1-E307A or ded1-R489A led to the accumulation of Pab1p-GFP-containing granules, but granule formation was faster with the ded1-E307A or ded1-R489A alleles (Figure 2C). These results argue that Ded1 can assemble a translationally repressed mRNP that accumulates in SGs independent of its ATPase function, but that release of the mRNP from SGs is ATP-dependent. Consistent with that interpretation, the GFP fusions of the ATPase deficient alleles (E307A and R489A) accumulate in granules upon glucose deprivation (Figure S3D). Alternatively, as ded1-E307A and ded1-R489A have an ATP binding defect, it is possible that ATP binding to Ded1 prevents SG assembly.
To determine the nature of the complex accumulating in SGs upon Ded1 over-expression, we examined the subcellular distribution of other GFP tagged initiation factors. We observed that Pub1 and eIF4A, 4B, 4G, and 4E accumulate in SGs upon DED1 over-expression, although eIF3, eIF2α, eIF1 and eIF5 do not (Figure 2D and Figure S3F). Moreover, Ded1, under the same conditions, co-purifies with MFA2 mRNA and a range of sizes of polyA+ RNAs (Figure 2E and Figure S3E), whereas another DEAD-box protein, Fal1 does not (Figure S3E). These results argue that in the absence of Ded1 ATP binding or ATPase function, or due to Ded1 over-expression, Ded1 accumulates in SGs with mRNAs and a subset of translation initiation factors, all of which function upstream of 43S joining to the mRNA.
If the accumulation of repressed mRNPs in SGs due to Ded1 over-expression contributes to the inhibition of growth, then mutations limiting the Ded1 over-expression growth defect should also reduce the formation of SGs by blocking upstream assembly steps. Indeed, we observed that while SGs accumulate upon over-expression of ded1-E307A, they are reduced upon over expression of ded1-dam1+E307A or ded1-dam2+E307A (Figure 2C), even though these mutant proteins are expressed at similar levels to wild type DED1 (Figure S2C). Moreover, ded1Δ497-504-GFP, which harbors a deletion adjacent to the ATP binding site, forms large, constitutive SGs, which are reduced in ded1Δ497-504+dam1-GFP (Figure S3B). These results suggest that Ded1's assembly domains contribute to the formation of an mRNP complex accumulating in SGs containing eIF4F, Ded1, Pab1, but not the 43S complex.
Ded1 can inhibit translation in vitro
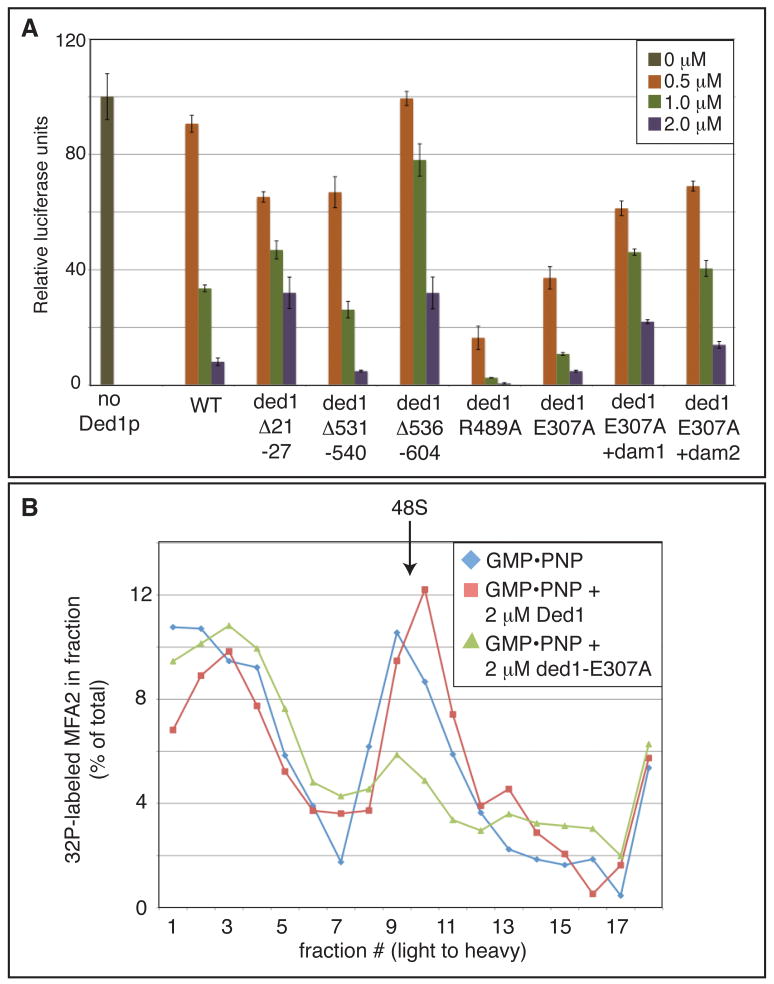
To test directly whether Ded1 could promote formation of a repressed mRNP, we purified recombinant wild type or mutant Ded1 and tested their effect on the translation of a capped, adenylated luciferase reporter in yeast extracts. We found that adding increasing amounts of wild type recombinant Ded1 (rDed1; Figure S5A) led to decreasing amounts of protein synthesis (Figure 3A). In contrast, equal concentrations of eIF4A do not, indicating that this repression is specific to rDed1 and is not a consequence of non-specific RNA helicase activity in the extract (Figure S4A). In addition, we verified that this rDed1-dependent decrease in protein production was not due to destabilization of the reporter mRNA (Figure S4B). These observations indicate rDed1 can repress translation in vitro.
