Abstract
In this study, we assessed association of genome-wide association studies (GWAS) “hits” by race with adjustment for potential population stratification (PS) in two large, diverse study populations; the Carolina Breast Cancer Study (CBCS; N total = 3693 individuals) and the University of Pennsylvania Study of Clinical Outcomes, Risk, and Ethnicity (SCORE; N total = 1135 individuals). In both study populations, 136 ancestry information markers and GWAS “hits” (CBCS: FGFR2, 8q24; SCORE: JAZF1, MSMB, 8q24) were genotyped. Principal component analysis was used to assess ancestral differences by race. Multivariable unconditional logistic regression was used to assess differences in cancer risk with and without adjustment for the first ancestral principal component (PC1) and for an interaction effect between PC1 and the GWAS “hit” (SNP) of interest. PC1 explained 53.7% of the variance for CBCS and 49.5% of the variance for SCORE. European Americans and African Americans were similar in their ancestral structure between CBCS and SCORE and cases and controls were well matched by ancestry. In the CBCS European Americans, 9/11 SNPs were significant after PC1 adjustment, but after adjustment for the PC1 by SNP interaction effect, only one SNP remained significant (rs1219648 in FGFR2); for CBCS African Americans, 6/11 SNPs were significant after PC1 adjustment and after adjustment for the PC1 by SNP interaction effect, all six SNPs remained significant and an additional SNP now became significant. In the SCORE European Americans, 0/9 SNPs were significant after PC1 adjustment and no changes were seen after additional adjustment for the PC1 by SNP interaction effect; for SCORE African Americans, 2/9 SNPs were significant after PC1 adjustment and after adjustment for the PC1 by SNP interaction effect, only one SNP remained significant (rs16901979 at 8q24). We show that genetic associations by race are modified by interaction between individual SNPs and PS.
Keywords: population stratification, ancestry, prostate cancer, breast cancer, GWAS “hits”
Introduction
Multiple genome-wide association studies (GWAS) have now been performed for many complex diseases including breast and prostate cancers. The vast majority of these studies have been performed in individuals of European ancestry only; breast cancer (Easton et al., 2007; Hunter et al., 2007; Stacey et al., 2007; Gold et al., 2008; Ahmed et al., 2009; Thomas et al., 2009; Zheng et al., 2009c) and prostate cancer (Amundadottir et al., 2006; Freedman et al., 2006; Duggan et al., 2007; Gudmundsson et al., 2007, 2009; Haiman et al., 2007; Yeager et al., 2007, 2009; Sun et al., 2008; Thomas et al., 2008; Al Olama et al., 2009; Eeles et al., 2009; Hsu et al., 2009; Lou et al., 2009; Zheng et al., 2009a). Hence, further study of these GWAS “hits” in different racial/ethnic groups is warranted.
While it is now standard practice to test for population stratification (PS) and remove ancestral outliers from analysis in GWAS studies, further adjustment for PS may be necessary when studying a recently admixed population such as African Americans or Hispanic Americans (Barnholtz-Sloan et al., 2008; Tiwari et al., 2008). In addition, because risk allele frequencies can vary by ancestral group, interaction effects between a SNP of interest and PS may be needed in order to fully understand differences in potential genetic associations by race. The objective of the current study was to validate potential GWAS “hit” associations in two large, diverse study populations of breast cancer and prostate cancer, and to evaluate adjustments for potential PS in association testing of GWAS “hits” within each study.
Materials and Methods
Carolina breast cancer study study population
The CBCS is a population-based case–control study of breast cancer conducted in North Carolina (as described in Millikan et al., 2003). Briefly, eligible cases included women ages 20–74 who were diagnosed with primary invasive breast cancer from 1993 to 2001 and lived within a 24-county study area. Cases were identified using rapid case ascertainment in cooperation with the North Carolina Central Cancer Registry. Randomized recruitment was used to oversample African Americans and women younger than 50 years of age. Women diagnosed with breast carcinoma in situ (CIS) from 1996 to 2001 were also enrolled in the study. Eligible controls were women aged 20–74 years, residing within the study area, with no history of breast cancer and were identified using Division of Motor Vehicles lists (for women under 65) and Medicare records (for women age 65–74). Controls were frequency matched to cases according to race within 5 year age categories. Women who agreed to participate in the study provided informed consent and completed an in-home interview regarding known and suspected breast cancer risk factors. Women were also asked to provide a 30-ml blood sample. DNA was extracted from the blood samples and stored at −80°C. The interview participation rates for invasive cases and controls were 76 and 55%, respectively, and for CIS cases and controls were 83 and 65%, respectively. The final study population consisted of 3693 individuals, 2319 European Americans (62.8%), and 1374 African Americans (37.2%); 1946 (52.7%) cases and 1747 (47.3%) controls. Fifty-three (N = 53; <2%) individuals were excluded who reported as “other” race (Hispanic, mixed race or other). Age was defined as age in years at breast cancer diagnosis for cases or at the time of sampling for controls. Self-identified race was reported by each study participant during the study interview. An offset variable to account for the sampling design was included in the analysis. All study procedures involving human subjects were approved by the University of North Carolina (UNC) at Chapel Hill Institutional Review Board.
Study of clinical outcomes, risk, and ethnicity at the university of pennsylvania
Incident prostate cancer cases were identified through Urologic Oncology Clinics at multiple hospitals of the University of Pennsylvania Health System (UPHS) between 1995 and 2008 and included in the SCORE study. Controls were men attending UPHS general medicine clinics and were ascertained concurrently with the prostate cancer cases (i.e., between 1995 and 2008) and were frequency matched to cases according to race. All study participants provided informed consents. Three hundred five (N = 305; 20%) individuals were excluded as they were of “other” race, or they had missing genotype data for ancestry estimation. Our final study population consisted of 1135 individuals, 713 European Americans (62.8%), and 422 African Americans (37.2%); 808 (71.1%) were cases and 327 (28.9%) were controls. Age was defined as age at consent for both cases and controls. Self-identified race was reported by each study participant during the study interview. All study procedures involving human subjects were approved by the University of Pennsylvania Institutional Review Board.
Genotyping results
A panel of 200 ancestry informative markers (AIMs) was genotyped as part of a multiplex, custom candidate gene SNP panel assay using the Illumina Goldengate platform for all 3693 CBCS and all 1165 SCORE individuals (Illumina, Inc., San Diego, CA, USA; Barnholtz-Sloan et al., 2010); previous studies have shown that at least 50–100 AIMs are needed to accurately assign one’s individual ancestry; fewer markers when the average allele frequency difference between ancestral populations 0.6 and above (Risch et al., 2002; Tsai et al., 2005; Choudhry et al., 2006). AIMs were selected to maximize the difference in allele frequencies between ancestral populations and the Fisher’s information criterion (FIC; Pfaff et al., 2004) for distinguishing between African and European ancestry, based upon ancestral allele frequencies from African (YRI) and European (CEU) populations in HapMap (www.hapmap.org). AIMs were prioritized based on having the highest FIC values in the following order: 90% European/10% African, 10% European/90% African, and 50% European/50% African. This prioritization allowed the AIMs to be chosen to represent the whole expected ancestral distribution of this population. In the CBCS dataset, 42 AIMs SNPs were dropped by Illumina and 14 failed genotyping resulting in 144 genotyped AIMs. In the SCORE dataset, 42 SNPs were dropped by Illumina, and 9 failed genotyping, resulting in 149 genotyped AIMs. There were 136 AIMs in common between the final CBCS and SCORE datasets, and these AIMS were used in the analysis.
