Abstract
A three-generation resource population was constructed by crossing pigs from the Duroc and Pietrain breeds. In this study, 954 F2 animals were used to identify quantitative trait loci (QTL) affecting carcass and meat quality traits. Based on results of the first scan analyzed with a line-cross (LC) model using 124 microsatellite markers and 510 F2 animals, 9 chromosomes were selected for genotyping of additional markers. Twenty additional markers were genotyped for 954 F2 animals and 20 markers used in the first scan were genotyped for 444 additional F2 animals. Three different Mendelian models using least-squares for QTL analysis were applied for the second scan: a LC model, a half-sib (HS) model, and a combined LC and HS model. Significance thresholds were determined by false discovery rate (FDR). In total, 50 QTL using the LC model, 38 QTL using the HS model, and 3 additional QTL using the combined LC and HS model were identified (q < 0.05). The LC and HS models revealed strong evidence for QTL regions on SSC6 for carcass traits (e.g., 10th-rib backfat; q < 0.0001) and on SSC15 for meat quality traits (e.g., tenderness, color, pH; q < 0.01), respectively. QTL for pH (SSC3), dressing percent (SSC7), marbling score and moisture percent (SSC12), CIE a* (SSC16), and carcass length and spareribs weight (SSC18) were also significant (q < 0.01). Additional marker and animal genotypes increased the statistical power for QTL detection, and applying different analysis models allowed confirmation of QTL and detection of new QTL.
Keywords: pig, QTL, carcass merit, meat quality
Introduction
Quantitative trait loci (QTL) mapping has been conducted using numerous pig populations to identify genomic regions controlling phenotypic variation for hundreds of traits (http://www.animalgenome.org/cgi-bin/QTLdb/SS/index). Nevertheless, the implementation of QTL into breeding programs which is a major goal of QTL mapping has been limited not only due to insufficient numbers of identified causative mutations, but because of unknown linkage disequilibrium (LD) phase between markers and QTL resulting from cross breeding systems (Spelman and Bovenhuis, 1998; Hayes et al., 2009). We have developed a F2 Duroc × Pietrain resource population at Michigan State University (Edwards et al., 2008b) and reported QTL for carcass merit and meat quality traits (Edwards et al., 2008a). The Duroc and Pietrain breeds are used in breeding programs as sire breeds worldwide, and these breeds exhibit variation in carcass merit and meat quality phenotypes. Pietrain pigs have been shown to have less backfat (Affentranger et al., 1996; Edwards et al., 2003) and larger longissimus muscle area (LMA; Edwards et al., 2003). Duroc and Duroc-sired pigs generally have more favorable meat quality (Langlois and Minvielle, 1989; Affentranger et al., 1996; Jeremiah et al., 1999; Edwards et al., 2003), whereas Pietrain and Pietrain-sired pigs are leaner with average meat quality (Edwards et al., 2003).
A line-cross (LC) model, which assumes the founder lines to be fixed for alternative QTL alleles, has been most commonly used to identify QTL for F2 population designs (Haley et al., 1994). The first genome scan for our Duroc × Pietrain population was performed using a LC analysis (Edwards et al., 2008a, b). However, for crosses between outbred lines such as domestic animals, not all QTL alleles are completely fixed so effects under the LC model can be biased downwards (Pérez-Enciso and Varona, 2000). To identify QTL segregating within parental breeds, a half-sib (HS) model that does not assume fixation of QTL alleles in the founder lines was introduced by Knott et al. (1996), and Kim et al. (2005) developed a combined line-cross and half-sib (CB) model that accounts for both line and HS effects. We have recently utilized LC, HS, and CB models to identify QTL for growth traits in our Duroc × Pietrain population (Choi et al., 2010). The objective of this study was to confirm previously identified carcass merit and meat quality QTL regions with addition of new marker genotypes and additional F2 animals, and to detect new QTL for carcass merit and meat quality traits using three different least-squares models under different assumptions; (1) founders fixed for alternative QTL alleles (LC model), (2) segregation of QTL alleles at similar frequencies in founders (HS model), and (3) segregation of QTL alleles at different frequencies in founders (CB model).
Materials and Methods
Animals and phenotypic data
A three-generation resource population developed at Michigan State University was used for this study. A detailed description of the animals and phenotypic data was previously reported (Edwards et al., 2008a, b). All grandparents were confirmed to be homozygous normal for the polymorphism at position 1843 in the RYR1 gene (Edwards et al., 2008b). Animal protocols were approved by the Michigan State University All University Committee on Animal Use and Care (AUF# 09/03-114-00). A total of 954 F2 pigs were used which included the 510 animals evaluated in the first genome scan. These pigs were produced from 6 F1 boars and 50 F1 sows which were retained from 4 F0 Duroc sires and 15 F0 Pietrain dams. The F2 pigs were analyzed for 38 carcass and meat quality traits.
Details of carcass and meat quality phenotype collection were reported in Edwards et al., 2008a. Briefly, animals were slaughtered at the Michigan State University Meat Laboratory (East Lansing, MI, USA) or a federally inspected commercial plant (DeVries Meats, Coopersville, MI, USA). Slaughter age was 165.8 ± 9.2 days and the minimum off-farm body weight (BW) for slaughter was 82.54 kg. Hot carcass weight (HCW), and pH and temperature of the longissimus muscle (LM) at 45-min and 24-h postmortem were obtained. After overnight chilling, backfat thickness, number of ribs and carcass length were measured, and the weights of primal cuts were recorded. A single trained evaluator scored color, marbling, and firmness using two 2.54-cm thick chops cut from the LM, and objective color scores of CIE L*, a*, and b* were obtained using a Minolta colorimeter. The remaining section of the LM was used to determine drip loss, cook yield, Warner-Bratzler shear force, proximate analysis measures, and sensory attributes. A trained sensory panel evaluated juiciness, tenderness, overall tenderness, connective tissue, and off-flavor using an 8-point hedonic scale. Descriptive statistics for phenotypes used in this study are presented in Table 1.
Table 1.
Number of records, means, and SD for carcass and meat quality traits.
| Traits | N | Mean | SD |
|---|---|---|---|
| CARCASS MEASURE | |||
| Off-farm BW, kg | 948 | 112.08 | 8.56 |
| Hot carcass weight, kg | 948 | 81.84 | 6.81 |
| Dressing percent, % | 948 | 73.01 | 2.11 |
| 45 min carcass temperature, °C | 947 | 39.47 | 2.23 |
| 24 h carcass temperature, °C | 945 | 2.91 | 1.19 |
| 45 min pH | 934 | 6.37 | 0.22 |
| 24 h pH | 927 | 5.51 | 0.14 |
| 45 min–24 h pH decline | 914 | 0.86 | 0.22 |
| Carcass length, cm | 947 | 78.72 | 2.51 |
| Number of ribs | 669 | 14.83 | 0.85 |
| First-rib backfat, mm | 859 | 40.67 | 7.09 |
| Last-rib backfat, mm | 947 | 28.69 | 6.38 |
| Last lumbar vertebra backfat, mm | 946 | 22.25 | 6.23 |
| 10th-rib backfat, mm | 941 | 24.16 | 7.35 |
| Longissimus muscle area, cm2 | 942 | 40.61 | 4.74 |
| PRIMAL CUT WEIGHT | |||
| Ham weight, kg | 947 | 9.63 | 0.77 |
| Loin weight, kg | 947 | 8.28 | 0.83 |
| Boston shoulder weight, kg | 947 | 3.90 | 0.56 |
| Picnic shoulder weight, kg | 947 | 3.72 | 0.57 |
| Belly weight, kg | 947 | 5.03 | 0.67 |
| Spareribs weight, kg | 943 | 1.53 | 0.20 |
| MEAT QUALITY EVALUATION | |||
| Color, 1–6 | 945 | 3.16 | 0.82 |
| Marbling, 1–10 | 946 | 2.82 | 0.84 |
| Firmness, 1–5 | 932 | 2.86 | 0.79 |
| L* | 900 | 53.77 | 2.24 |
| a* | 900 | 17.25 | 1.83 |
| b* | 900 | 9.13 | 1.61 |
| PROXIMATE ANALYSIS | |||
| Moisture, % | 936 | 73.94 | 1.53 |
| Fat, % | 936 | 3.18 | 1.40 |
| Protein, % | 935 | 23.44 | 1.13 |
| LABORATORY ANALYSES | |||
| Drip loss, % | 946 | 1.85 | 1.18 |
| Cook yield, % | 936 | 77.26 | 2.83 |
| Warner-Bratzler shear force, kg | 935 | 3.21 | 0.69 |
| SENSORY PANEL ANALYSES | |||
| Juiciness, 1–8 | 942 | 5.23 | 0.59 |
| Tenderness, 1–8 | 942 | 5.55 | 0.62 |
| Overall tenderness, 1–8 | 942 | 5.63 | 0.55 |
| Connective tissue, 1–8 | 942 | 6.39 | 0.39 |
| Off-flavor, 1–8 | 942 | 1.14 | 0.21 |
Genotypic data
Nine chromosomes (SSC3–7, 12, 15, 16, and 18) were selected based on results of the first genome scan (Edwards et al., 2008a, b) which had been completed using 510 F2 animals and 124 microsatellite markers. For the second scan 20 additional microsatellite markers were selected on these chromosomes (1–4 markers per chromosome; Choi et al., 2010) in order to increase the power of QTL detection and to narrow the QTL locations. All F0, F1, and the 954 F2 pigs were genotyped for the 20 new markers, and the 444 additional F2 pigs were also genotyped for 20 markers flanking the QTL regions on the 9 selected chromosomes. Sex-averaged genetic linkage maps were estimated for all autosomes using CRI-MAP version 2.4 (Green et al., 1990) and converted to the Haldane map function (Choi et al., 2010).