Figure 3. Ded1 represses translation initiation via the assembly domains and prior to 48S accumulation.
A. In vitro translation in yeast extract with capped, polyadenylated luciferase mRNA and increasing amounts of recombinant wild type or mutant Ded1. Translation in the absence of additional rDed1 was normalized to 100. Error bars represent the high and low values of duplicate reactions of a representative experiment. These mutants were tested in parallel at least three times with three independent yeast extract and recombinant protein preparations. B. Yeast extract, capped and polyadenylated radiolabeled MFA2 mRNA, and GMP•PNP were incubated with or without recombinant wild type rDed1 or rded1-E307A before separation by sucrose gradient. Fractions were measured for the presence of radiolabeled mRNA to assess 48S formation. See also Figure S4.
Several observations indicate that the translation repression by rDed1 in vitro is related to Ded1 function in vivo. First, the ded1 assembly mutants rded1Δ21-27 and rded1Δ536-604 reduce translation repression in vitro compared to wild type (Figure 3A); ded1-P24A, -L26A, and Δ536-604 also reduce the growth inhibition due to DED1 over-expression in vivo (Figure 1B). The ded1Δ531-540 protein, which reduces growth inhibition in vivo, does not reduce translation repression in vitro, suggesting there is some difference between the in vivo and in vitro function of ded1Δ531-540. Second, the rded1-E307A and rded1-R489A mutant proteins functioned as hyper-repressors in vitro (Figure 3A), consistent with their stronger inhibition of growth when over-expressed in vivo (Figure 1D). Finally, we observed that the dam1 or dam2 deletions reduced the hyper-repression conferred by the rded1-E307A protein in vitro (Figure 3A), consistent with the assembly mutants suppressing the ded1-E307A hyper-repression phenotypes in vivo (Figures 1D and 2C).
The ability of rDed1 to repress translation in vitro allowed us to use biochemical assays to define the step in translation initiation blocked by excess Ded1 or Ded1 that is unable to utilize ATP. Based on the accumulation of specific factors in SGs in vivo upon over-expression of DED1, rDed1 is anticipated to cause the accumulation of a repressed mRNP that contains eIF4F, Pab1, and Pub1, but has not yet docked with the 43S pre-initiation complex (Figures 2D and S3F), and therefore is blocked upstream of 48S complex formation. To test this prediction, we trapped 48S complexes in the presence of GMP•PNP and determined if rDed1 or rded1-E307A blocked translation upstream of 48S complex formation as assayed by the position of radiolabeled capped, adenylated MFA2 mRNA on sucrose gradients.
We observed that 2 μM of the hyper-repressor rded1-E307A diminished the size of the 48S peak formed in the presence of GMP•PNP (Figure 3B). Higher concentrations of wild type rDed1 (10 μM; Figure S4C) also reduced the accumulation of 48S, suggesting that wild type Ded1 and ded1-E307A repress translation via the same mechanism. Consistent with Ded1 inhibiting an early step in translation initiation, addition of wild type rDed1 or rded1-E307A lowers the levels of 80S complexes that accumulate in the presence of cycloheximide (Figure S4D). These results indicate that excess rDed1, or defects in Ded1 ATP binding or turnover, inhibits translation upstream of 48S complex formation, which is consistent with the composition of SGs we observe in vivo upon over-expression of wild type or ATPase deficient Ded1 (Figures 2C,D and S3F).
Ded1 interacts with eIF4G1
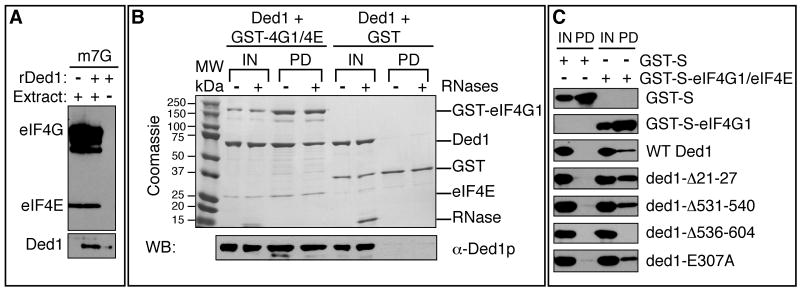
The accumulation of Ded1 with eIF4F in SGs raised the possibility that Ded1 inhibits translation due to interactions with mRNPs that include eIF4F components. Moreover, Ded1 co-immunopurifies with eIF4G1 from yeast (Collins et al., 2007; Krogan et al., 2004; Tarassov et al., 2008), and DDX3, Ded1's mammalian ortholog, interacts with eIF4E (Shih et al., 2008). Thus we hypothesized that Ded1 could interact with eIF4F when it is bound to the cap structure. To test this possibility, we asked if rDed1 was affinity purified by eIF4F bound to a cap column. Under the same conditions where rDed1 represses translation in extracts, we observed that exogenous rDed1 protein was detected on cap column along with eIF4E and eIF4G1 (Figure 4A). In the absence of extracts to provide eIF4F, only a small amount of rDed1 associated with the cap-sepharose (Figure 4A). Thus, rDed1 can associate with eIF4F bound to the cap structure.