In addition, for CBCS the custom Illumina SNP panel included many SNPs in breast cancer candidate genes including SNPs in known GWAS hit genes/regions, FGFR2 and 8q24, and for SCORE the custom Illumina SNP panel included many SNPs in prostate cancer candidate genes including SNPs in known GWAS hit genes/regions, MSMB (rs7920517 only), and 8q24. Some SNPs for SCORE were genotyped using Taqman (JAZF1, MSMB). For CBCS all 1946 cases and 1747 controls were genotyped for 11 breast cancer GWAS SNPs. For SCORE, only 597 cases and 322 controls were genotyped for 9 prostate cancer GWAS SNPs (88.8% of the total SCORE sample). For each of these GWAS hit genes/regions, 10/11 SNPs in FGFR2 and 1/1 SNPs in 8q24 for CBCS and 1/1 SNPs in JAZF1, 5/5 SNPs in MSMB and 3/3 SNPs in 8q24 for SCORE passed each study’s respective quality control criteria for successful genotyping and were included for analysis. Further details regarding the genotyping call rates for both datasets have been published elsewhere (Barnholtz-Sloan et al., 2010; Chang et al., 2011).
Statistical analysis
Population stratification for each study population was assessed using a multi-pronged approach using the 136 AIM panel stratified by study site (CBCS or SCORE). First, principal components analysis was used to assess overall similarities of European Americans and Africans Americans by study site via scatter plots of the first principal component (PC1) versus the second principal component (PC2) using R. Second, principal components analysis was used to assess PS by race and case–control status via scatter plots of PC1 versus PC2 using R-based programs. Finally, we conducted a set of analyses using multivariable unconditional logistic regression for top breast cancer GWAS “hits,” in the CBCS case–control data, and for top prostate cancer GWAS “hits,” in the SCORE case–control data, with and without adjustment for PC1 and also for PC1 by GWAS “hit” interaction effects, generating odds ratios (OR) and 95% confidence intervals (95% CI) using R-based programs and SAS version 9.2.
Results
We first compared the allele frequencies for the tested GWAS “hits” in the CBCS and SCORE studies by race to the HapMap African Yorubans (YRI; as compared to African Americans) and CEPH Europeans (CEU; as compared to European Americans; Table 1). For CBCS African Americans, allele frequencies for six of the SNPs varied from YRI by more than 5%, while for CBCS European Americans, none of the allele frequencies varied from CEU by more than 5%. For SCORE African Americans, allele frequencies for six of the SNPs varied from YRI by more than 5%, while for SCORE European Americans, allele frequencies for three of the SNPs varied from CEU by more than 5%.
Table 1.
Carolina Breast Cancer Study and SCORE GWAS “hit” allele frequencies as compared to HapMap CEPH Europeans (CEU) and African Yorubans (YRI).
| SNP | Gene or region | Risk allele | YRI allele frequency | CEU allele frequency | CBCS allele frequency | SCORE allele frequency | |||
|---|---|---|---|---|---|---|---|---|---|
| African American | European American | African American | European American | ||||||
| CBCS | rs2981582 | FGFR2 | C | 0.49 | 0.54 | 0.53 | 0.59 | ||
| rs1219648 | A | 0.54 | 0.54 | 0.57 | 0.59 | ||||
| rs2420946 | C | 0.40 | 0.54 | 0.48 | 0.59 | ||||
| rs11200014 | G | 0.76 | 0.53 | 0.80 | 0.57 | ||||
| rs2981579 | C | 0.33 | 0.54 | 0.40 | 0.57 | ||||
| rs2162540 | A | 0.43 | 0.55 | 0.49 | 0.59 | ||||
| rs3135718 | A | 0.37 | 0.57 | 0.44 | 0.59 | ||||
| rs2936870 | C | 0.36 | 0.58 | 0.43 | 0.58 | ||||
| rs2912774 | C | 0.38 | 0.54 | 0.44 | 0.58 | ||||
| rs1896395 | C | 0.75 | 1.00 | 0.80 | 0.99 | ||||
| rs13281615 | 8q24 | A | 0.57 | 0.54 | 0.56 | 0.58 | |||
| SCORE | rs1447295 | 8q24 | C | 0.61 | 0.93 | 0.70 | 0.99 | ||
| rs4242384 | A | 0.82 | 0.93 | 0.84 | 0.90 | ||||
| rs16901979 | C | 0.46 | 0.98 | 0.55 | 0.95 | ||||
| rs10486567 | JAZF1 | C | 0.10 | 0.75 | 0.73 | 0.78 | |||
| rs7920517 | MSMB | G | 0.19 | 0.42 | 0.72 | 0.51 | |||
| rs10993994 | T | 0.29 | 0.34 | 0.63 | 0.45 | ||||
| rs7904463 | C | 0.27 | 0.62 | 0.33 | 0.66 | ||||
| rs10740051 | G | 0.94 | 0.66 | 0.89 | 0.70 | ||||
| rs10826223 | G | 0.86 | 0.91 | 0.88 | 0.92 | ||||
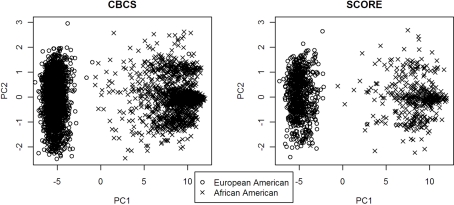
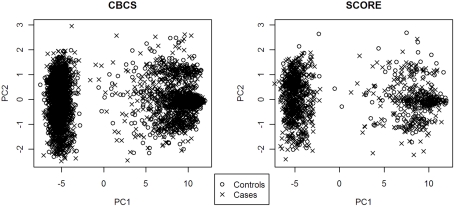
PC1 explained 53.7% of the variance in ancestry for CBCS and 49.5% of the variance in ancestry for SCORE. As seen with the joint analysis, the remaining PCs each accounted for <1% of the variance for each study site individually. Figure 1 shows the ancestral structure for CBCS and SCORE separately by race via principal components analysis. PC1 in both samples can be interpreted as the African axis of ancestral variation. Both panels of Figure 1 showed that in this joint analysis, European Americans and African Americans were similar in terms of their ancestral structure between CBCS and SCORE. Figure 2 shows the ancestral structure for CBCS and SCORE separately by case–control status via principal component analysis, showing that cases and controls seemed to be fairly well matched by ancestry.
Figure 1.
Scatter plots of PC1 vs. PC2 for CBCS and SCORE.
Figure 2.
Scatter plots of PC1 vs. PC2 for CBCS and SCORE by case–control status.
Eleven breast cancer GWAS “hits” were tested in CBCS overall and stratified by race with and without PS adjustment (Table 2). Overall CBCS results are shown Table A1 in Appendix. In the CBCS European Americans, nine of 11 GWAS “hits” were significant after adjustment for PC1 only, but after adjustment for the PC1 by SNP interaction effect, only one SNP remained significant [rs1219648 in FGFR2; OR = 1.93, 95% CI (1.12,3.36)]; for CBCS African Americans, six of 11 GWAS “hits” were significant after adjustment for PC1 only and after adjustment for the PC1 by SNP interaction effect, all six SNPs remained significant (although the p-value for significance changed from 0.003 to 0.02–0.04 for some SNPs), and an additional SNP now became significant. Interestingly, only six of the nine SNPs significant in European Americans after adjustment for PC1 were significant in African Americans; there was no overlap by race for the GWAS “hits” that were statistically significant after adjustment for the PC1 by SNP interaction effect. Although in general for CBCS the p-values for the PC1 by SNP interaction effects were all non-significant, except one SNP for African Americans (rs13281615; p-value for interaction = 0.01).
Table 2.