Statistical analysis
Three different models using least-squares (LC, HS, and CB models) were adopted for QTL analysis (Kim et al., 2005) and analyses were performed using the methods described in Choi et al. (2010). Significance thresholds were determined by false discovery rate (FDR; Weller et al., 1998).
The LC analysis assumes the QTL to be fixed for alternative alleles in the founder lines. Probabilities of each F2 individual being homozygous for two Duroc alleles (P11), homozygous for two Pietrain alleles (P22), or heterozygous (P12 or P21) were estimated at fixed 1-cM intervals across the genome using the QTL Express software (Seaton et al., 2002). By denoting the mean of homozygous animals for the Duroc allele as positive additive (a), the mean of heterozygous animals as dominance (d) and the mean of homozygous animals for the Pietrain allele as negative additive (−a), the following linear model was fitted at every cM across the genome.
Where yj is the phenotype of F2 progeny j, Xj, and b are the design matrix and solution vector for the fixed effects, respectively, a and d are the estimated additive and dominance effects of a putative QTL at the given location, respectively, Paj = P11 − P22 is the conditional expectation of the number of Duroc alleles carried by animal j, Pdj = P12 + P21 is the conditional probability of animal j to be heterozygous, and ej is the residual error.
The HS analysis assumes the QTL to be segregating in the parental breeds, and the 6 F1 sires were regarded as common parents. QTL Express (Seaton et al., 2002) was used to calculate the probabilities of individuals inheriting allele (A1) or allele (A2) from the common F1 sire (A1 or A2) at fixed 1-cM intervals (Knott et al., 1996). In these analyses contrasts were made between the two haplotypes of every F1 sire.
Where yij is the phenotype of F2 progeny j of F1 sire i, Xij, and b are the design matrix and the solution vector for fixed effects, respectively, si is the effect of the ith F1 sire, αHSi is the substitution effect for the two putative QTL alleles (A1 or A2) carried by the ith F1 sire, PSij is the probability that the F2 individuals inherited the arbitrary allele (Ai1) from F1 sire i, and eij is the residual error.
The CB model assumes the QTL to be segregating in the parental breeds.
Where yij is the phenotype of F2 progeny j of F1 sire i, Xij, and b are the design matrix and the solution vector for fixed effects, respectively, si is the effect of the ith F1 sire, a and d are the additive and dominance effects of breed-origin alleles, respectively, Paij and Pdij are the corresponding breed-origin coefficients as described above, αCBi is the substitution effect for the two putative QTL alleles carried by the ith F1 sire, PSij is the probability that the F2 individuals inherited the arbitrary allele (Ai1) from F1 sire i, and eijis the residual error. In this model, a and d account for the average effects of breed-origin alleles through both the F1 sire and the F1 dam and αCBi represents the difference between the two QTL alleles that a given F1 sire received from the two parental breeds as a deviation from their average additive effect (Kim et al., 2005). To avoid increasing Type I error rate due to multiple testing, a significance threshold of q < 0.05 was used, where q is the FDR corrected p-value. QTL detected using the LC, HS, or CB models were declared using the following criteria:
-
(1)
LC QTL declared if qLC = min(qLC, qHS) < 0.05
-
(2)
HS QTL declared if qHS = min(qLC, qHS) < 0.05
-
(3)
CB QTL declared if qCB < 0.05 and qLC > 0.05 and qHS > 0.05
A QTL was declared under the CB model only if it had not been previously detected using the LC or HS models.
Results
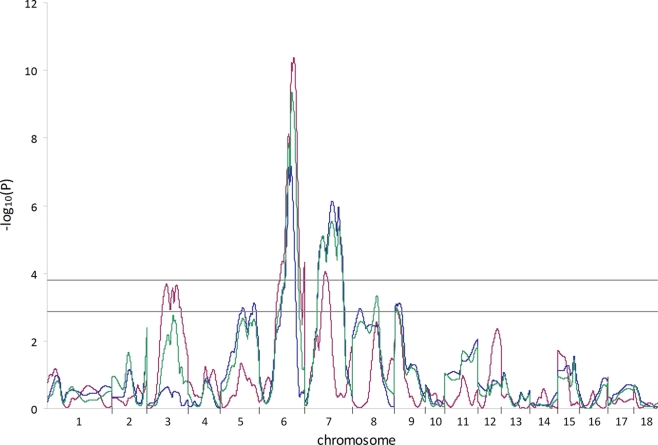
Three different models for QTL analysis revealed a total of 91 QTL for carcass and meat quality traits on all autosomes except SSC11 and 17. The LC analysis revealed 50 QTL (Table 2) including 14 new QTL on 6 chromosomes (SSC3, 6, 7, 12, 16, and 18) which had not been identified in the first genome scan of this population (Edwards et al., 2008a). The HS analysis revealed 38 QTL, and 3 additional QTL were detected using the CB model (Table 2). The thresholds used in this study were −log10(P) = 3.78 and −log10(P) = 2.88 at the 1 and 5% FDR levels, respectively. As an example, the genome scan for ham weight is shown in Figure 1. At the 1% FDR level, two QTL were identified using the LC model on SSC6 and 7, and one QTL was identified using the HS model on SSC7. At the 5% FDR level, additional QTL were revealed on SSC3 with the LC model and on SSC5, 8, and 9 with the HS model.
Table 2.