Figure 4. The C terminal assembly domain of Ded1 is critical for interaction with eIF4E/eIF4G.
A. Yeast translation extract and rDed1 were incubated with m7G-sepharose and bound proteins identified by immunoblotting with antibodies against Ded1, eIF4E, or eIF4G. B. Interactions between recombinant Ded1 and GST or co-purified GST-eIF4G/eIF4E were tested by immunoprecipitation of eIF4G by glutathione in the presence of RNases. Proteins were detected by coomassie staining (top) and western blot (for Ded1, bottom). The input lanes show 5% of the reaction volume; pull-down lanes show the entire sample. C. As in Figure 4B, except that bound proteins were analyzed by western blot with anti-S antibody (for GST and GST-eIF4G1) or anti-Ded1 antibody. See also Figure S5, S6.
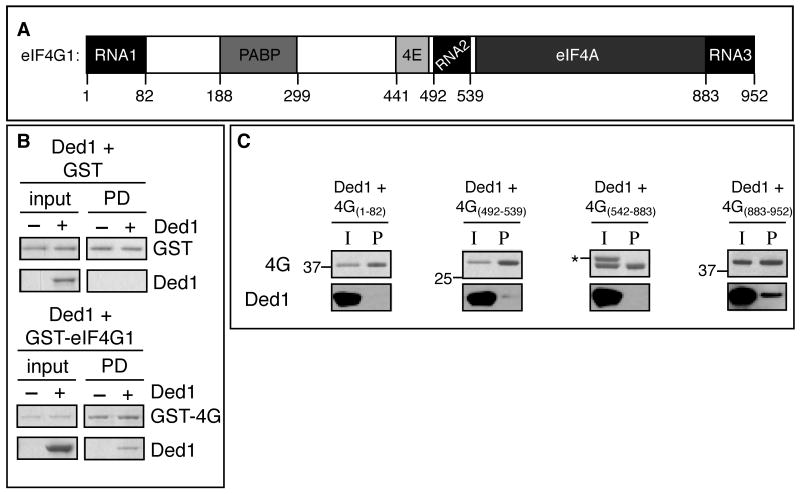
To determine if Ded1 directly bound eIF4F we examined if recombinant Ded1 could interact with a complex of yeast eIF4E and eIF4G1 co-purified from E. coli (Figure S6A). We observed that rDed1 directly binds the purified eIF4E/eIF4G1 complex and this interaction persists after extensive RNase treatment (Figures 4B,S6C). Moreover, the Ded1-eIF4E/eIF4G1 interaction was greatly reduced by the C-terminal truncation rded1Δ536-604 (Figure 4C), which also affects the ability of rDed1 to inhibit translation in vitro (Figure 3A). The Ded1 interaction with eIF4E/eIF4G1 was not disrupted by mutations in the ATP binding site, or by the ded1Δ21-27 (Figure 4C), which is the motif in DDX3 identified as binding eIF4E (Shih et al., 2008).
To determine if Ded1 directly bound to eIF4G1 (Figure 5A), we examined the ability of rDed1 to interact with GST-eIF4G1 in E. coli lysates. E. coli lysate was used for this experiment because of the instability of eIF4G1 in the absence of eIF4E (Berset et al., 2003). Affinity chromatography shows that rDed1p can interact directly with eIF4G1 (Figure 5B). However, we cannot detect a direct interaction between recombinant eIF4E and Ded1 under the same conditions (data not shown), suggesting that the interaction observed between rDed1 and eIF4E/4G (Figure 4B) is mediated largely by eIF4G. As rded1Δ536-604 disrupts the interaction with eIF4E/4G complex (Figure 4C), we conclude that the C-terminus of Ded1 is important for the interaction with eIF4G. This same region of Ded1 is required for translation repression, which argues that the direct interaction of Ded1 with eIF4G1 contributes to the formation of a translationally repressed mRNP accumulating in SGs.
Figure 5. Ded1 interacts directly with eIF4G1 via its third RNA binding motif.
A. Schematic of eIF4G1 and its protein binding motifs. B. Purified GST or full-length GST-eIF4G1 (in absence of eIF4E) in E. coli cell lysate was bound to gutathione resin prior to the addition of control buffer or 1 uM Ded1p. RNases were included in each reaction. Samples were resolved by PAGE and stained with coomassie. The input lanes showed 11.7% of the reactions in pull-down lanes. C. 1 uM of GST-eIF4G1 fragment was incubated overnight with 1 uM Ded1p and RNases before addition of glutathione sepharose. 11.1% of total input was shown for each reaction. Recombinant Ded1p was analyzed by western blot (bottom). eIF4G1 fragments were analyzed by coomassie (top); nearby ladder size in kDa is marked. The asterisk marks recombinant Ded1. See also Figure S6.
To determine what portion of eIF4G1 interacts with Ded1, we looked for an interaction between recombinant Ded1 and various recombinant eIF4G1 fragments (Figure S6B). We tested whether rDed1 could interact with any of the three RNA binding domains, the PABP/4E binding domains, or the eIF4A binding domain of eIF4G1 (Figure 5A). To test whether the eIF4G1 fragments were functional, we confirmed that eIF4G542-883 could interact with recombinant eIF4A by affinity chromatography (Figure S6D). Only fragment eIF4G883-952 was able to interact with recombinant Ded1, suggesting that the third RNA binding domain of eIF4G is sufficient to form a binding site for Ded1 (Figure 5C). We cannot rule out whether there are other Ded1 binding domains within eIF4G that involve more than one of the fragments we tested.