Assessment of population stratification adjustment on GWAS hit association results for CBCS stratified by race.
| Race | SNP | Gene or region | Risk allele | N cases | N controls | Age and offset | Age, PC1, and offset | Age, PC1, PC1*SNP, and offset | p-Value for PC1*SNP interaction | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | OR (95% CI) | p-Value | |||||||
| European American | rs2981582 | FGFR2 | C | 1204 | 1089 | 1.31 (1.15,1.48) | <0.0001 | 1.31 (1.15,1.48) | <0.0001 | 1.36 (0.77,2.40) | 0.29 | 0.88 |
| rs1219648 | A | 1203 | 1089 | 1.31 (1.15,1.48) | <0.0001 | 1.31 (1.16,1.48) | <0.0001 | 1.93 (1.12,3.36) | 0.02 | 0.15 | ||
| rs2420946 | C | 1202 | 1086 | 1.30 (1.15,1.37) | <0.0001 | 1.30 (1.15,1.48) | <0.0001 | 1.54 (0.96,2.49) | 0.07 | 0.47 | ||
| rs11200014 | G | 1204 | 1089 | 1.31 (1.15,1.48) | <0.0001 | 1.31 (1.15,1.48) | <0.0001 | 0.97 (0.58,1.62) | 0.90 | 0.24 | ||
| rs2981579 | C | 1204 | 1089 | 1.33 (1.18,1.51) | <0.0001 | 1.33 (1.18,1.51) | <0.0001 | 1.22 (0.79,1.90) | 0.37 | 0.70 | ||
| rs2162540 | A | 1202 | 1089 | 1.31 (1.15,1.48) | <0.0001 | 1.31 (1.15,1.48) | <0.0001 | 1.56 (0.96,2.51) | 0.07 | 0.46 | ||
| rs3135718 | A | 1204 | 1089 | 1.31 (1.15,1.48) | <0.0001 | 1.31 (1.16,1.48) | <0.0001 | 1.56 (0.97,2.53) | 0.07 | 0.45 | ||
| rs2936870 | C | 1204 | 1089 | 1.30 (1.15,1.47) | <0.0001 | 1.30 (1.15,1.47) | <0.0001 | 1.59 (0.98,2.57) | 0.06 | 0.40 | ||
| rs2912774 | C | 1202 | 1087 | 1.30 (1.15,1.47) | <0.0001 | 1.30 (1.15,1.47) | <0.0001 | 1.60 (0.99,2.59) | 0.06 | 0.38 | ||
| rs1896395 | C | 1204 | 1089 | 1.86 (0.53,6.48) | 0.33 | 2.07 (0.59,7.20) | 0.25 | 2.53 (0.18,36.03) | 0.49 | 0.51 | ||
| rs13281615 | 8q24 | A | 1198 | 1088 | 1.11 (0.98,1.26) | 0.09 | 1.11 (0.98,1.26) | 0.09 | 1.16 (0.64,2.10) | 0.63 | 0.89 | |
| African American | rs2981582 | FGFR2 | C | 742 | 658 | 1.19 (1.02,1.39) | 0.03 | 1.19 (1.02,1.39) | 0.03 | 1.64 (0.97,2.76) | 0.06 | 0.21 |
| rs1219648 | A | 742 | 658 | 1.19 (1.02,1.39) | 0.03 | 1.19 (1.02,1.39) | 0.03 | 1.62 (0.99,2.66) | 0.06 | 0.20 | ||
| rs2420946 | C | 742 | 658 | 1.17 (1.00,1.37) | 0.05 | 1.17 (1.00,1.37) | 0.05 | 1.84 (1.11,3.05) | 0.02 | 0.06 | ||
| rs11200014 | G | 742 | 658 | 1.04 (0.86,1.26) | 0.69 | 1.05 (0.86,1.27) | 0.64 | 1.16 (0.71,1.91) | 0.55 | 0.65 | ||
| rs2981579 | C | 742 | 658 | 1.22 (1.04,1.42) | 0.01 | 1.22 (1.04,1.42) | 0.01 | 1.84 (1.10,3.07) | 0.02 | 0.09 | ||
| rs2162540 | A | 742 | 657 | 1.23 (1.05,1.44) | 0.01 | 1.23 (1.05,1.44) | 0.01 | 1.93 (1.16,3.22) | 0.01 | 0.06 | ||
| rs3135718 | A | 742 | 656 | 1.26 (1.08,1.46) | 0.003 | 1.26 (1.08,1.46) | 0.003 | 1.73 (1.04,2.89) | 0.04 | 0.20 | ||
| rs2936870 | C | 741 | 658 | 1.27 (1.09,1.48) | 0.003 | 1.27 (1.08,1.48) | 0.003 | 1.82 (1.08,3.08) | 0.02 | 0.15 | ||
| rs2912774 | C | 742 | 657 | 1.27 (1.09,1.49) | 0.003 | 1.27 (1.09,1.49) | 0.003 | 1.86 (1.10,3.14) | 0.02 | 0.14 | ||
| rs1896395 | C | 742 | 658 | 1.01 (0.84,1.23) | 0.89 | 1.01 (0.83,1.23) | 0.93 | 1.18 (0.50,2.78) | 0.71 | 0.72 | ||
| rs13281615 | 8q24 | A | 736 | 658 | 1.00 (0.86,1.18) | 0.97 | 1.00 (0.86,1.18) | 0.96 | 0.50 (0.28,0.88) | 0.02 | 0.01 | |
Nine prostate cancer GWAS “hits” were tested in SCORE overall and stratified by race with and without PS adjustment (Table 3). Overall SCORE results are shown Table A1 in Appendix. In the SCORE European Americans, zero out of nine GWAS “hits” were significant after adjustment for PC1 only and no changes were seen after additional adjustment for the PC1 by SNP interaction effect; for SCORE African Americans, two of the nine GWAS “hits” were significant after adjustment for PC1 only and after adjustment for the PC1 by SNP interaction effect, only one SNP remained significant [rs16901979 at 8q24; OR = 3.24, 95% CI (1.19,8.78)]. Although in general for SCORE the p-values for the PC1 by SNP interaction effects were all non-significant.
Table 3.
Assessment of population stratification adjustment on GWAS hit association results for SCORE stratified by race.
| Race | SNP | Gene or region | Risk allele | N cases | N controls | Age | Age and PC1 | Age, PC1, and PC1*SNP | p-Value for PC1*SNP interaction | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | OR (95% CI) | p-Value | |||||||
| European American | rs1447295 | 8q24 | C | 575 | 127 | 1.46 (0.88,2.46) | 0.14 | 1.47 (0.89,2.42) | 0.14 | 1.94 (0.53,7.09) | 0.31 | 0.63 |
| rs4242384 | A | 576 | 127 | 1.38 (0.84,2.27) | 0.20 | 1.37 (0.83,2.25) | 0.22 | 2.07 (0.32,13.25) | 0.44 | 0.65 | ||
| rs16901979 | C | 575 | 127 | 1.56 (0.76,3.18) | 0.22 | 1.76 (0.85,3.64) | 0.13 | 3.31 (0.67,16.46) | 0.14 | 0.32 | ||
| rs10486567 | JAZF1 | C | 572 | 127 | 1.10 (0.78,1.55) | 0.60 | 1.10 (0.78,1.55) | 0.59 | 1.49 (0.35,6.27) | 0.59 | 0.70 | |
| rs7920517 | MSMB | G | 577 | 127 | 0.84 (0.64,1.10) | 0.20 | 0.83 (0.63,1.09) | 0.17 | 0.73 (0.22,2.42) | 0.61 | 0.84 | |
| rs10993994 | T | 571 | 124 | 0.79 (0.60,1.04) | 0.10 | 0.79 (0.60,1.05) | 0.10 | 1.50 (0.41,5.45) | 0.54 | 0.33 | ||
| rs7904463 | C | 567 | 125 | 0.89 (0.66,1.20) | 0.46 | 0.90 (0.67,1.21) | 0.49 | 0.46 (0.09,2.38) | 0.36 | 0.42 | ||
| rs10740051 | G | 572 | 125 | 0.84 (0.62,1.12) | 0.23 | 0.83 (0.62,1.12) | 0.23 | 0.38 (0.09,1.55) | 0.18 | 0.26 | ||
| rs10826223 | G | 567 | 127 | 1.38 (0.80,2.38) | 0.25 | 1.39 (0.80,2.41) | 0.24 | 2.85 (0.44,18.41) | 0.27 | 0.42 | ||
| African American | rs1447295 | 8q24 | C | 215 | 195 | 1.27 (0.93,1.75) | 0.14 | 1.28 (0.93,1.76) | 0.13 | 0.94 (0.36,2.47) | 0.90 | 0.51 |
| rs4242384 | A | 216 | 195 | 1.32 (0.901.92) | 0.15 | 1.32 (0.90,1.92) | 0.15 | 1.48 (0.45,4.87) | 0.52 | 0.84 | ||
| rs16901979 | C | 214 | 195 | 1.39 (1.06,1.83) | 0.02 | 1.44 (1.08,1.91) | 0.01 | 3.24 (1.19,8.78) | 0.02 | 0.09 | ||
| rs10486567 | JAZF1 | C | 203 | 188 | 0.80 (0.58,1.09) | 0.16 | 0.80 (0.58,1.10) | 0.17 | 1.24 (0.46,3.37) | 0.67 | 0.36 | |
| rs7920517 | MSMB | G | 216 | 195 | 0.70 (0.51,0.95) | 0.02 | 0.69 (0.50,0.94) | 0.02 | 0.52 (0.24,1.14) | 0.10 | 0.44 | |
| rs10993994 | T | 207 | 191 | 0.84 (0.63,1.12) | 0.23 | 0.84 (0.63,1.11) | 0.22 | 0.54 (0.26,1.11) | 0.09 | 0.19 | ||
| rs7904463 | C | 206 | 191 | 1.04 (0.79,1.37) | 0.78 | 1.05 (0.79,1.40) | 0.73 | 1.93 (0.88,4.25) | 0.10 | 0.10 | ||
| rs10740051 | G | 208 | 190 | 1.18 (0.75,1.85) | 0.48 | 1.17 (0.74,1.86) | 0.50 | 0.79 (0.33,1.91) | 0.60 | 0.30 | ||
| rs10826223 | G | 202 | 188 | 1.24 (0.79,1.95) | 0.35 | 1.24 (0.79,1.96) | 0.34 | 1.13 (0.27,4.62) | 0.87 | 0.88 | ||
Discussion