Position and significance level of carcass and meat quality trait QTL.
| Chr1 | Position2 | Trait | Type3 | −log10p4 | FDR5 | Flanking markers | Additive6 | Dominance7 |
|---|---|---|---|---|---|---|---|---|
| 1 | 12 | LM area, cm2 | LC | 3.48 | 0.0177 | SW1514–SW1515 | −1.27 (0.32) | 0.45 (0.59) |
| 236 | Spareribs wt, kg | LC | 5.36 | 0.0005 | SW974–S0056 | 0.02 (0.02) | −0.12 (0.02) | |
| 2 | 81 | Juiciness, 1–8 | HS | 2.98 | 0.0471 | S0170–SW1026 | ||
| 100 | 45-m carcass temperature, °C | HS | 3.00 | 0.0460 | SW1026–S0370 | |||
| 3 | 47 | 45-m carcass temperature, °C | HS | 4.73 | 0.0020 | SW2021–S0206 | ||
| 97 | Ham wt, kg | LC | 3.68 | 0.0121 | S0206–SWR978 | 0.11 (0.03) | 0.03 (0.05) | |
| 117 | 45-min to 24-h pH decline | CB | 3.21 | 0.0469 | ACTG2–SW2141 | |||
| 135 | 45-min pH | LC | 3.92 | 0.0076 | SW2047–SW2408 | −0.04 (0.01) | 0.00 (0.02) | |
| 151 | Loin wt, kg | LC | 3.36 | 0.0215 | SW2047–SW2408 | 0.02 (0.03) | −0.20 (0.05) | |
| 4 | 19 | Off-farm BW, kg | LC | 2.91 | 0.0473 | SW2509–S0301 | −1.58 (0.45) | 0.78 (0.77) |
| 21 | HCW, kg | LC | 3.11 | 0.0334 | SW2509–S0301 | −1.30 (0.35) | 0.56 (0.61) | |
| 5 | 94 | First-rib backfat, mm | HS | 3.95 | 0.0083 | SWR453–SW2 | ||
| 151 | 24-h carcass temperature, °C | LC | 3.14 | 0.0317 | S0005–S0018 | 0.04 (0.03) | 0.22 (0.06) | |
| 173 | Ham wt, kg | HS | 3.13 | 0.0364 | S0018–IGF1 | |||
| 6 | 103 | Picnic shoulder wt, kg | LC | 3.39 | 0.0204 | SW2525–S0087 | −0.05 (0.02) | 0.05 (0.02) |
| 114 | Moisture, % | LC | 5.12 | 0.0008 | S0087–S0220 | −0.25 (0.08) | 0.48 (0.13) | |
| 124 | Firmness, 1–5 | LC | 4.31 | 0.0037 | S0220–SW122 | 0.17 (0.04) | −0.03 (0.06) | |
| 141 | Fat, % | LC | 19.03 | 0.0000 | SW2173–SW1647 | 0.56 (0.06) | −0.23 (0.09) | |
| 146 | Marbling, 1–10 | LC | 16.18 | 0.0000 | SW2173–SW1647 | 0.34 (0.04) | −0.14 (0.06) | |
| 152 | a* | LC | 4.40 | 0.0031 | SW2173–SW1647 | 0.12 (0.05) | 0.24 (0.07) | |
| 160 | First-rib backfat, mm | LC | 6.71 | 0.0000 | SW1647–SW1881 | 1.54 (0.30) | −1.19 (0.47) | |
| 162 | 10th-rib backfat, mm | LC | 35.82 | 0.0000 | SW1647–SW1881 | 3.20 (0.25) | −1.65 (0.38) | |
| 162 | Carcass length, cm | LC | 10.42 | 0.0000 | SW1647–SW1881 | −0.46 (0.11) | 0.55 (0.16) | |
| 163 | Loin wt, kg | LC | 19.86 | 0.0000 | SW1647–SW1881 | −0.21 (0.02) | 0.18 (0.04) | |
| 164 | Last lumbar vertebra backfat, mm | LC | 14.87 | 0.0000 | SW1647–SW1881 | 1.95 (0.25) | −1.51 (0.39) | |
| 168 | Belly wt, kg | LC | 3.78 | 0.0101 | SW1881–SW322 | 0.06 (0.02) | −0.06 (0.02) | |
| 174 | Ham wt, kg | LC | 10.38 | 0.0000 | SW1881–SW322 | −0.16 (0.02) | 0.08 (0.04) | |
| 174 | Last-rib backfat, mm | LC | 7.47 | 0.0000 | SW1881–SW322 | 1.28 (0.27) | −1.62 (0.43) | |
| 175 | LM area, cm2 | LC | 8.30 | 0.0000 | SW1881–SW322 | −1.30 (0.21) | 0.61 (0.35) | |
| 179 | Protein, % | LC | 5.41 | 0.0005 | SW1881–SW322 | −0.30 (0.06) | 0.09 (0.10) | |
| 182 | HCW, kg | LC | 3.10 | 0.0341 | SW1881–SW322 | 0.44 (0.35) | −2.23 (0.61) | |
| 183 | 24-h carcass temperature, °C | LC | 4.98 | 0.0010 | SW1881–SW322 | 0.13 (0.03) | −0.11 (0.05) | |
| 7 | 15 | Protein, % | LC | 3.44 | 0.0186 | S0025–S0064 | −0.27 (0.07) | −0.23 (0.15) |
| 75 | Spareribs wt, kg | CB | 3.40 | 0.0339 | SW2019–SW859 | |||
| 84 | Dressing percent, % | LC | 12.04 | 0.0000 | SW2019–SW859 | −0.81 (0.11) | −0.05 (0.19) | |
| 86 | Carcass length, cm | LC | 11.50 | 0.0000 | SW2019–SW859 | 0.88 (0.13) | 0.29 (0.23) | |
| 97 | LM area, cm2 | LC | 8.25 | 0.0000 | SW2019–SW859 | −1.60 (0.26) | 0.21 (0.46) | |
| 104 | Ham wt, kg | LC | 4.05 | 0.0060 | SW2019–SW859 | −0.12 (0.03) | −0.02 (0.04) | |
| 130 | Marbling, 1–10 | HS | 2.97 | 0.0480 | SW859–SW2040 | |||
| 139 | Ham wt, kg | HS | 6.13 | 0.0002 | SW859–SW2040 | |||
| 141 | Loin wt, kg | HS | 3.83 | 0.0102 | SW859–SW2040 | |||
| 178 | Number of ribs | HS | 9.23 | 0.0000 | S0115–SW632 | |||
| 8 | 39 | Ham wt, kg | HS | 2.96 | 0.0490 | SW905–SWR1101 | ||
| 126 | LM area, cm2 | LC | 2.97 | 0.0429 | S0017–SW2160 | −0.84 (0.24) | −0.40 (0.37) | |
| 205 | Off-flavor, 1–8 | HS | 4.48 | 0.0031 | SW1085–S0178 | |||
| 214 | Cook yield, % | HS | 3.73 | 0.0122 | SW1085–S0178 | |||
| 9 | 0 | Drip loss, % | LC | 2.94 | 0.0449 | SW21 | −0.04 (0.07) | 0.35 (0.10) |
| 25 | Ham wt, kg | HS | 3.11 | 0.0380 | SW983–SW911 | |||
| 10 | 0 | Overall tenderness, 1–8 | LC | 2.90 | 0.0484 | SWR136 | 0.04 (0.04) | 0.19 (0.05) |
| 21 | Protein, % | HS | 3.06 | 0.0414 | SWR136–SW249 | |||
| 72 | Connective tissue, 1–8 | LC | 2.99 | 0.0410 | SW1041–SW920 | 0.08 (0.04) | 0.17 (0.07) | |
| 12 | 47 | Marbling, 1–10 | LC | 5.50 | 0.0004 | SW957–SW874 | 0.23 (0.04) | 0.03 (0.07) |
| 50 | Belly wt, kg | LC | 4.14 | 0.0051 | SW957–SW874 | 0.09 (0.02) | −0.01 (0.03) | |
| 50 | Fat, % | LC | 3.97 | 0.0070 | SW957–SW874 | 0.30 (0.07) | −0.11 (0.12) | |
| 69 | Moisture, % | LC | 5.80 | 0.0002 | SW37–S0090 | −0.36 (0.07) | 0.08 (0.11) | |
| 93 | b* | LC | 3.72 | 0.0112 | S0090–SWC23 | 0.17 (0.04) | 0.08 (0.07) | |
| 110 | a* | LC | 3.07 | 0.0360 | SWC23–SW2180 | 0.19 (0.05) | 0.08 (0.08) | |
| 13 | 122 | Last-rib backfat, mm | HS | 2.96 | 0.0488 | SW398–SW2440 | ||
| 14 | 62 | a* | LC | 3.30 | 0.0239 | SW210–SW886 | −0.29 (0.07) | 0.06 (0.13) |
| 73 | Boston shoulder wt, kg | HS | 3.18 | 0.0336 | SW210–SW886 | |||
| 136 | Belly wt, kg | LC | 4.26 | 0.0041 | SW1557–SWC27 | 0.06 (0.03) | −0.26 (0.06) | |
| 15 | 70 | Loin wt, kg | HS | 3.57 | 0.0160 | S0088–SW1683 | ||
| 71 | First-rib backfat, mm | HS | 3.11 | 0.0383 | S0088–SW1683 | |||