Ded1 stimulates translation in vitro via its assembly and ATPase domains
An unresolved issue is how Ded1 promotes translation and how that would relate to its repression of translation by forming a complex with eIF4F. These could be separate functions of Ded1, which would predict that the mutations in the “assembly” motifs would have no effect on the ability of Ded1 to promote translation. Alternatively, the assembly motifs may promote translation by affecting the formation of an eIF4F-Ded1-mRNA complex, which is primed for subsequent entry into translation. This latter model predicts that mutations in the assembly motifs should also block the ability of Ded1 to promote translation. To distinguish between these models, we examined the role of Ded1 in promoting translation in vitro, using a system adapted from previous work (Chuang et al., 1997).
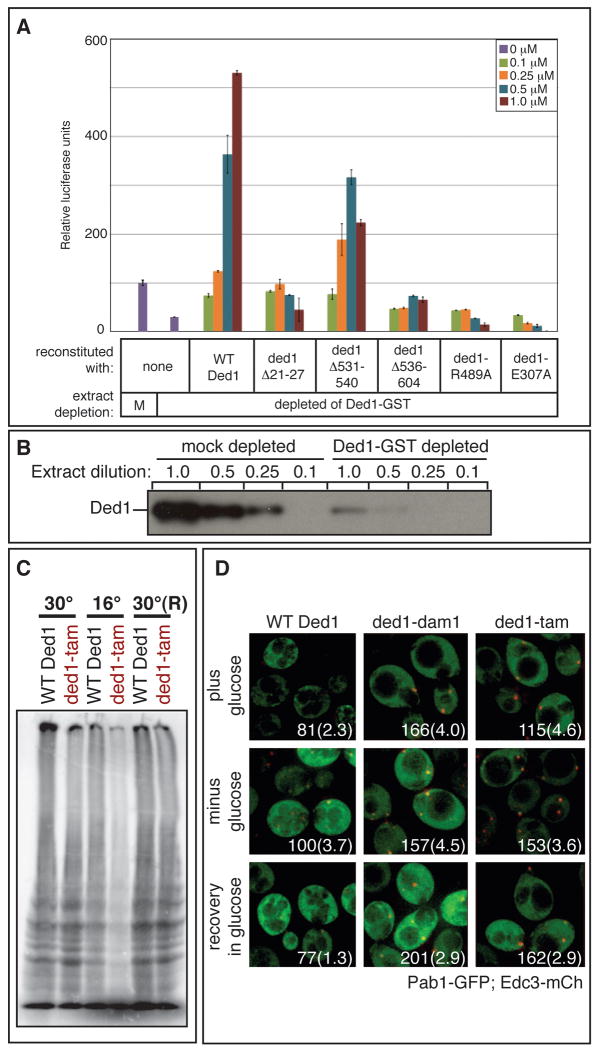
We prepared extracts from a yeast strain with a Ded1-GST fusion protein where we could deplete Ded1-GST by ∼80%, yielding a corresponding ∼80% drop in translation (Figure 6A&B). This loss of translation could be rescued by the addition of recombinant HIS-tagged Ded1, indicating the depletion does not remove critical amounts of other important factors (Figure 6A; Chuang et al., 1997). In fact, depletion of Ded1-GST did not dramatically alter the levels of eFI4E, eIF4G, or Pab1 in the extracts (Figure S4F). We observed that reconstitution with His-tagged rDed1 elevated translation above the starting levels, probably due to the Ded1-GST fusion protein being only partially functional, as extracts from a Ded1-GST strain are consistently less active than extracts from wild-type cells (data not shown). Consistent with our results with non-depleted extracts (Figure 3A), higher levels of rDed1 reconstitution in depleted extract eventually led to repression (Figure S4E). Moreover, we observed that the ATPase deficient alleles, rded1-E307A and rded1-R489A, failed to restore translation activity (Figure 6A), and functioned as hyper-repressors. Thus, Ded1 is required for efficient translation in vitro, and this stimulatory role is ATP-dependent.
Figure 6. Ded1 ATPase and assembly domains promote translation.
A. Extracts that were mock or Ded1 depleted were used for in vitro translation reactions as in Figure 3A. B. Sequential dilution of mock and Ded1 depleted extracts analyzed by western blot with α-Ded1 antibody. C. yRP2799 containing either wild type DED1 or ded1-tam as the sole copy of DED1 was assayed for translation by 35S incorporation during growth at 30°C, after a 2 hour shift to 16°C, and after recovery for 1 hour at 30°C(R). D. yRP2799 containing either wild type DED1, ded1-dam1, or ded1-tam as the sole copy of DED1 were transformed with plasmid pRP1657, which encodes Edc3-mCh (a P-body marker) and Pab1-GFP (a SG marker). Strains were grown to mid-log phase and then shifted to 16°C for 2 hours. Localization was assessed in the presence of 2% glucose, after 15 minutes of glucose deprivation, and after a subsequent 15-minute recovery in 2% glucose. Each image includes quantitation of P-body intensity, normalized to wild type Ded1 after glucose deprivation, and the average number of P-bodies per cell, shown in parentheses. See also Figure S4.