We investigated the importance of adjustment for PS effects using principal components analysis on two large epidemiologic datasets of breast and prostate cancer that include both European and African Americans. In addition, we show the importance of additionally adjusting for a potential SNP by PS interaction effect. Our results show that PS, in particular PS by GWAS “hit” interaction effects, can greatly change the significance of these GWAS “hits” by racial group. We also show that the first principal component explains the majority of the ancestral variation by race in each study population; for CBCS PC1 explained 53.7% of the variance and for SCORE PC1 explained 49.5% of the variance and the remaining PCs each accounted for <1% of the variance for each study site. In a completely random matrix with no structure, having 136 dimensions (i.e., 136 AIM SNPs), each PC would account for ∼0.7% of the variance, which is what we see for all PC’s beyond PC1 for the joint analysis and for each specific study site individually. PS can cause both false positive and false negative associations in epidemiologic studies of individuals with mixed ancestry. There are now multiple examples in the literature about the importance of adjustment for PS and how PS can affect study inference in recently admixed populations such as African Americans or Hispanic Americans (e.g., Kittles et al., 2002; Ziv et al., 2006).
Multiple GWAS have now been performed for many complex diseases including breast and prostate cancers. The vast majority of these studies have been performed in individuals of European ancestry only; breast cancer (Easton et al., 2007; Hunter et al., 2007; Stacey et al., 2007; Gold et al., 2008; Ahmed et al., 2009; Thomas et al., 2009; Zheng et al., 2009c) and prostate cancer (Amundadottir et al., 2006; Freedman et al., 2006; Duggan et al., 2007; Gudmundsson et al., 2007, 2009; Haiman et al., 2007; Yeager et al., 2007, 2009; Sun et al., 2008; Thomas et al., 2008; Al Olama et al., 2009; Eeles et al., 2009; Hsu et al., 2009; Lou et al., 2009; Zheng et al., 2009a). We showed in this study that association statistics for different racial groups can be impacted when adjustment for PS and an interaction between PS and the SNP of interest is performed. Further replication studies of the GWAS “hits” included in this analysis and others have been performed in African Americans with breast cancer (Zheng et al., 2009b; Barnholtz-Sloan et al., 2010) and prostate cancer (Robbins et al., 2007; Hooker et al., 2009; Waters et al., 2009; Xu et al., 2009; Chang et al., 2011; Table 4). One of these studies was based on the CBCS population (Barnholtz-Sloan et al., 2010) and another included the SCORE population as one of many studies used for analysis (Chang et al., 2011). Although these studies used different AIMs panels and different techniques for ancestry/PS estimation, all studies showed that PS adjustment did affect the magnitude of association statistics and was therefore a necessary adjustment factor. Additionally, Chang et al. (2011) showed that for those study sites included in their analysis that had available PS ancestry information that the average ancestry varied significantly by study site. Hence, they concluded that adjustment for study site would also serve as a partial proxy for PS adjustment.
Table 4.
Comparison of common GWAS “hit” results with previously published results in African Americans with breast or prostate cancer.
| Cancer type | SNP | Gene or region tested in the current study | Study (odds ratio with 95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Zheng et al. (2009b)* | Barnholtz-Sloan et al. (2010)* | Robbins et al. (2007)# | Waters et al. (2009)# | Xu et al. (2009)# | Hooker et al. (2009)# | Chang et al. (2011)# | |||
| Breast | rs2981582 | FGFR2 | 1.03 (0.91,1.18) | 1.19 (1.02,1.39) | |||||
| rs1219648 | 1.21 (1.06,1.37) | 1.19 (1.02,1.39) | |||||||
| rs2420946 | 1.10 (0.97,1.25) | 1.17 (1.00,1.37) | |||||||
| rs11200014 | 1.04 (0.86,1.26) | ||||||||
| rs2981579 | 1.08 (0.95,1.23) | 1.22 (1.04,1.42) | |||||||
| rs2162540 | 1.23 (1.05,1.44) | ||||||||
| rs3135718 | 1.26 (1.08,1.46) | ||||||||
| rs2936870 | 1.27 (1.09,1.48) | ||||||||
| rs2912774 | 1.27 (1.09,1.49) | ||||||||
| rs1896395 | 1.01 (0.83,1.23) | ||||||||
| rs13281615 | 8q24 | 1.02 (0.91,1.15) | 1.00 (0.86,1.18) | ||||||
| Prostate | rs1447295 | 8q24 | 1.40 (0.70,1.30) | 1.06 (0.91,1.23) | |||||
| rs4242384 | 1.23 (0.95,1.56) | ||||||||
| rs16901979 | 1.50 (1.10,2.20) | 1.38 (1.19,1.60) | |||||||
| rs10486567 | JAZF1 | 1.18 (1.00,1.40) | 0.91 (0.69,1.18) | 1.18 (1.08,1.29) | |||||
| rs7920517 | MSMB | 1.09 (0.82,1.44) | 1.13 (0.93,1.36) | ||||||
| rs10993994 | 1.05 (0.90,1.24) | 1.11 (0.96,1.28) | 0.95 (0.74,1.21) | 1.12 (1.03,1.21) | |||||
| rs7904463 | 1.13 (0.92,1.39) | ||||||||
| rs10740051 | |||||||||
| rs10826223 | |||||||||
*Zheng et al. (2009b) adjusted for age, education, study site, and PC1; Barnholtz-Sloan et al. (2010) was adjusted for age, offset, and European ancestry proportion.
#Robbins et al. (2007) adjusted for age and West African ancestry proportion; Waters et al. (2009) adjusted for age, European ancestry proportion, and age-ethnicity strata; Xu et al. (2009) adjusted for age, STRUCTURE ancestry proportion and study population; Hooker et al. (2009) adjusted for age and West African ancestry proportion; Chang et al. (2011) adjusted for age and study site.
In this study we show that the additional adjustment for a PS by SNP interaction effect changes the magnitude and significance of most association statistics in both racial groups studied, particularly for the CBCS study, although the p-values for this interaction were non-significant. For CBCS European Americans, all but one SNP were non-significant after adjustment for the PS by SNP interaction effect where the magnitude of the interaction OR was much higher than previously reported in other studies at 1.93. For the six of the seven SNPs that remained significant in CBCS African Americans after adjustment for the PS by SNP interaction effect their association statistics corroborated with previously published studies in African Americans (Table 4) in terms of their direction and significance of effect, however the magnitude of the ORs was much higher. Interestingly, in previous studies rs13281615 has been shown to be non-significant in African Americans and in this study showed a significant protective effect for breast cancer development after interaction adjustment. In the SCORE study none of the SNPs were significant in European Americans; while in African Americans only one SNP remained significant after PS by SNP interaction effect adjustment (rs16901979). Interestingly, in previous prostate cancer case–control studies of African Americans, rs16901979 was the only SNP that showed a consistent significant effect in multiple studies (Table 4).