| 72 | 10th-rib backfat, mm | HS | 3.77 | 0.0115 | S0088–SW1683 | |||
| 74 | Color, 1–6 | HS | 4.43 | 0.0033 | SW1683–SW906 | |||
| 76 | L* | HS | 4.74 | 0.0020 | SW1683–SW906 | |||
| 78 | Juiciness, 1–8 | HS | 4.56 | 0.0027 | SW1683–SW906 | |||
| 78 | Moisture, % | HS | 5.59 | 0.0004 | SW1683–SW906 | |||
| 80 | Warner-Bratzler shear force, kg | HS | 5.51 | 0.0005 | SW1683–SW906 | |||
| 80 | Overall tenderness, 1–8 | HS | 10.22 | 0.0000 | SW1683–SW906 | |||
| 80 | Protein, % | HS | 27.55 | 0.0000 | SW1683–SW906 | |||
| 80 | Tenderness, 1–8 | HS | 9.72 | 0.0000 | SW1683–SW906 | |||
| 81 | a* | HS | 4.51 | 0.0029 | SW1683–SW906 | |||
| 81 | 24-h pH | HS | 10.48 | 0.0000 | SW1683–SW906 | |||
| 82 | Firmness, 1–5 | HS | 3.35 | 0.0240 | SW1683–SW906 | |||
| 83 | Drip loss, % | HS | 10.36 | 0.0000 | SW906–SW1983 | |||
| 85 | Cook yield, % | HS | 16.06 | 0.0000 | SW906–SW1983 | |||
| 87 | Connective tissue, 1–8 | HS | 4.89 | 0.0015 | SW906–SW1983 | |||
| 90 | Belly wt, kg | HS | 3.38 | 0.0226 | SW906–SW1983 | |||
| 16 | 42 | LM area, cm2 | LC | 3.58 | 0.0145 | SW419–SW1454 | 0.80 (0.20) | −0.23 (0.32) |
| 66 | L* | HS | 3.48 | 0.0191 | SW1454 | |||
| 70 | a* | HS | 4.91 | 0.0015 | SW1454–SW2517 | |||
| 97 | Moisture, % | HS | 3.45 | 0.0199 | SW2517–SW1897 | |||
| 143 | Fat, % | LC | 3.24 | 0.0265 | S0061 | 0.18 (0.06) | 0.22 (0.09) | |
| 143 | Moisture, % | LC | 3.35 | 0.0219 | S0061 | −0.14 (0.07) | −0.31 (0.09) | |
| 18 | 0 | 10th-rib backfat, mm | LC | 3.00 | 0.0408 | SW1808 | 0.96 (0.26) | 0.04 (0.35) |
| 4 | Carcass length, cm | LC | 5.67 | 0.0003 | SW2540–SW1023 | −0.49 (0.10) | −0.23 (0.14) | |
| 24 | Spareribs wt, kg | LC | 5.13 | 0.0008 | SW2540–SW1023 | −0.04 (0.01) | 0.00 (0.02) | |
| 30 | Last lumbar vertebra backfat, mm | LC | 4.40 | 0.0031 | SW2540–SW1023 | 1.60 (0.35) | 0.01 (0.64) | |
| 33 | 24-h carcass temperature, °C | CB | 3.40 | 0.0339 | SW2540–SW1023 | |||
| 70 | Spareribs wt, kg | HS | 5.34 | 0.0007 | SW1023–SW1984 |
1Chr, chromosome. 2Position in Haldane cM. 3LC, QTL declared as line-cross type; HS, half-sib type; CB, combined type. 4Negative logarithm of the comparison-wise p-value of the test statistic against the null hypothesis of no QTL at the most likely position for the inferred QTL model. 5FDR, false discovery rate. 6Estimates of additive effects with SE for LC QTL. The effects are expressed as (DD-PP)/2, where D, Duroc allele and P, Pietrain allele. 7Estimates of dominance effects with SE for LC QTL. The effects are expressed as DP-PD, where D, Duroc allele and P, Pietrain allele.
Figure 1.
Genome scan results for ham weight determined using different analysis models. A whole genome scan to identify QTL for the trait ham weight was performed using three different analysis models (line-cross, red line; half-sib, blue line; combined line-cross and half-sib, green line). The X-axis indicates positions of chromosomes 1–18. Horizontal lines indicate significance thresholds (lower line, 5% FDR; upper line, 1% FDR).
Line-cross analysis
A total of 50 significant QTL were identified on SSC1, 3–10, 12, 14, 16, and 18 using the LC model (Table 2). Of these, 29 QTL were below the 1% FDR threshold on SSC1, 3, 6, 7, 12, 14, and 18. On SSC1, QTL affecting LMA and spareribs weight detected at 12 and 236 cM supported our previous results, but a QTL for dressing percent which was significant at the 1% chromosome-wise level in the first scan of this population (Edwards et al., 2008a) did not reach significance in the second scan. On SSC3, a QTL for 45-min pH was significant at the 1% FDR level, also confirming results from our first scan (Edwards et al., 2008a), and a QTL for ham weight was newly identified at the 5% FDR level.
On SSC6, QTL for moisture and firmness were located in the S0087–S0220 interval, QTL influencing meat quality traits were mapped to the SW2173–SW1647 interval, and QTL affecting fat deposition and carcass traits were identified in the SW1647–SW1881–SW322 interval (Figure 2). The QTL detected in these marker intervals showed additive pleiotropic effects indicating that the Duroc allele contributed to increased fat deposition and reduced muscularity. In contrast to SSC6, QTL affecting muscle mass located in the SW2019–SW859 interval on SSC7 showed negative additive effects, and the Pietrain allele was associated with higher muscularity. The incorporation of the new SSC7 marker SW2019 in the SW1369–SW850 marker interval allowed refining the QTL position detected in the first scan, as well as increasing the statistical power and narrowing the QTL interval. A QTL for LMA detected in the SW859–S0115 interval in the first scan was repositioned at 86 cM in the SW2019–SW859 interval in the second scan.
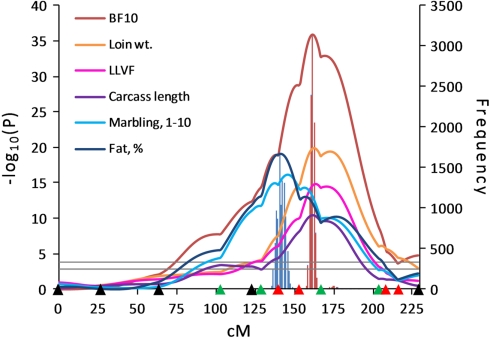
Figure 2.
Line-cross analysis indicated strong evidence for QTL influencing fat deposition traits on SSC6. Highly significant QTL for traits related to fat deposition were identified on SSC6. Confidence intervals for fat percentage and 10th-rib backfat (BF10) were estimated using 10,000 bootstrap permutations as 136–146 cM (blue bar) and 159–165 cM (red bar), respectively. Marker positions are shown as triangles on the X-axis (black, markers used for both QTL scans and genotyped only in 510 animals; green, markers used for both QTL scans and genotyped in all animals; red, markers used for second scan only and genotyped in all animals). Horizontal lines indicate significance thresholds (lower line, 5% FDR; upper line, 1% FDR).