We then examined how rded1Δ21-27, rded1Δ531-540, and rded1Δ536-604 affected Ded1-dependent stimulation of translation. A striking result was that all of these mutants were reduced in their ability to restore translation to Ded1-depleted extracts (Figure 6A). These purified mutant proteins are functional in vitro; rded1Δ21-27 and rded1Δ531-540 have RNA helicase activity comparable to wild type levels, although rded1Δ536-604 has reduced helicase activity (Figure S5). The inability of the mutant proteins to restore translation suggests that the same regions that allow Ded1 to inhibit translation are also required for Ded1's role in translation activation. Thus, formation of the eIF4F-Ded1-mRNA complex by Ded1 appears to be a prerequisite for activation of translation by Ded1.
Ded1's assembly motifs promote translation in vivo
The above results suggested that the ability of Ded1 to promote translation in vitro requires the “assembly domains.” Since mutations in these regions are not essential during growth at 30°C, it suggests that under optimal conditions Ded1 can perform its function in promoting translation independent of these domains. However, mutations in the assembly domains are cold sensitive, particularly when combined in double or triple mutants (Figure 1C; Table S4), suggesting that Ded1 may be required for optimal translation at low temperatures in a manner dependent on these assembly domains. To test this possibility, we assayed translation in strains containing either wild type or ded1-tam as the sole copy of ded1, monitoring incorporation of 35S-labeled methionine and cysteine after a three hour shift to 16°C.
An important result was that cells containing ded1-tam showed reduced translation at 16°C as compared to wild type (Figure 6C). After a 1 hour recovery at 30°C, both wild type and ded1-tam strains were able to translate with equal efficiency. These results demonstrate that the assembly domains are required for efficient translation in vivo at low temperatures, thus providing additional evidence that the ability of Ded1 to promote translation is enhanced by its assembly motifs.
These cold-sensitive mutations also cause the accumulation of P-bodies, where translationally repressed mRNAs accumulate. In strains containing either wild type DED1, ded1-dam1, ded1-dam2, or ded1-tam as the sole copy of ded1, we shifted the cells to 16°C for two hours and then assessed P-body and SG formation before and after a 15 minute glucose deprivation stress, as well as recovery from that stress. We observed that the mutant strains, compared to wild type yeast, accumulated brighter P-bodies and often more numerous P-bodies in non-stress conditions, after stress, and after recovery from stress (Figure 6D and data not shown). These trends were observed at the permissive temperature (30°C; data not shown), but were more apparent at the restrictive temperature, where translation is inhibited and likely contributes to the increase in P-bodies (Teixeira et al., 2005). These results suggest that Ded1, via its assembly domains, either antagonizes P-body formation or promotes P-body disassembly. As the assembly domains are important for inhibiting translation in vitro (Figure 3A), for promoting translation in vivo and in vitro, and for SG induction (Figure 2C), we hypothesize that Ded1 remodels mRNPs accumulating in P-bodies into a complex minimally containing Ded1, eIF4F, and mRNA.
Discussion
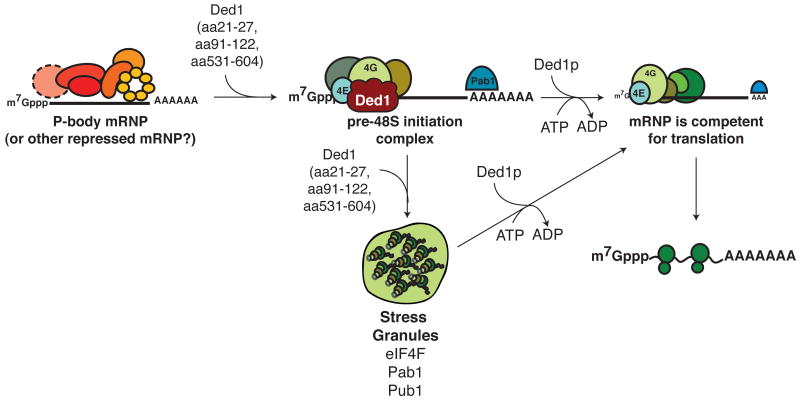
Our data (summarized in Table S5) suggest that Ded1 functions to modulate translation by first forming a complex minimally containing Ded1, eIF4F, and mRNA that is stalled in translation initiation upstream of 43S joining, and in a subsequent step, by utilizing ATP hydrolysis and turnover to allow that complex to progress into translation (Figure 7).
Figure 7. Working model of Ded1 function in translation.
We propose that Ded1 first assembles a complex, minimally containing Ded1, mRNA, and eIF4F, via its assembly domains and through direct interactions with eIF4G. This complex is translationally inactive and can accumulate in SGs. Subsequently, Ded1 functions in a ATP-dependent step to remodel the Ded1-mRNA- eIF4F complex and return the mRNA to translation; this second step correlates with exit from SGs.
The conclusion that a Ded1-eIF4F-mRNA complex first forms and is stalled in translation is supported by multiple lines of evidence. First, lethal deletions near the ATPase domain of Ded1 act dominantly to drive constitutive SG accumulation (Figure 2B). Second, over-expression of Ded1 inhibits translation in vivo (Beckham et al., 2008) and leads to the accumulation of Ded1 in SGs along with eIF4F and Pab1 (Figures 2C, 2D, S3F), and under these same conditions, Ded1 co-purifies with MFA2 mRNA and a diversity of poly(A)+ RNAs (Figures S3E, 2E). Third, the accumulation of such granules is enhanced by the over-expression of Ded1 variants that are unable to utilize ATP (Figure 2C), indicating the formation of these complexes does not depend on ATP hydrolysis and turnover. Fourth, exogenous rDed1 represses translation in vitro upstream of 48S joining to the mRNA (Figure 3B). Under these conditions, rDed1 is recruited to the m7G cap. Fifth, rded1 mutants defective in ATPase activity function as hyper-repressors in vitro (Figure 3A), providing biochemical evidence that formation of a translationally repressed complex is independent of Ded1's ATPase activity.