The SNPs used in this analysis were all highly selected given they had been previously shown to be associated with breast or prostate cancer in previous GWAS studies, irrespective of this fact, this analysis showed that PS can cause inflation and deflation of the significance of association statistics. Additionally, some of the affects on association statistics after adjustment for PS or for the PS by SNP interaction effect could have also been due to issues related to study design, since cases and controls cannot be perfectly matched for allele frequency within each racial group. We did not perform haplotype analyses in this study as out goal was limited to single SNP replication of GWAS hits previously reported in European and African Americans; however haplotype analyses could have been informative to show further differences in genetic associations by race. There may be other relevant SNPs within these loci that are associated with breast or prostate cancer, particularly for different racial groups that were not examined. In addition, our sample size was relatively small for African Americans, particularly in the SCORE study. We also realize that there are many additional factors that may potentially influence a GWAS replication study that were not adjusted for in this analysis; we adjusted for the same factors as previous studies in order to replicate previous findings. Future research will require pooling of data from different breast and prostate cancer studies for different racial groups in order to gain a more complete understanding of differences in risk alleles by race and in order to study gene–gene and gene–environment interaction.
In conclusion, we demonstrated that genetic associations by race are modified by interactions between individual SNPs and PS and that significance of particular GWAS “hits” is not the same between racial groups. Our results and results of previously published studies in African Americans as shown in Table 4 highlight the need to conduct GWAS and GWAS replication studies in a variety of racial groups.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all participants in the CBCS and SCORE studies. We also thank Priya Shetty, Yanwen Chen, PhD, MS, Lynette Phillips, PhD for their technical assistance. This work was supported by the Case Comprehensive Cancer Center Core Grant (NIH/NCI P30-CA043703 to Jill S. Barnholtz-Sloan), the Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223 to Robert C. Millikan); Center for Environmental Health and Susceptibility (NIEHS P30-ES10126 to Robert C. Millikan); Lineberger Comprehensive Cancer Center Core Grant (NIH/NCI P30-CA16086 to Robert C. Millikan); NIH/NCI grants R01-CA085074 and P50-CA105641 (both to Timothy R. Rebbeck).
Appendix
Table A1.
Assessment of population stratification adjustment on GWAS hit association results for CBCS and SCORE.
| Gene or region | Risk allele | N cases | N controls | Age and race | Age, race, and PC1 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Dataset | SNP | OR (95% CI) | p-Value | OR (95% CI) | p-Value | ||||
| CBCS* | rs2981582 | FGFR2 | C | 1946 | 1747 | 1.26 (1.14,139) | <0.0001 | 1.26 (1.14,1.39) | <0.0001 |
| rs1219648 | A | 1945 | 1747 | 1.26 (1.15,1.39) | <0.0001 | 1.26 (1.15,1.39) | <0.0001 | ||
| rs2420946 | C | 1944 | 1744 | 1.25 (1.13,1.38) | <0.0001 | 1.25 (1.13,1.38) | <0.0001 | ||
| rs11200014 | G | 1946 | 1747 | 1.22 (1.10,1.36) | <0.0001 | 1.23 (1.10,1.36) | 0.0001 | ||
| rs2981579 | C | 1946 | 1747 | 1.29 (1.17,1.42) | <0.0001 | 1.29 (1.17,1.42) | <0.0001 | ||
| rs2162540 | A | 1944 | 1746 | 1.28 (1.16,1.41) | <0.0001 | 1.28 (1.16,1.41) | <0.0001 | ||
| rs3135718 | A | 1946 | 1745 | 1.29 (1.17,1.42) | <0.0001 | 1.29 (1.17,1.42) | <0.0001 | ||
| rs2936870 | C | 1945 | 1747 | 1.29 (1.17,1.42) | <0.0001 | 1.29 (1.17,1.42) | <0.0001 | ||
| rs2912774 | C | 1944 | 1744 | 1.29 (1.17,1.42) | <0.0001 | 1.29 (1.17,1.42) | <0.0001 | ||
| rs1896395 | C | 1946 | 1747 | 1.03 (0.85,1.24) | 0.77 | 1.03 (0.85,1.25) | 0.77 | ||
| rs13281615 | 8q24 | A | 1934 | 1746 | 1.07 (0.97,1.18) | 0.19 | 1.07 (0.97,1.18) | 0.19 | |
| SCORE | rs1447295 | 8q24 | C | 790 | 322 | 1.33 (1.02,1.73) | 0.04 | 1.33 (1.02,1.74) | 0.03 |
| rs4242384 | A | 792 | 322 | 1.34 (0.99,1.81) | 0.06 | 1.34 (0.99,1.81) | 0.06 | ||
| rs16901979 | C | 789 | 322 | 1.42 (1.10,1.82) | 0.007 | 1.49 (1.14,1.94) | 0.003 | ||
| rs10486567 | JAZF1 | C | 775 | 315 | 0.92 (0.73,1.16) | 0.50 | 0.93 (0.74,1.17) | 0.52 | |
| rs7920517 | MSMB | G | 793 | 322 | 0.77 (0.63,0.94) | 0.01 | 0.76 (0.62,0.94) | 0.01 | |
| rs10993994 | T | 778 | 315 | 0.81 (0.67,0.99) | 0.04 | 0.81 (0.66,0.99) | 0.04 | ||
| rs7904463 | C | 773 | 316 | 0.97 (0.79,1.19) | 0.76 | 0.98 (0.80,1.20) | 0.83 | ||
| rs10740051 | G | 780 | 315 | 0.92 (0.72,1.18) | 0.53 | 0.91 (0.71,1.17) | 0.48 | ||
| rs10826223 | G | 769 | 315 | 1.29 (0.91,1.82) | 0.15 | 1.29 (0.91,1.83) | 0.14 | ||
*Additionally adjusted for a study design offset term.