On SSC12, QTL for fat related traits including marbling score, belly weight, and intramuscular fat percent detected in the SW874–S0090 interval in the first scan were identified in the second scan in the SW957–SW874 interval at the 1% FDR level. In addition, at the 5% FDR level, QTL for a* and b* not identified in the first scan were mapped to 93 and 110 cM of SSC12, respectively. A QTL for LMA mapped to 42 cM and QTL for intramuscular fat and moisture percent located at 143 cM were newly discovered on SSC16 in the second scan. In the SW2540–SW1023 interval of SSC18, not only was a QTL for spareribs weight confirmed from the first scan, but QTL for carcass length and last-lumber backfat were also newly identified in the second scan.
Half-sib analysis
Half-sib analysis revealed a total of 38 QTL on SSC2, 3, 5, 7–10, 13–16, and 18 (Table 2). Of these, 20 QTL identified on SSC3, 5, 7, 8, 15, 16, and 18 were significant at the 1% FDR level including 13 QTL detected on SSC15.
A QTL affecting 45-min carcass temperature (q < 0.01) was detected at 47 cM on SSC3. On SSC5, a QTL for first-rib backfat was declared as a HS QTL (q < 0.01) in the second scan, whereas a first-rib backfat QTL had previously been identified in this location with the LC analysis in the first scan (Edwards et al., 2008a). On SSC8, a QTL affecting ham weight was identified at 39 cM, and QTL for off-flavor and cook yield were mapped to the distal region of SSC8 near S0178. In addition, a QTL for ham weight on SSC9 and a QTL for protein percent on SSC10 were identified (q < 0.01). On SSC16, HS QTL were identified for L* (q < 0.02), a* (q < 0.01), and moisture percent (q < 0.02). A highly significant HS QTL influencing spareribs weight (q < 0.0007) was detected on SSC18 with an estimated location at 70 cM. The location of the LC QTL for spareribs weight on SSC18 was estimated at 24 cM so these QTL were considered to be unique QTL.
On SSC7, QTL affecting ham weight and number of ribs were identified in the SW859–SW2040–S0115 interval (q < 0.01), and QTL for marbling score and loin weight significant at the 5% FDR level were located in the same interval. For ham weight, the QTL identified with the HS analysis was mapped to 139 cM (q < 0.0002), whereas the ham weight QTL revealed with the LC analysis was mapped to 104 cM (q < 0.006). Since these QTL detected by the different models mapped to distinct locations, they were considered to be separate unique QTL.
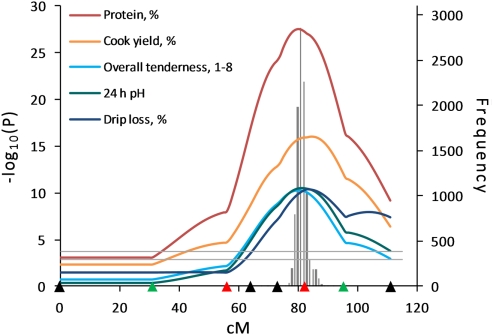
The HS analysis revealed evidence for QTL influencing meat quality traits in the SW1683–SW906–SW1983 interval on SSC15 (Figure 3). In the SW1683–SW906 interval, a QTL for protein percent had the highest test statistic [–log10(P) = 27.55; q < 0.0001] among the QTL detected on SSC15. In addition, a QTL for 24 h pH, a trait that is associated with many other meat quality traits, was highly significant (q < 0.0001; Figure 3). The LC analysis also revealed significant QTL for these traits in the same interval, but the HS model showed much higher statistical evidence.
Figure 3.
Half-sib analysis indicated strong evidence for QTL influencing meat quality traits on SSC15. Highly significant QTL for meat quality traits were identified on SSC15. Confidence interval for protein percentage was estimated by 10,000 bootstrap permutations as 77–85 cM (gray bar). Marker positions are shown as triangles on the X-axis (black, markers used for both QTL scans and genotyped only in 510 animals; green, markers used for both QTL scans and genotyped in all animals; red, markers used for second scan only and genotyped in all animals). Horizontal lines indicate significance thresholds (lower line, 5% FDR; upper line, 1% FDR).
Combined analysis
In addition to QTL identified with the LC and HS analyses, three additional QTL exceeded the 5% FDR significance threshold using the CB analysis. A QTL for pH decline from 45-min to 24-h was mapped to 117 cM on SSC3, a QTL for spareribs weight was detected in the SW2019–SW859 interval on SSC7 and a QTL for 24-h carcass temperature was found in the SW2540–SW1023 interval on SSC18. Although the statistical power was sufficient to detect QTL, the CB model revealed a small number of additional QTL because most QTL had been declared using either the LC or HS models due to higher test statistics with these analyses.
Effect of additional markers and animals on QTL detection
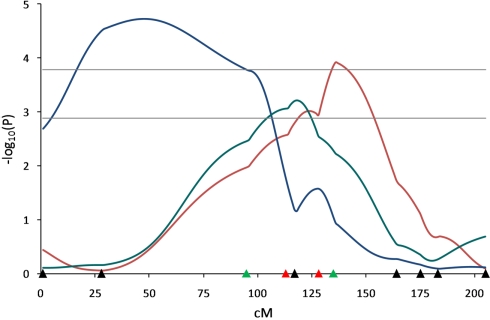
The QTL analyses under three different models revealed QTL for pH associated traits at different positions on SSC3 (Figure 4). The first scan of this chromosome using the LC model had revealed QTL for 45-min pH and pH decline from 45-min to 24-h postmortem (Edwards et al., 2008a). The second scan included two additional markers and genotypes for additional F2 pigs. A QTL for 45-min pH using the LC model (q ≤ 0.0076) was mapped at 135 cM near marker SW2047 (134.8 cM) and a QTL for pH decline from 45-min to 24-h was detected using the CB model (q ≤ 0.0469) at 117 cM near marker ACTG2 (116.5 cM) which did not reach the significance threshold in the LC analysis (q ≤ 0.065). In addition, a QTL for 45-min carcass temperature was detected using the HS model (q ≤ 0.002) located at 47 cM in the SW2021–S0206 marker interval. These results confirm results of the first scan for 45-min pH and pH decline, and add new results for 45-min carcass temperature.
Figure 4.
Quantitative trait loci results determined by different models for pH related traits on SSC3. Line-cross model detected a QTL (q < 0.01) for 45 min pH (red line) and half-sib model detected a QTL (q < 0.01) for 45 min carcass temperature (blue line). Combined model identified a QTL (q < 0.01) affecting pH decline from 45 min to 24 h (green line). Marker positions are shown as triangles on the X-axis (black, markers used for both QTL scans and genotyped only in 510 animals; green, markers used for both QTL scans and genotyped in all animals; red, markers used for second scan only and genotyped in all animals). Horizontal lines indicate significance thresholds (lower line, 5% FDR; upper line, 1% FDR).
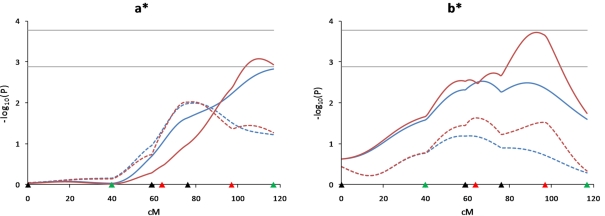
We have recently used LC, HS, and CB models to identify QTL for growth traits in our Duroc × Pietrain resource population, and we reported that additional markers and animals contributed to reduce the confidence intervals and increase the test statistics for QTL detection (Choi et al., 2010). For the present study, QTL affecting the a* and b* objective color measures were newly detected on SSC12 (q < 0.04). In order to determine how the QTL peaks for these traits were changed, analyses were performed under 4 different scenarios; 5 and 7 markers with 510 and 948 animals (Figure 5). The results indicated that increasing the number of animals or increasing the number of markers was effective in increasing the power to detect QTL on this chromosome, and that increasing the numbers of both animals and markers allowed detection of the a* and b* QTL.
Figure 5.