An unresolved issue is whether Ded1 directly acts as a translational repressor in vivo. Our data show that over-expression of Ded1 causes translation repression and that ATPase activity antagonizes this repression. While we have not yet shown that endogenous levels of Ded1 repress translation of mRNAs, our results suggest that inhibition of Ded1 ATPase activity could maintain mRNA in a translationally inactive state. Consistent with this, Ded1 and its orthologs are found in mRNA storage granules.
Our data suggest that a direct interaction of Ded1 with eIF4G contributes to the formation of a translationally repressed Ded1-eIF4F-mRNA intermediate. rDed1 binds directly to the RNA3 domain of eIF4G1 (Figure 5B,C) and this interaction is dependent on the Ded1 C terminal region (Figure 4C), which is also required for translation repression in vitro (Figure 3A) and for over-expressed based growth inhibition in vivo (Figure 1B). This deletion of the C terminal region of Ded1 also causes a decrease in helicase activity (Figure S5), but this decreased helicase activity is unlikely to influence eIF4G binding, since Ded1-eIF4E/4G interactions does not depend on RNA (Figure 4B). Moreover, rded1-E307A, which has no detectable helicase activity (Iost et al., 1999), shows normal association with eIF4G and increased translational repression in vitro (Figures 4C, 3A). Consistent with these direct interactions in vitro, Ded1 co-immunopurifies with eIF4G from yeast in numerous experiments (Collins et al., 2007; Krogan et al., 2004; Tarassov et al., 2008).
We mapped three regions of Ded1 that contribute to translation repression (Figure 1B, 3A). The 21-27 region does not affect Ded1 binding to eIF4G (Figure 4C), suggesting that interactions with other factors may contribute to the formation of the Ded1-eIF4F-mRNA complex. Notably, Ded1 residues 21-27 correspond to the region in the mammalian ortholog of Ded1, DDX3, that have been suggested to bind directly to eIF4E (Shih et al., 2008). Although we have not detected a direct Ded1-eIF4E interaction (data not shown), it is possible that Ded1-eIF4E interact weakly and contribute to the formation of the Ded1-eIF4F-mRNA complex.
Evidence from the literature and our experiments (Figure 5) indicate that Ded1 also promotes translation initiation (Chuang et al., 1997) in an ATP-dependent manner (Figure 6). Several of our observations now indicate that Ded1's ability to form the complex minimally containing Ded1, eIF4F, and mRNA is critical to its stimulation of translation. First, mutations in Ded1's “assembly domains” are defective in both translation repression and translation initiation in vitro (Figures 3A and 6A). Second, combining assembly mutations with an ATPase-deficient mutation reverses several phenotypes of the ATPase-deficient mutation alone, including hyper-repression in vivo and in vitro and the formation of large SGs (Figures 1D, 2C, and 3A) indicating that the assembly function works upstream of the ATP-dependent function. Finally, ded1-tam, which contains deletions in the three assembly domains, shows a defect in global translation in vivo at low temperatures (Figure 6C). We suggest that this phenotype is only observed at low temperature either because ded1-tam is a partial loss-of-function mutation, as ded1-tam still inhibits growth to some extent when over-expressed (Figure 1B), and/or because the role of Ded1 in translation initiation becomes more critical at low temperatures. Thus, we suggest Ded1 promotes translation initiation in a two-step process by first forming a complex minimally containing eIF4F-Ded1 and mRNA. Subsequently, Ded1, in an ATP-dependent fashion, allows entry of the mRNA into active translation. Since ATP turnover by Ded1 weakens its interaction with RNA (Iost et al., 1999; Liu et al., 2008) and Ded1 is not seen in polysomes (Beckham et al., 2008), we hypothesize that ATP turnover by Ded1 promotes its release from the pre-initiation mRNP, allowing the mRNP to proceed into translation initiation.
Ded1 likely interacts on a range of mRNAs, as Ded1 co-purifies with a range of polyA+ RNA in vivo (Figure 2E). Additionally, the cold-sensitive ded1 assembly mutants cause accumulation of P-bodies (Figure 6D), suggesting Ded1 can recruit mRNAs from P-bodies into translation. Since Ded1 co-purifies with Xrn1 (data not shown; Krogan et al., 2004), one possibility is that Ded1 is recruited to P-body mRNPs via Xrn1, and Ded1 then recruits eIF4F to that complex to promote a transition from a P-body mRNP into a Ded1-eIF4F-mRNA complex. In addition, since Ded1 and its orthologs shuttle between the nucleus and cytoplasm and interact with mRNA export factors (Lai et al., 2008; Yedavalli et al., 2004), it is possible that Ded1 might also assemble eIF4F-Ded1 complexes on newly exported mRNPs.