References
- Ahmed S., Thomas G., Ghoussaini M., Healey C. S., Humphreys M. K., Platte R., Morrison J., Maranian M., Pooley K. A., Luben R., Eccles D., Evans D. G., Fletcher O., Johnson N., Dos Santos Silva I., Peto J., Stratton M. R., Rahman N., Jacobs K., Prentice R., Anderson G. L., Rajkovic A., Curb J. D., Ziegler R. G., Berg C. D., Buys S. S., Mccarty C. A., Feigelson H. S., Calle E. E., Thun M. J., Diver W. R., Bojesen S., Nordestgaard B. G., Flyger H., Dork T., Schurmann P., Hillemanns P., Karstens J. H., Bogdanova N. V., Antonenkova N. N., Zalutsky I. V., Bermisheva M., Fedorova S., Khusnutdinova E., Search, Kang D., Yoo K. Y., Noh D. Y., Ahn S. H., Devilee P., Van Asperen C. J., Tollenaar R. A., Seynaeve C., Garcia-Closas M., Lissowska J., Brinton L., Peplonska B., Nevanlinna H., Heikkinen T., Aittomaki K., Blomqvist C., Hopper J. L., Southey M. C., Smith L., Spurdle A. B., Schmidt M. K., Broeks A., Van Hien R. R., Cornelissen S., Milne R. L., Ribas G., Gonzalez-Neira A., Benitez J., Schmutzler R. K., Burwinkel B., Bartram C. R., Meindl A., Brauch H., Justenhoven C., Hamann U., Consortium G., Chang-Claude J., Hein R., Wang-Gohrke S., Lindblom A., Margolin S., Mannermaa A., Kosma V. M., Kataja V., Olson J. E., Wang X., Fredericksen Z., Giles G. G., Severi G., Baglietto L., English D. R., Hankinson S. E., Cox D. G., Kraft P., Vatten L. J., Hveem K., Kumle M., Sigurdson A., Doody M., Bhatti P., Alexander B. H., Hooning M. J., van den Ouweland A. M., Oldenburg R. A., Schutte M., Hall P., Czene K., Liu J., Li Y., Cox A., Elliott G., Brock I., Reed M. W., Shen C. Y., Yu J. C., Hsu G. C., Chen S. T., Anton-Culver H., Ziogas A., Andrulis I. L., Knight J. A., kConFab, Australian Ovarian Cancer Study Group. Beesley J., Goode E. L., Couch F., Chenevix-Trench G., Hoover R. N., Ponder B. A., Hunter D. J., Pharoah P. D., Dunning A. M., Chanock S. J., Easton D. F. (2009). Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 41, 585–590 10.1038/ng.354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama A. A., Kote-Jarai Z., Giles G. G., Guy M., Morrison J., Severi G., Leongamornlert D. A., Tymrakiewicz M., Jhavar S., Saunders E., Hopper J. L., Southey M. C., Muir K. R., English D. R., Dearnaley D. P., Ardern-Jones A. T., Hall A. L., O’brien L. T., Wilkinson R. A., Sawyer E., Lophatananon A., Oncology, U. K. G. P. C. S. C. B. A. O. U. S. S. O., Cancer, U. K. P. T. F. Treatment Study C., Horwich A., Huddart R. A., Khoo V. S., Parker C. C., Woodhouse C. J., Thompson A., Christmas T., Ogden C., Cooper C., Donovan J. L., Hamdy F. C., Neal D. E., Eeles R. A., Easton D. F. (2009). Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 41, 1058–1060 10.1038/ng.452 [DOI] [PubMed] [Google Scholar]
- Amundadottir L. T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B. A., Sigurdsson A., Benediktsdottir K. R., Cazier J. B., Sainz J., Jakobsdottir M., Kostic J., Magnusdottir D. N., Ghosh S., Agnarsson K., Birgisdottir B., Le Roux L., Olafsdottir A., Blondal T., Andresdottir M., Gretarsdottir O. S., Bergthorsson J. T., Gudbjartsson D., Gylfason A., Thorleifsson G., Manolescu A., Kristjansson K., Geirsson G., Isaksson H., Douglas J., Johansson J. E., Balter K., Wiklund F., Montie J. E., Yu X., Suarez B. K., Ober C., Cooney K. A., Gronberg H., Catalona W. J., Einarsson G. V., Barkardottir R. B., Gulcher J. R., Kong A., Thorsteinsdottir U., Stefansson K. (2006). A common variant associated with prostate cancer in European and African populations. Nat. Genet. 38, 652–658 10.1038/ng1808 [DOI] [PubMed] [Google Scholar]
- Barnholtz-Sloan J. S., Mcevoy B., Shriver M. D., Rebbeck T. R. (2008). Ancestry estimation and correction for population stratification in molecular epidemiologic association studies. Cancer Epidemiol. Biomarkers Prev. 17, 471–477 10.1158/1055-9965.EPI-07-0491 [DOI] [PubMed] [Google Scholar]
- Barnholtz-Sloan J. S., Shetty P. B., Guan X., Nyante S. J., Luo J., Brennan D. J., Millikan R. C. (2010). FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis 31, 1417–1423 10.1093/carcin/bgq128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. L., Spangler E., Gallagher S., Haiman C. A., Henderson B., Isaacs W., Benford M. L., Kidd L. R., Cooney K., Strom S., Ingles S. A., Stern M. C., Corral R., Joshi A. D., Xu J., Giri V. N., Rybicki B., Neslund-Dudas C., Kibel A. S., Thompson I. M., Leach R. J., Ostrander E. A., Stanford J. L., Witte J., Casey G., Eeles R., Hsing A. W., Chanock S., Hu J. J., John E. M., Park J., Stefflova K., Zeigler-Johnson C., Rebbeck T. R. (2011). Validation of genome-wide prostate cancer associations in men of African descent. Cancer Epidemiol. Biomarkers Prev. 20, 23–32 10.1158/1055-9965.EPI-11-0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry S., Coyle N. E., Tang H., Salari K., Lind D., Clark S. L., Tsai H. J., Naqvi M., Phong A., Ung N., Matallana H., Avila P. C., Casal J., Torres A., Nazario S., Castro R., Battle N. C., Perez-Stable E. J., Kwok P. Y., Sheppard D., Shriver M. D., Rodriguez-Cintron W., Risch N., Ziv E., Burchard E. G. (2006). Population stratification confounds genetic association studies among Latinos. Hum. Genet. 118, 652–664 10.1007/s00439-005-0071-3 [DOI] [PubMed] [Google Scholar]
- Duggan D., Zheng S. L., Knowlton M., Benitez D., Dimitrov L., Wiklund F., Robbins C., Isaacs S. D., Cheng Y., Li G., Sun J., Chang B. L., Marovich L., Wiley K. E., Balter K., Stattin P., Adami H. O., Gielzak M., Yan G., Sauvageot J., Liu W., Kim J. W., Bleecker E. R., Meyers D. A., Trock B. J., Partin A. W., Walsh P. C., Isaacs W. B., Gronberg H., Xu J., Carpten J. D. (2007). Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J. Natl. Cancer Inst. 99, 1836–1844 10.1093/jnci/djm250 [DOI] [PubMed] [Google Scholar]
- Easton D. F., Pooley K. A., Dunning A. M., Pharoah P. D., Thompson D., Ballinger D. G., Struewing J. P., Morrison J., Field H., Luben R., Wareham N., Ahmed S., Healey C. S., Bowman R., Collaborators S., Meyer K. B., Haiman C. A., Kolonel L. K., Henderson B. E., Le Marchand L., Brennan P., Sangrajrang S., Gaborieau V., Odefrey F., Shen C. Y., Wu P. E., Wang H. C., Eccles D., Evans D. G., Peto J., Fletcher O., Johnson N., Seal S., Stratton M. R., Rahman N., Chenevix-Trench G., Bojesen S. E., Nordestgaard B. G., Axelsson C. K., Garcia-Closas M., Brinton L., Chanock S., Lissowska J., Peplonska B., Nevanlinna H., Fagerholm R., Eerola H., Kang D., Yoo K. Y., Noh D. Y., Ahn S. H., Hunter D. J., Hankinson S. E., Cox D. G., Hall P., Wedren S., Liu J., Low Y. L., Bogdanova N., Schurmann P., Dork T., Tollenaar R. A., Jacobi C. E., Devilee P., Klijn J. G., Sigurdson A. J., Doody M. M., Alexander B. H., Zhang J., Cox A., Brock I. W., Macpherson G., Reed M. W., Couch F. J., Goode E. L., Olson J. E., Meijers-Heijboer H., Van Den Ouweland A., Uitterlinden A., Rivadeneira F., Milne R. L., Ribas G., Gonzalez-Neira A., Benitez J., Hopper J. L., Mccredie M., Southey M., Giles G. G., Schroen C., Justenhoven C., Brauch H., Hamann U., Ko Y. D., Spurdle A. B., Beesley J., Chen X., Kconfab, AOCS Management Group. Mannermaa A., Kosma V. M., Kataja V., Hartikainen J., Day N. E., Cox D. R., Ponder B. A. (2007). Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093 10.1038/nature05887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles R. A., Kote-Jarai Z., Al Olama A. A., Giles G. G., Guy M., Severi G., Muir K., Hopper J. L., Henderson B. E., Haiman C. A., Schleutker J., Hamdy F. C., Neal D. E., Donovan J. L., Stanford J. L., Ostrander E. A., Ingles S. A., John E. M., Thibodeau S. N., Schaid D., Park J. Y., Spurdle A., Clements J., Dickinson J. L., Maier C., Vogel W., Dork T., Rebbeck T. R., Cooney K. A., Cannon-Albright L., Chappuis P. O., Hutter P., Zeegers M., Kaneva R., Zhang H. W., Lu Y. J., Foulkes W. D., English D. R., Leongamornlert D. A., Tymrakiewicz M., Morrison J., Ardern-Jones A. T., Hall A. L., O’brien L. T., Wilkinson R. A., Saunders E. J., Page E. C., Sawyer E. J., Edwards S. M., Dearnaley D. P., Horwich A., Huddart R. A., Khoo V. S., Parker C. C., Van as N., Woodhouse C. J., Thompson A., Christmas T., Ogden C., Cooper C. S., Southey M. C., Lophatananon A., Liu J. F., Kolonel L. N., Le Marchand L., Wahlfors T., Tammela T. L., Auvinen A., Lewis S. J., Cox A., Fitzgerald L. M., Koopmeiners J. S., Karyadi D. M., Kwon E. M., Stern M. C., Corral R., Joshi A. D., Shahabi A., Mcdonnell S. K., Sellers T. A., Pow-Sang J., Chambers S., Aitken J., Gardiner R. A., Batra J., Kedda M. A., Lose F., Polanowski A., Patterson B., Serth J., Meyer A., Luedeke M., Stefflova K., Ray A. M., Lange E. M., Farnham J., Khan H., Slavov C., Mitkova A., Cao G., UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology, UK ProtecT Study Collaborators, PRACTICAL Consortium. Easton D. F. (2009). Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 41, 1116–1121 10.1038/ng.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman M. L., Haiman C. A., Patterson N., Mcdonald G. J., Tandon A., Waliszewska A., Penney K., Steen R. G., Ardlie K., John E. M., Oakley-Girvan I., Whittemore A. S., Cooney K. A., Ingles S. A., Altshuler D., Henderson B. E., Reich D. (2006). Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl. Acad. Sci. U.S.A. 103, 14068–14073 10.1073/pnas.0608127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B., Kirchhoff T., Stefanov S., Lautenberger J., Viale A., Garber J., Friedman E., Narod S., Olshen A. B., Gregersen P., Kosarin K., Olsh A., Bergeron J., Ellis N. A., Klein R. J., Clark A. G., Norton L., Dean M., Boyd J., Offit K. (2008). Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl. Acad. Sci. U.S.A. 105, 4340–4345 10.1073/pnas.0800441105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J., Sulem P., Gudbjartsson D. F., Blondal T., Gylfason A., Agnarsson B. A., Benediktsdottir K. R., Magnusdottir D. N., Orlygsdottir G., Jakobsdottir M., Stacey S. N., Sigurdsson A., Wahlfors T., Tammela T., Breyer J. P., Mcreynolds K. M., Bradley K. M., Saez B., Godino J., Navarrete S., Fuertes F., Murillo L., Polo E., Aben K. K., Van Oort I. M., Suarez B. K., Helfand B. T., Kan D., Zanon C., Frigge M. L., Kristjansson K., Gulcher J. R., Einarsson G. V., Jonsson E., Catalona W. J., Mayordomo J. I., Kiemeney L. A., Smith J. R., Schleutker J., Barkardottir R. B., Kong A., Thorsteinsdottir U., Rafnar T., Stefansson K. (2009). Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 41, 1122–1126 10.1038/ng.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J., Sulem P., Steinthorsdottir V., Bergthorsson J. T., Thorleifsson G., Manolescu A., Rafnar T., Gudbjartsson D., Agnarsson B. A., Baker A., Sigurdsson A., Benediktsdottir K. R., Jakobsdottir M., Blondal T., Stacey S. N., Helgason A., Gunnarsdottir S., Olafsdottir A., Kristinsson K. T., Birgisdottir B., Ghosh S., Thorlacius S., Magnusdottir D., Stefansdottir G., Kristjansson K., Bagger Y., Wilensky R. L., Reilly M. P., Morris A. D., Kimber C. H., Adeyemo A., Chen Y., Zhou J., So W. Y., Tong P. C., Ng M. C., Hansen T., Andersen G., Borch-Johnsen K., Jorgensen T., Tres A., Fuertes F., Ruiz-Echarri M., Asin L., Saez B., Van Boven E., Klaver S., Swinkels D. W., Aben K. K., Graif T., Cashy J., Suarez B. K., Van Vierssen Trip O., Frigge M. L., Ober C., Hofker M. H., Wijmenga C., Christiansen C., Rader D. J., Palmer C. N., Rotimi C., Chan J. C., Pedersen O., Sigurdsson G., Benediktsson R., Jonsson E., Einarsson G. V., Mayordomo J. I., Catalona W. J., Kiemeney L. A., Barkardottir R. B., Gulcher J. R., Thorsteinsdottir U., Kong A., Stefansson K. (2007). Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 39, 977–983 10.1038/ng2062 [DOI] [PubMed] [Google Scholar]
- Haiman C. A., Patterson N., Freedman M. L., Myers S. R., Pike M. C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., Mcdonald G. J., Greenway S. C., Stram D. O., Le Marchand L., Kolonel L. N., Frasco M., Wong D., Pooler L. C., Ardlie K., Oakley-Girvan I., Whittemore A. S., Cooney K. A., John E. M., Ingles S. A., Altshuler D., Henderson B. E., Reich D. (2007). Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 39, 638–644 10.1038/ng2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker S., Hernandez W., Chen H., Robbins C., Torres J. B., Ahaghotu C., Carpten J., Kittles R. A. (2009). Replication of prostate cancer risk loci on 8q24, 11q13, 17q12, 19q33, and Xp11 in African Americans. Prostate 70, 270–275 [DOI] [PubMed] [Google Scholar]
- Hsu F. C., Sun J., Wiklund F., Isaacs S. D., Wiley K. E., Purcell L. D., Gao Z., Stattin P., Zhu Y., Kim S. T., Zhang Z., Liu W., Chang B. L., Walsh P. C., Duggan D., Carpten J. D., Isaacs W. B., Gronberg H., Xu J., Zheng S. L. (2009). A novel prostate cancer susceptibility locus at 19q13. Cancer Res. 69, 2720–2723 10.1158/0008-5472.CAN-08-4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D. J., Kraft P., Jacobs K. B., Cox D. G., Yeager M., Hankinson S. E., Wacholder S., Wang Z., Welch R., Hutchinson A., Wang J., Yu K., Chatterjee N., Orr N., Willett W. C., Colditz G. A., Ziegler R. G., Berg C. D., Buys S. S., Mccarty C. A., Feigelson H. S., Calle E. E., Thun M. J., Hayes R. B., Tucker M., Gerhard D. S., Fraumeni J. F., Jr., Hoover R. N., Thomas G., Chanock S. J. (2007). A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 10.1038/ng2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittles R. A., Chen W., Panguluri R. K., Ahaghotu C., Jackson A., Adebamowo C. A., Griffin R., Williams T., Ukoli F., Adams-Campbell L., Kwagyan J., Isaacs W., Freeman V., Dunston G. M. (2002). CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum. Genet. 110, 553–560 10.1007/s00439-002-0731-5 [DOI] [PubMed] [Google Scholar]
- Lou H., Yeager M., Li H., Bosquet J. G., Hayes R. B., Orr N., Yu K., Hutchinson A., Jacobs K. B., Kraft P., Wacholder S., Chatterjee N., Feigelson H. S., Thun M. J., Diver W. R., Albanes D., Virtamo J., Weinstein S., Ma J., Gaziano J. M., Stampfer M., Schumacher F. R., Giovannucci E., Cancel-Tassin G., Cussenot O., Valeri A., Andriole G. L., Crawford E. D., Anderson S. K., Tucker M., Hoover R. N., Fraumeni J. F., Jr., Thomas G., Hunter D. J., Dean M., Chanock S. J. (2009). Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. Proc. Natl. Acad. Sci. U.S.A. 106, 7933–7938 10.1073/pnas.0813135106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan R., Eaton A., Worley K., Biscocho L., Hodgson E., Huang W. Y., Geradts J., Iacocca M., Cowan D., Conway K., Dressler L. (2003). HER2 codon 655 polymorphism and risk of breast cancer in African Americans and whites. Breast Cancer Res. Treat. 79, 355–364 10.1023/A:1024068525763 [DOI] [PubMed] [Google Scholar]