Effect of additional markers and animals for detecting meat color QTL on SSC12. Effects of additional marker genotypes and animals for detecting QTL for a* and b* objective meat color measures on SSC12 were compared under 4 different scenarios (5 and 7 markers with 510 F2 animals, 5 and 7 markers with 948 F2 animals). Blue lines indicate 5 markers (black and green triangles on the X-axis) and red lines indicate 7 markers (black, green and red triangles on the X-axis). Solid lines indicate 948 animals and dotted lines indicate 510 animals. Marker positions are shown as triangles on the X-axis (black, markers used for both QTL scans and genotyped only in 510 animals; green, markers used for both QTL scans and genotyped in all animals; red, markers used for second scan only and genotyped in all animals). Horizontal lines indicate significance thresholds (lower line, 5% FDR; upper line, 1% FDR).
Discussion
This study identified 91 QTL for pig carcass and meat quality traits located on all autosomes except SSC11 and 17 using three least-squares Mendelian analysis models. The LC analysis, which detected QTL segregating between breeds, revealed 50 QTL including 13 new QTL on 6 chromosomes (SSC3, 6, 7, 12, 16, and 18) that had not been identified in the first genome scan of this population (Edwards et al., 2008a). The HS analysis, which detected QTL segregating within breeds, revealed 38 QTL including 18 on SSC15. Three additional QTL were detected using the CB model (Kim et al., 2005).
Application of the three different models for SSC3 identified not only QTL influencing muscularity under the LC model, but also QTL affecting pH and carcass temperature using all three models. The LC QTL for 45-min pH detected at 135 cM near SW2047 confirmed the 45-min pH QTL observed in the first scan (Edwards et al., 2008a). Beeckmann et al. (2003) reported a QTL for 45-min pH at the same interval in a Wild boar × Meishan F2 population. Several studies (Óvilo et al., 2002a; de Koning et al., 2003; Evans et al., 2003; Wimmers et al., 2006) reported QTL affecting muscle pH in the SW2021–S0206 marker interval, a region where we identified a QTL for 45-min carcass temperature under the HS model. In addition, Duan et al. (2009) reported a QTL for pH decline from 45-min to 3-h in the SW2021–S0206 interval in a White Duroc × Chinese Erhualian population. We also detected a QTL for pH decline from 45-min to 24-h under the CB model, however, our QTL was located at 117 cM near ACTG2.
Significant QTL affecting backfat thickness were located on SSC6 within the SW1647–SW1881–SW322 marker interval at 160–174 cM. A 10,000 bootstrap permutation analysis showed the average QTL positions for each backfat trait to be located in the 160.12–167.96 cM region. The 95% confidence interval for 10th-rib backfat was estimated to be 159–165 cM (6 Haldane cM), which was considerably narrowed from the 38.5 Haldane cM interval observed for the first scan (Edwards et al., 2008a). Not only were QTL affecting fat deposition traits observed in this region, but QTL influencing muscularity were also identified at the same marker interval since Duroc alleles contributed to both fat accumulation and reduced muscle content.
Our results for backfat thickness traits were in agreement with other studies (Malek et al., 2001b; Óvilo et al., 2002b; Varona et al., 2002) that identified QTL for fatness traits in this region of SSC6. This region includes the leptin receptor (LEPR) gene which is considered as a potential candidate gene for fatness (Ernst et al., 1997; Óvilo et al., 2005; Mohrmann et al., 2006), and studies to identify a causal mutation in LEPR have been conducted (Mackowski et al., 2005; Muñoz et al., 2009). We also observed a QTL for intramuscular fat percent using the LC model in a position more proximal to this region of SSC6 at 141 cM, which coincided with a backfat thickness QTL detected with the HS model in a Duroc × Pietrain population by Liu et al. (2008). The SSC6 region affecting intramuscular fat percent also included QTL for marbling score and a*, which were all mapped to 141–152 cM in the SW2173–SW1647 interval. The confidence interval for these QTL did not overlap with the confidence interval for 10th-rib backfat. This result was consistent with previous studies (Szyda et al., 2003; Óvilo et al., 2005) which reported that QTL effects for backfat and intramuscular fat content resulted from different closely linked loci on SSC6. QTL affecting intramuscular fat content have been reported (de Koning et al., 2000; Grindflek et al., 2001) in the same region where we detected a QTL for marbling score, although no other reports of subjective marbling score QTL in this SSC6 region have been reported. Also, Harmegnies et al. (2006) identified QTL for a* as well as fat thickness in this same region.
The different models revealed distinct QTL regions on SSC7 with LC and HS QTL identified at 84–104 cM and at 130–178 cM, respectively. A highly significant QTL influencing muscle mass identified in the SW2019–SW859 interval had an additive effect for which Duroc alleles increased carcass length and decreased dressing percent, LMA and ham weight. In this region, Yue et al. (2003) found a 1% genome-wide level significant QTL influencing carcass composition traits such as carcass length in a Wild boar × Meishan population. Liu et al. (2008) reported QTL for carcass length and dressing percent with similar allelic substitution effect in their Duroc × Pietrain population as we observed in our study. However, Nezer et al. (2002) identified a QTL for carcass length at the more distal position from our QTL in a Pietrain × Large White population. In addition, Sato et al. (2003) detected a QTL for dressing percent in a Duroc × Meishan population in the same region as our study. A QTL for number of ribs was detected using the HS analysis. A QTL for number of ribs had been detected in this position at the 1% genome-wise significance level using the LC analysis in the first scan (Edwards et al., 2008a), however, evidence from the second scan suggests the HS model better describes the QTL allele frequency in the parental breeds. Also on SSC7, analyses using both the LC and HS models identified QTL for ham weight at different locations, which were in the SW2019–SW859 interval with the LC model and in the SW859–SW2040 interval with the HS model. Similarly, Milan et al. (2002) also reported suggestive QTL for ham weight at different positions using LC and HS models, and their LC QTL detected in the SLA–S0102 marker interval was in a similar region to our LC QTL.
We have recently used LC, HS, and CB models to identify QTL for growth traits in our Duroc × Pietrain resource population, and we reported that additional markers and animals contributed to reduce the confidence intervals and increase the test statistics for QTL detection (Choi et al., 2010). In the present study, genotyping of additional markers and animals increased the statistical power and facilitated discovery of new QTL which had not been observed in the first scan (Edwards et al., 2008a). For example, QTL for the objective color measures of a* and b* were identified on SSC12 with the addition of more F2 pigs and more marker genotypes using the LC analysis. The LC analysis also identified QTL on SSC12 related to intramuscular fat percent and moisture at 47–50 cM and at 69 cM, respectively. A QTL for marbling was located in the SW957–SW874 marker interval, whereas the position of this QTL had been more distal for the first scan (Edwards et al., 2008a). The additive effects of these QTL indicated that Duroc alleles increased marbling and intramuscular fat percent, and decreased moisture percent. Harmegnies et al. (2006) also reported a QTL for a* although at a more distal position than our current result, and Malek et al. (2001a) detected a QTL for subjective color score in the same region as our result.
The HS analysis revealed strong evidence for QTL affecting meat quality traits on SSC15 at 74–90 cM in the SW1683–SW1983 marker interval where 13 and 2 QTL were significant at the 1 and 5% FDR levels, respectively, including a highly significant QTL for protein percent. Significant QTL had been identified in this region using the LC analysis in the first scan (Edwards et al., 2008a), and a negative additive effect had been seen for protein percent, color, and tenderness traits suggesting contributions from segregation of Pietrain alleles. The pleiotropic effects of Pietrain alleles contributing to leanness resulted in effects on other meat quality traits resulting in more muscularity, paler muscle color, and less tenderness. QTL for 24-h pH, L*, and tenderness significant at the 1% genome-wise level were identified in this region of SSC15 in a Berkshire × Yorkshire population (Malek et al., 2001a; Thomsen et al., 2004; Kim et al., 2005). Very few studies have measured the trait of protein percent and no QTL for protein percent have been reported on SSC15.