Given the role of Ded1 in promoting P-body disassembly and in forming a eIF4F-Ded1-mRNA complex, we suggest that Ded1 is a prototype of a unique class of translation factors that remodels repressed or nascent mRNPs to promote entry into translation. Ded1 function could affect this transition by one of two mechanisms. First, Ded1 could promote the recruitment of eIF4E/eIF4G to the mRNA. This would be analogous to the function of other factors that load critical machines onto nucleic acids, such as Prp28, another DEAD-box protein, which promotes spliceosome assembly in an ATP-dependent matter by facilitating the switch of U1 snRNA for U6 snRNA on pre-mRNA (Staley and Guthrie, 1999). Another example is provided by the clamp loader, RCF, which loads the clamp PCNA onto DNA, hydrolyzes ATP and is then ejected from the complex (reviewed in O'Donnell and Kuriyan, 2006). Analogous to RCF, Ded1 may hydrolyze ATP to release the Ded1-eIF4F-mRNA pre-initiation complex, allowing it to proceed into translation initiation. Such an assembly role for Ded1 in translation would explain why it affects translation initiation, yet is not a component of polysomes (Beckham et al., 2008), unlike eIF4E and eIF4G (Hinton et al., 2007). Over-expression of DED1, which we show promotes formation of the Ded1-eIF4F-mRNA complex, also suppresses defects in eIF4E (de la Cruz et al., 1997), consistent with Ded1 promoting eIF4F interaction with the cap structure. Note that Ded1 could affect the specificity of translation in this manner, by preferentially interacting with some mRNAs and thereby increasing their ability to recruit eIF4F when this complex is limiting.
A second, but not mutually exclusive, hypothesis is that Ded1 could stall translation initiation after eIF4F recruitment and prevent assembly of the multifactor complex (MFC) and 43S ribosomal subunit, perhaps by interfering with interactions between eIF4G and the MFC. Thus, analogous to eIF4AIII (Ballut et al., 2005), Ded1 may maintain the Ded1-eIF4F-mRNA complex while ATP hydrolysis is repressed. Upon ATP hydrolysis and dissociation of the hydrolysis products, we propose that Ded1 would release or remodel this complex, acting as an ATP-dependent switch to allow mRNAs to proceed through translation initiation. This scenario implies that Ded1 orthologs involved in the storage of mRNAs in neuronal or germinal granules may form analogous complexes to control the release of mRNAs in an ATP-dependent manner. By acting as a switch, Ded1 may be able to function to both promote translation and repress translation depending on the regulation of its ATP-dependent functions. Ded1 may be controlled at the level of ATP binding or turnover, as the ATPase mutants used in this study show defects both in ATP binding (data not shown) and ATP hydrolysis (data not shown; Iost et al., 1999). Thus, an important goal of future work will be to determine protein-protein interactions or modifications that modulate the ability of Ded1 to bind or turnover ATP.
Experimental Procedures
Yeast strains, plasmids, and growth assays
Yeast strains and plasmids are listed in Tables S1-3. Details of their construction and of yeast growth assays can be found in Supplemental Information.
Microscopy
To analyze P-body and/or SG formation, the cells were analyzed as described (Buchan et al., 2008), except for a few modifications (See Supplemental Information). All images are collapsed Z-series and are representative of at least three independent experiments that include images of at least 100 cells. Whenever the levels of the red and green channels were adjusted for print quality all figure panels were changed equivalently.
Immunopurification of Ded1 in vivo
Yeast containing either an empty vector, DED1, or FAL1 under the control of a galactose-inducible promoter and tagged with protein A were lysed by bead beating at 4°C. Protein A-tagged complexes were immunopurified by IgG Sepharose 6 fast flow resin. RNA from complexes was isolated by phenol-chloroform extraction and analyzed by Northern blot. See Supplemental Information for details.
Protein purification
Recombinant Ded1 proteins were purified as described (Iost et al., 1999) with the following modifications. For Figures 3, 4A, 6A and S4, BL21(DE3) E. coli cells were induced at 28°C over-night. Cells were resuspended in 50 mM NaH2PO4, 300 mM NaCl, 10% glycerol, 0.5% NP-40, 2 mM imidazole and protease inhibitors. The cellular lysate was bound to TALON metal affinity resin (Clontech). The eluted protein was dialyzed into 10 mM Hepes, pH 8.2, 150 mM potassium acetate, 50% glycerol, and 2 mM DTT to increase protein stability in a buffer compatible with translation assays. For Figures 4B,C, 5, and S5B,C, wild type and mutant Ded1p were purified as described (Fairman et al., 2004; Yang and Jankowsky, 2005), which includes an additional purification through a P11 phosphocellulose column to remove any contaminating RNAs. Note that both purification methods yield comparable translation inhibition in vitro (Figure S4A).
See Supplemental Information for details on purification of eIF4E/eIF4G1 complex and eIF4G1 fragments.
In vitro translation assays
Yeast extracts and translation reactions (Figure 3A,S4A) were prepared as described (Iizuka and Sarnow, 1997; Wu et al., 2007), with some modifications (Supplemental Information). To assess 40S and 80S complex formation (Figure 3B and S4C,D), translation reactions were assembled as described in Supplemental Information, except that capped, adenylated radiolabeled MFA2p(G) mRNA was used. MFA2 mRNA was transcribed from XbaI-linearized pRP803 with MAXIScript (Ambion) and adenylated with Poly(A) Tailing kit (Ambion). The reactions were supplemented with 5 mM GMP•PNP and 7.5 mM magnesium acetate to analyze 48S formation or 0.5 g/L cycloheximide to analyze 80S formation. The extracts were sedimented in 15-50% sucrose gradients for 15 hours at 27000 rpm at 4°C in a SW40-Ti rotor. Fractions were hand collected and amount of radioactive MFA2 mRNA was assayed by Cerenkov counting.