- Pfaff C. L., Barnholtz-Sloan J., Wagner J. K., Long J. C. (2004). Information on ancestry from genetic markers. Genet. Epidemiol. 26, 305–315 10.1002/gepi.10319 [DOI] [PubMed] [Google Scholar]
- Risch N., Burchard E., Ziv E., Tang H. (2002). Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 3, 1–12 10.1186/gb-2002-3-7-comment2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins C., Torres J. B., Hooker S., Bonilla C., Hernandez W., Candreva A., Ahaghotu C., Kittles R., Carpten J. (2007). Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 17, 1717–1722 10.1101/gr.6782707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey S. N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S. A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., Aben K. K., Strobbe L. J., Albers-Akkers M. T., Swinkels D. W., Henderson B. E., Kolonel L. N., Le Marchand L., Millastre E., Andres R., Godino J., Garcia-Prats M. D., Polo E., Tres A., Mouy M., Saemundsdottir J., Backman V. M., Gudmundsson L., Kristjansson K., Bergthorsson J. T., Kostic J., Frigge M. L., Geller F., Gudbjartsson D., Sigurdsson H., Jonsdottir T., Hrafnkelsson J., Johannsson J., Sveinsson T., Myrdal G., Grimsson H. N., Jonsson T., Von Holst S., Werelius B., Margolin S., Lindblom A., Mayordomo J. I., Haiman C. A., Kiemeney L. A., Johannsson O. T., Gulcher J. R., Thorsteinsdottir U., Kong A., Stefansson K. (2007). Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 39, 865–869 10.1038/ng2064 [DOI] [PubMed] [Google Scholar]
- Sun J., Zheng S. L., Wiklund F., Isaacs S. D., Purcell L. D., Gao Z., Hsu F. C., Kim S. T., Liu W., Zhu Y., Stattin P., Adami H. O., Wiley K. E., Dimitrov L., Li T., Turner A. R., Adams T. S., Adolfsson J., Johansson J. E., Lowey J., Trock B. J., Partin A. W., Walsh P. C., Trent J. M., Duggan D., Carpten J., Chang B. L., Gronberg H., Isaacs W. B., Xu J. (2008). Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet. 40, 1153–1155 10.1038/ng0308-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Jacobs K. B., Kraft P., Yeager M., Wacholder S., Cox D. G., Hankinson S. E., Hutchinson A., Wang Z., Yu K., Chatterjee N., Garcia-Closas M., Gonzalez-Bosquet J., Prokunina-Olsson L., Orr N., Willett W. C., Colditz G. A., Ziegler R. G., Berg C. D., Buys S. S., Mccarty C. A., Feigelson H. S., Calle E. E., Thun M. J., Diver R., Prentice R., Jackson R., Kooperberg C., Chlebowski R., Lissowska J., Peplonska B., Brinton L. A., Sigurdson A., Doody M., Bhatti P., Alexander B. H., Buring J., Lee I. M., Vatten L. J., Hveem K., Kumle M., Hayes R. B., Tucker M., Gerhard D. S., Fraumeni J. F., Jr., Hoover R. N., Chanock S. J., Hunter D. J. (2009). A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet. 41, 579–584 10.1038/ng.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Jacobs K. B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., Crenshaw A., Cancel-Tassin G., Staats B. J., Wang Z., Gonzalez-Bosquet J., Fang J., Deng X., Berndt S. I., Calle E. E., Feigelson H. S., Thun M. J., Rodriguez C., Albanes D., Virtamo J., Weinstein S., Schumacher F. R., Giovannucci E., Willett W. C., Cussenot O., Valeri A., Andriole G. L., Crawford E. D., Tucker M., Gerhard D. S., Fraumeni J. F., Jr., Hoover R., Hayes R. B., Hunter D. J., Chanock S. J. (2008). Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 40, 310–315 10.1038/ng.91 [DOI] [PubMed] [Google Scholar]
- Tiwari H. K., Barnholtz-Sloan J., Wineinger N., Padilla M. A., Vaughan L. K., Allison D. B. (2008). Review and evaluation of methods correcting for population stratification with a focus on underlying statistical principles. Hum. Hered. 66, 67–86 10.1159/000119107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. J., Choudhry S., Naqvi M., Rodriguez-Cintron W., Burchard E. G., Ziv E. (2005). Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum. Genet. 118, 424–433 10.1007/s00439-005-0067-z [DOI] [PubMed] [Google Scholar]
- Waters K. M., Le Marchand L., Kolonel L. N., Monroe K. R., Stram D. O., Henderson B. E., Haiman C. A. (2009). Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol. Biomarkers Prev. 18, 1285–1289 10.1158/1055-9965.EPI-09-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Kibel A. S., Hu J. J., Turner A. R., Pruett K., Zheng S. L., Sun J., Isaacs S. D., Wiley K. E., Kim S. T., Hsu F. C., Wu W., Torti F. M., Walsh P. C., Chang B. L., Isaacs W. B. (2009). Prostate cancer risk associated loci in African Americans. Cancer Epidemiol. Biomarkers Prev. 18, 2145–2149 10.1158/1055-9965.EPI-08-0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M., Chatterjee N., Ciampa J., Jacobs K. B., Gonzalez-Bosquet J., Hayes R. B., Kraft P., Wacholder S., Orr N., Berndt S., Yu K., Hutchinson A., Wang Z., Amundadottir L., Feigelson H. S., Thun M. J., Diver W. R., Albanes D., Virtamo J., Weinstein S., Schumacher F. R., Cancel-Tassin G., Cussenot O., Valeri A., Andriole G. L., Crawford E. D., Haiman C. A., Henderson B., Kolonel L., Le Marchand L., Siddiq A., Riboli E., Key T. J., Kaaks R., Isaacs W., Isaacs S., Wiley K. E., Gronberg H., Wiklund F., Stattin P., Xu J., Zheng S. L., Sun J., Vatten L. J., Hveem K., Kumle M., Tucker M., Gerhard D. S., Hoover R. N., Fraumeni J. F., Jr., Hunter D. J., Thomas G., Chanock S. J. (2009). Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 41, 1055–1057 10.1038/ng.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M., Orr N., Hayes R. B., Jacobs K. B., Kraft P., Wacholder S., Minichiello M. J., Fearnhead P., Yu K., Chatterjee N., Wang Z., Welch R., Staats B. J., Calle E. E., Feigelson H. S., Thun M. J., Rodriguez C., Albanes D., Virtamo J., Weinstein S., Schumacher F. R., Giovannucci E., Willett W. C., Cancel-Tassin G., Cussenot O., Valeri A., Andriole G. L., Gelmann E. P., Tucker M., Gerhard D. S., Fraumeni J. F., Jr., Hoover R., Hunter D. J., Chanock S. J., Thomas G. (2007). Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649 10.1038/ng2022 [DOI] [PubMed] [Google Scholar]
- Zheng S. L., Stevens V. L., Wiklund F., Isaacs S. D., Sun J., Smith S., Pruett K., Wiley K. E., Kim S. T., Zhu Y., Zhang Z., Hsu F. C., Turner A. R., Johansson J. E., Liu W., Kim J. W., Chang B. L., Duggan D., Carpten J., Rodriguez C., Isaacs W., Gronberg H., Xu J. (2009a). Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol. Biomarkers Prev. 18, 1815–1820 10.1158/1055-9965.EPI-09-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Cai Q., Signorello L. B., Long J., Hargreaves M. K., Deming S. L., Li G., Li C., Cui Y., Blot W. J. (2009b). Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol. Biomarkers Prev. 18, 2761–2764 10.1158/1055-9965.EPI-08-0983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Long J., Gao Y. T., Li C., Zheng Y., Xiang Y. B., Wen W., Levy S., Deming S. L., Haines J. L., Gu K., Fair A. M., Cai Q., Lu W., Shu X. O. (2009c). Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 41, 324–328 10.1038/ng.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E., John E. M., Choudhry S., Kho J., Lorizio W., Perez-Stable E. J., Burchard E. G. (2006). Genetic ancestry and risk factors for breast cancer among Latinas in the San Francisco Bay Area. Cancer Epidemiol. Biomarkers Prev. 15, 1878–1885 10.1158/1055-9965.EPI-06-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]