Several candidate genes such as myostatin (MSTN), Titin (TTN), and protein kinase AMP-activated gamma 3 (PRKAG3) are located in the SW1683–SW1983 interval (Sonstegard et al., 1998; Milan et al., 2000; Davoli et al., 2003). Stinckens et al. (2008) reported that pigs of the Pietrain breed had higher muscularity as a result of association between a polymorphism in the MSTN MEF3 binding site and muscle mass. Wimmers et al. (2007) reported that a polymorphism in TTN was associated with leanness in a Pietrain population. Also, Milan et al. (2000) mapped the PRKAG3 gene in the SW1683–SW1983 marker interval and Ciobanu et al. (2001) identified three polymorphic sites in the PRKAG3 gene that affect meat quality traits including 24-h pH.
Conclusion
This study used a Duroc × Pietrain F2 resource population and identified a total of 91 QTL for carcass merit and meat quality phenotypes. Three different least-squares models were applied under different assumptions; (1) founders fixed for alternative QTL alleles (LC model), (2) segregation of QTL alleles at similar frequencies in founders (HS model), and (3) segregation of QTL alleles at different frequencies in founders (combined model). The addition of new marker and animal genotypes contributed to increasing the statistical power for QTL detection, and the application of alternative models allowed confirmation of QTL and detection of new QTL segregating either between or within breeds.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was supported by National Research Initiative Competitive Grant no. 2004-35604-14580 from the USDA National Institute of Food and Agriculture. We thank D. Edwards for technical contributions in development of the pig resource population.
References
- Affentranger P., Gerwig C., Seewer G. J. F., Schworer D., Kunzi N. (1996). Growth and carcass characteristics as well as meat and fat quality of three types of pigs under different feeding regimens. Livest. Prod. Sci. 45, 187–196 10.1016/0301-6226(96)00011-5 [DOI] [Google Scholar]
- Beeckmann P., Schröffel J., Moser G., Bartenschlager H., Reiner G., Geldermann H. (2003). Linkage and QTL mapping for Sus scrofa chromosome 3. J. Anim. Breed. Genet. 120, 20–27 10.1046/j.0931-2668.2003.00420.x [DOI] [Google Scholar]
- Choi I., Steibel J. P., Bates R. O., Raney N. E., Rumph J. M., Ernst C. W. (2010). Application of alternative models to identify QTL for growth traits in an F2 Duroc x Pietrain pig resource population. BMC Genet. 11, 97. 10.1186/1471-2156-11-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanu D., Bastiaansen J., Malek M., Helm J., Woollard J., Plastow G., Rothschild M. (2001). Evidence for new alleles in the protein kinase adenosine monophosphate-activated (3-subunit gene associated with low glycogen content in pig skeletal muscle and improved meat quality. Genetics 159, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli R., Braglia S., Lama B., Fontanesi L., Buttazzoni L., Baiocco C., Russo V. (2003). Mapping, identification of polymorphisms and analysis of allele frequencies in the porcine skeletal muscle myopalladin and titin genes. Cytogenet. Genome Res. 102, 152–156 10.1159/000075741 [DOI] [PubMed] [Google Scholar]
- de Koning D. J., Pong-Wong R., Varona L., Evans G. J., Giuffra E., Sanchez A., Plastow G., Noguera J. L., Andersson L., Haley C. S. (2003). Full pedigree quantitative trait locus analysis in commercial pigs using variance components. J. Anim. Sci. 81, 2155–2163 [DOI] [PubMed] [Google Scholar]
- de Koning D. J., Rattink A. P., Harlizius B., van Arendonk J. A., Brascamp E. W., Groenen M. A. (2000). Genome-wide scan for body composition in pigs reveals important role of imprinting. Proc. Natl. Acad. Sci. U.S.A. 97, 7947–7950 10.1073/pnas.140216397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y. Y., Ma J. W., Yuan F., Huang L. B., Yang K. X., Xie J. P., Wu G. Z., Huang L. S. (2009). Genome-wide identification of quantitative trait loci for pork temperature, pH decline, and glycolytic potential in a large-scale White Duroc x Chinese Erhualian resource population. J. Anim. Sci. 87, 9–16 10.2527/jas.2008-1128 [DOI] [PubMed] [Google Scholar]
- Edwards D. B., Bates R. O., Osburn W. N. (2003). Evaluation of Duroc- vs. Pietrain-sired pigs for carcass and meat quality measures. J. Anim. Sci. 81, 1895–1899 [DOI] [PubMed] [Google Scholar]
- Edwards D. B., Ernst C. W., Raney N. E., Doumit M. E., Hoge M. D., Bates R. O. (2008a). Quantitative trait locus mapping in an F2 Duroc x Pietrain resource population: II. Carcass and meat quality traits. J. Anim. Sci. 86, 254–266 10.2527/jas.2006-626 [DOI] [PubMed] [Google Scholar]
- Edwards D. B., Ernst C. W., Tempelman R. J., Rosa G. J. M., Raney N. E., Hoge M. D., Bates R. O. (2008b). Quantitative trait loci mapping in an F2 Duroc x Pietrain resource population: I. Growth traits. J. Anim. Sci. 86, 241–253 [DOI] [PubMed] [Google Scholar]
- Ernst C. W., Kapke P. A., Yerle M., Rothschild M. F. (1997). The leptin receptor gene (LEPR) maps to porcine chromosome 6. Mamm. Genome 8, 226. 10.1007/s003359900397 [DOI] [PubMed] [Google Scholar]
- Evans G. J., Giuffra E., Sanchez A., Kerje S., Davalos G., Vidal O., Illán S., Noguera J. L., Varona L., Velander I., Southwood O. I., de Koning D. J., Haley C. S., Plastow G. S., Andersson L. (2003). Identification of quantitative trait loci for production traits in commercial pig populations. Genetics 164, 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P., Fallis K., Crooks S. (1990). Documentation for CRIMAP, Version 2.4. Washington University School of Medicine, St. Louis, MO [Google Scholar]
- Grindflek E., Szyda J., Liu Z., Lien S. (2001). Detection of quantitative trait loci for meat quality in a commercial slaughter pig cross. Mamm. Genome 12, 299–304 10.1007/s003350010278 [DOI] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., Elsen J. M. (1994). Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics 136, 1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmegnies N., Davin F., De Smet S., Buys N., Georges M., Coppieters W. (2006). Results of a whole-genome quantitative trait locus scan for growth, carcass composition and meat quality in a porcine four-way cross. Anim. Genet. 37, 543–553 10.1111/j.1365-2052.2006.01523.x [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Goddard M. E. (2009). Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92, 433–443 10.3168/jds.2008-1646 [DOI] [PubMed] [Google Scholar]
- Jeremiah L. E., Gibson J. P., Gibson L. L., Ball R. O., Aker C., Fortin A. (1999). The influence of breed, gender, and PSS (Halothane) genotype on meat quality, cooking loss, and palatability of pork. Food Res. Int. 32, 59–71 10.1016/S0963-9969(99)00077-0 [DOI] [Google Scholar]
- Kim J. J., Zhao H., Thomsen H., Rothschild M. F., Dekkers J. C. (2005). Combined line-cross and half-sib QTL analysis of crosses between outbred lines. Genet. Res. 85, 235–248 10.1017/S0016672305007597 [DOI] [PubMed] [Google Scholar]
- Knott S. A., Elsen J. M., Haley C. S. (1996). Methods for multiple-marker mapping of quantitative trait loci in half-sib populations. Theor. Appl. Genet. 93, 71–80 10.1007/BF00225729 [DOI] [PubMed] [Google Scholar]
- Langlois A., Minvielle F. (1989). Comparisons of three-way and backcross swine: II. Wholesale cuts and meat quality. J. Anim. Sci. 67, 2025–2032 [Google Scholar]