To assess Ded1's role in translation (Figures 6A and S4E,F), translation extracts were made from strain yRP2805, where chromosomal DED1 has a C-terminal GST tag. Ded1-GST was depleted (or mock depleted) as detailed in Supplemental Information. Ded1 depletion was assayed by western blot with an anti-Ded1 antibody (Figure 6B). Translation reactions were set up as described above.
35S Incorporation
Strains expressing wild type DED1 (pRP1555) or ded1-tam (pRP2048) were grown in liquid rich media to mid-log phase at 30°C. Part of the culture was washed and resuspended in –met synthetic complete (SC) media with 2% glucose and labeled with 35S methionine/cysteine for 10 minutes. Cells were pelleted and frozen on dry ice. The rest of the culture was shifted to 16°C and an equal O.D. of cells was 35S labeled after 2 hours. After a 1 hour recovery at 30°C the same O.D. of cells was labeled with 35S. Proteins were extracted by boiling the pellets in 50 μl lysis buffer (2% SDS, 90 mM Hepes pH 7.5, 30 mM DTT) for 10 minutes.
Cap binding experiment
In Figure 4A, yeast translation extract was incubated with or without 2.5 μg of recombinant Ded1 in binding buffer (50 mM Hepes pH 7, 100 mM NaCl, 1 mM DTT, 2 mM MnCl2, 2 mM MgCl2, 0.1% triton-X, 10% glycerol) for 10 minutes on ice. The reactions were then incubated with 5% v/v of either m7G-sepharose or protein G-sepharose, washed three times and associated proteins eluted by boiling and identified by immunoblotting.
Pull-down reactions
See full details in Supplemental Information. Briefly, either co-purified eIF4E/eIF4G1 or purified eIF4G1 fragments were incubated with purified Ded1p (720nM), 5 mg/ml BSA (Roche), and RNases (unless indicated otherwise) overnight at 4°C before affinity purification on glutathione-sepharose resin. For experiments with full length eIF4G only (Figure 5B), eIF4G in E. coli lysate was incubated with glutathione-sepharose resin; the resin was washed, and incubated as described above. Bound proteins were eluted, resolved on 6-10% gradient SDS-PAGE, and subjected to western blot analysis with either primary antibodies against Ded1p or against the S-tag antibody.
Supplementary Material
Highlights.
Ded1 interacts directly with eIF4G to promote formation of a pre-translation complex
Ded1 utilizes ATP to allow the mRNA-eIF4F-Ded1 complex to enter translation
Ded1 can both repress and promote translation, depending on ATPase activity
Acknowledgments
T.H. Chang (Ded1 antibody); Elizabeth Grayhack (plasmid pRP1827); Micheal Altmann (eIF4G1 fragment and eIF4A expression constructs); John McCarthy and Allan Jacobson (antibodies); Drs. Carolyn Decker, Tracy Nissan, and Carla Beckham and members of the Parker lab for discussion; Heath Bowers (Case Western) for initial experiments on the Ded1p-eIF4G1 interaction, Andrea Putnam (Case Western) for the unwinding assays; funding from NIH (GM45443) and HHMI (RP), Leukemia and Lymphoma Society (AH), NIH (GM067700 to EJ) for EJ and ZG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10(6):430–6. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Beckham C, Hilliker A, Cziko AM, Noueiry A, Ramaswami M, Parker R. The DEAD-Box RNA Helicase Ded1p Affects and Accumulates in Saccharomyces cerevisiae P-Bodies. Mol Biol Cell. 2008;19:984–993. doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset C, Zurbriggen A, Djafarzadeh S, Altmann M, Trachsel H. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA. 2003;9:871–880. doi: 10.1261/rna.5380903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers HA, Maroney PA, Fairman ME, Kastner B, Luhrmann R, Nilsen TW, Jankowsky E. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Boisvenue S, Mokas S, Mazroui R, Cote J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and - independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton TM, Coldwell MJ, Carpenter GA, Morley SJ, Pain VM. Functional analysis of individual binding activities of the scaffold protein eIF4G. J Biol Chem. 2007;282:1695–1708. doi: 10.1074/jbc.M602780200. [DOI] [PubMed] [Google Scholar]
- Iizuka N, Sarnow P. Translation-competent extracts from Saccharomyces cerevisiae: effects of L-A RNA, 5′ cap, and 3′ poly(A) tail on translational efficiency of mRNAs. Methods. 1997;11:353–360. doi: 10.1006/meth.1996.0433. [DOI] [PubMed] [Google Scholar]
- Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry AO, Chen J, Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc Natl Acad Sci U S A. 2000;97:12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M, Arndt GM. Dominant genetic screen for cofactors that enhance antisense RNA-mediated gene silencing in fission yeast. Nucleic Acids Res. 2002;30:2546–2554. doi: 10.1093/nar/30.11.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, Wu Lee YH. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Ramet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- Wu C, Amrani N, Jacobson A, Sachs MS. The use of fungal in vitro systems for studying translational regulation. Methods Enzymol. 2007;429:203–225. doi: 10.1016/S0076-6879(07)29010-X. [DOI] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.