- Liu G., Kim J. J., Jonas E., Wimmers K., Ponsuksili S., Murani E., Phatsara C., Tholen E., Juengst H., Tesfaye D., Chen J. L., Schellander K. (2008). Combined line-cross and half-sib QTL analysis in Duroc-Pietrain population. Mamm. Genome 19, 429–438 10.1007/s00335-008-9132-y [DOI] [PubMed] [Google Scholar]
- Mackowski M., Szymoniak K., Szydlowski M., Kamyczek M., Eckert R., Rozycki M., Switonski M. (2005). Missense mutations in exon 4 of the porcine LEPR gene encoding extracellular domain and their association with fatness traits. Anim. Genet. 36, 135–137 10.1111/j.1365-2052.2005.01247.x [DOI] [PubMed] [Google Scholar]
- Malek M., Dekkers J. C., Lee H. K., Baas T. J., Prusa K., Huff-Lonergan E., Rothschild M. F. (2001a). A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. II. Meat and muscle composition. Mamm. Genome 12, 637–645 10.1007/s003350020019 [DOI] [PubMed] [Google Scholar]
- Malek M., Dekkers J. C., Lee H. K., Baas T. J., Rothschild M. F. (2001b). A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. I. Growth and body composition. Mamm. Genome 12, 630–636 10.1007/s003350020018 [DOI] [PubMed] [Google Scholar]
- Milan D., Bidanel J. P., Iannuccelli N., Riquet J., Amigues Y., Gruand J., Le Roy P., Renard C., Chevalet C. (2002). Detection of quantitative trait loci for carcass composition traits in pigs. Genet. Sel. Evol. 34, 705–728 10.1186/1297-9686-34-6-705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan D., Jeon J. T., Looft C., Amarger V., Robic A., Thelander M., Rogel-Gaillard C., Paul S., Iannuccelli N., Rask L., Ronne H., Lundstrom K., Reinsch N., Gellin J., Kalm E., Roy P. L., Chardon P., Andersson L. (2000). A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science 288, 1248–1251 10.1126/science.288.5469.1248 [DOI] [PubMed] [Google Scholar]
- Mohrmann M., Roehe R., Knap P. W., Looft H., Plastow G. S., Kalm E. (2006). Quantitative trait loci associated with AutoFOM grading characteristics, carcass cuts and chemical body composition during growth of Sus scrofa Anim. Genet. 37, 435–443 10.1111/j.1365-2052.2006.01492.x [DOI] [PubMed] [Google Scholar]
- Muñoz G., Óvilo C., Silió L., Tomás A., Noguera J. L., Rodríguez M. C. (2009). Single- and joint-population analyses of two experimental pig crosses to confirm quantitative trait loci on Sus scrofa chromosome 6 and leptin receptor effects on fatness and growth traits. J. Anim. Sci. 87, 459–468 [DOI] [PubMed] [Google Scholar]
- Nezer C., Moreau L., Wagenaar D., Georges M. (2002). Results of a whole genome scan targeting QTL for growth and carcass traits in a Pietrain x Large White intercross. Genet. Sel. Evol. 34, 371–387 10.1186/1297-9686-34-3-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Óvilo C., Clop A., Noguera J. L., Oliver M. A., Barragán C., Rodríguez C., Silió L., Toro M. A., Coll A., Folch J. M., Sánchez A., Babot D., Varona L., Pérez-Enciso M. (2002a). Quantitative trait locus mapping for meat quality traits in an Iberian x Landrace F2 pig population. J. Anim. Sci. 80, 2801–2808 [DOI] [PubMed] [Google Scholar]
- Óvilo C., Oliver A., Noguera J. L., Clop A., Barragán C., Varona L., Rodríguez C., Toro M., Sánchez A., Pérez-Enciso M., Silió L. (2002b). Test for positional candidate genes for body composition on pig chromosome 6. Genet. Sel. Evol. 34, 465–479 10.1186/1297-9686-34-4-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Óvilo C., Fernández A., Noguera J. L., Barragán C., Letón R., Rodríguez C., Mercadé A., Alves E., Folch J. M., Varona L., Toro M. (2005). Fine mapping of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross. Genet. Res. 85, 57–67 10.1017/S0016672305007330 [DOI] [PubMed] [Google Scholar]
- Pérez-Enciso M., Varona L. (2000). Quantitative trait loci mapping in F2 crosses between outbred lines. Genetics 155, 391–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Oyamada Y., Atsuji K., Nade T., Kobayashi E., Mitsuhashi T., Nirasawa K., Komatsuda A., Saito Y., Terai S., Hayashi T., Sugimoto Y. (2003). Quantitative trait loci analysis for growth and carcass traits in a Meishan x Duroc F2 resource population. J. Anim. Sci. 81, 2938–2949 [DOI] [PubMed] [Google Scholar]
- Seaton G., Haley C. S., Knott S. A., Kearsey M., Visscher P. M. (2002). QTL Express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics 18, 339–340 10.1093/bioinformatics/18.2.339 [DOI] [PubMed] [Google Scholar]
- Sonstegard T. S., Rohrer G. A., Smith T. P. L. (1998). Myostatin maps to porcine chromosome 15 by linkage and physical analyses. Anim. Genet. 29, 19–22 10.1046/j.1365-2052.1998.00229.x [DOI] [PubMed] [Google Scholar]
- Spelman R. J., Bovenhuis H. (1998). Moving from QTL experimental results to the utilization of QTL in breeding programmes. Anim. Genet. 29, 77–84 10.1046/j.1365-2052.1998.00238.x [DOI] [PubMed] [Google Scholar]
- Stinckens A., Luyten T., Bijttebier J., Van den Maagdenberg K., Dieltiens D., Janssens S., De Smet S., Georges M., Buys N. (2008). Characterization of the complete porcine MSTN gene and expression levels in pig breeds differing in muscularity. Anim. Genet. 39, 586–596 10.1111/j.1365-2052.2008.01774.x [DOI] [PubMed] [Google Scholar]
- Szyda J., Grindflek E., Liu Z., Lien S. (2003). Multivariate mixed inheritance models for QTL detection on porcine chromosome 6. Genet. Res. 81, 65–73 10.1017/S0016672302006043 [DOI] [PubMed] [Google Scholar]
- Thomsen H., Lee H. K., Rothschild M. F., Malek M., Dekkers J. C. (2004). Characterization of quantitative trait loci for growth and meat quality in a cross between commercial breeds of swine. J. Anim. Sci. 82, 2213–2228 [DOI] [PubMed] [Google Scholar]
- Varona L., Ovilo C., Clop A., Noguera J. L., Perez-Enciso M., Coll A., Folch J. M., Barragan C., Toro M. A., Babot D., Sanchez A. (2002). QTL mapping for growth and carcass traits in an Iberian by Landrace pig intercross: additive, dominant and epistatic effects. Genet. Res. 80, 145–154 10.1017/S0016672302005803 [DOI] [PubMed] [Google Scholar]
- Weller J. I., Song J. Z., Heyen D. W., Lewin H. A., Ron M. (1998). A new approach to the problem of multiple comparisons in the genetic dissection of complex traits. Genetics 150, 1699–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers K., Fiedler I., Hardge T., Murani E., Schellander K., Ponsuksili S. (2006). QTL for microstructural and biophysical muscle properties and body composition in pigs. BMC Genet. 7, 15. 10.1186/1471-2156-7-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers K., Murani E., Te Pas M. F. W., Chang K. C., Davoli R., Merks J. W. M., Henne H., Muraniova M., Da Costa N., Harlizius B., Schellander K., Böll I., Braglia S., De Wit A. A. C., Cagnazzo M., Fontanesi L., Prins D., Ponsuksili S. (2007). Associations of functional candidate genes derived from gene-expression profiles of prenatal porcine muscle tissue with meat quality and muscle deposition. Anim. Genet. 38, 474–484 10.1111/j.1365-2052.2007.01639.x [DOI] [PubMed] [Google Scholar]
- Yue G., Stratil A., Cepica S., Schrffel J., Schrffelova D., Fontanesi L., Cagnazzo M., Moser G., Bartenschlager H., Reiner G., Geldermann H. (2003). Linkage and QTL mapping for Sus scrofa chromosome 7. J. Anim. Breed. Genet. 120, 56–65 10.1046/j.0931-2668.2003.00424.x [DOI] [Google Scholar]