Abstract
Nitric oxide (NO) is known to depress ribosome biogenesis in vitro. In this study we analyzed the influence of exogenous NO on ribosome biogenesis in vivo using a proven antihypertensive model of perinatal NO administration in genetically hypertensive rats. Fawn-hooded hypertensive rat (FHH) dams were supplied with the NO-donor molsidomine in drinking water from 2 weeks before to 4 weeks after birth, and the kidneys were subsequently collected from 2 day, 2 week, and 9 to 10-month-old adult offspring. Although the NO-donor increased maternal NO metabolite excretion, the NO status of juvenile renal (and liver) tissue was unchanged as assayed by EPR spectroscopy of NO trapped with iron-dithiocarbamate complexes. Nevertheless, microarray analysis revealed marked differential up-regulation of renal ribosomal protein genes at 2 days and down-regulation at 2 weeks and in adult males. Such differential regulation of renal ribosomal protein genes was not observed in females. These changes were confirmed in males at 2 weeks by expression analysis of renal ribosomal protein L36a and by polysome profiling, which also revealed a down-regulation of ribosomes in females at that age. However, renal polysome profiles returned to normal in adults after early exposure to molsidomine. No direct effects of molsidomine were observed on cellular proliferation in kidneys at any age, and the changes induced by molsidomine in renal polysome profiles at 2 weeks were absent in the livers of the same rats. Our results suggest that the previously found prolonged antihypertensive effects of perinatal NO administration may be due to epigenetically programmed alterations in renal ribosome biogenesis during a critical fetal period of renal development, and provide a salient example of a drug-induced reduction of ribosome biogenesis that is accompanied by a beneficial long-term health effect in both males and females.
Keywords: nitric oxide, ribosomal biogenesis, microarray, polysome profiling, perinatal, epigenetic, kidney
Introduction
Plasticity of organogenesis provides an opportunity for interventions in a specific window of early development that may have long-term beneficial or detrimental effects on adult health and disease (McMillen and Robinson, 2005). One critical regulation of such plasticity is protein synthesis. Upstream factors affecting protein synthesis include tight regulations at multiple stages of ribosome biogenesis. For example, it is well known that epigenetic silencing of ribosomal DNA (rDNA) regularly occurs, even in proliferating cells (McStay and Grummt, 2008; Sanij and Hannan, 2009). One exogenous factor that has been shown to affect rDNA and ribosome biogenesis is nitric oxide (NO). Exposure of cells to high levels of NO, using either NO-donors, or inducing expression of inducible NO synthase (iNOS), results in inhibition of the 80S ribosomal complex (Kim et al., 1998) and enhanced rRNA cleavage resulting in a reduction of both 60S and 80S ribosomal particles (Cai et al., 2000).
Hypertension is associated with decreased NO availability (Wilcox, 2005). The fawn-hooded hypertensive rat (FHH) is a genetic model of hypertension susceptible to progressive renal injury. In FHH hypertension is aggravated and the development of renal injury is accelerated when NOS is chronically inhibited, revealing partial NO dependency of the adult FHH phenotype (Van Dokkum et al., 1998). Renal transplantation under different conditions has shown that blood pressure regulation is intricately linked to the kidney (Smallegange et al., 2004; Crowley et al., 2005), and we hypothesized that this is also the case in the perinatal phase (Koeners et al., 2008a). Recently, we observed that perinatal supplementation of FHH dams with molsidomine, an NO-releasing prodrug (Feelisch, 1998; Singh et al., 1999), persistently lowered the blood pressure and attenuated the development of renal injury in male and female FHH (Koeners et al., 2008b).
Long-term regulation of blood pressure is determined by the relationship between renal perfusion pressure and NaCl excretion, and this relationship is facilitated by renal NO availability (Cowley, 2008; Garvin et al., 2011). Thus our interest is directed at mechanisms in the kidney that link availability of NO in early development to regulation blood pressure in adult life. Conceivably temporal changes in the regulation of renal protein synthesis via ribosomal control of gene translation could constitute such a link (Kasinath et al., 2006). For instance, compensatory renal hypertrophy involves a global increase in polysome profiles within less than 1 day after uninephrectomy (Chen et al., 2005). Based on the known effects of high levels of NO on the ribosomal elements in cultured cells, we hypothesized that ribosome biogenesis in vivo in the neonatal FHH kidney may also be regulated by NO availability. This perinatal regulation of ribosome biogenesis may then affect kidney organogenesis in a manner that impacts the long-term regulation of blood pressure and renal integrity.
Here we demonstrate that the perinatal administration of NO results in a dramatic biphasic change of ribosomal protein gene expression in FHH rats at 2 days and 2 weeks of age. This results in decreased post-translational levels of certain ribosomal proteins, and a remarkable reduction of assembled ribosome structures at the 2-week point. Intriguingly, we did not find an increase in renal NO content at 2 weeks in the offspring of NO-donor-treated rats. Our results suggest that the increased availability of NO in gestation epigenetically alters renal ribosome biogenesis during a critical period of renal development. In conjunction with previously published findings, we conclude that this effect by NO may alter renal organogenesis in a manner that alleviates the hypertension phenotype normally experienced by FHH rats.
Materials and Methods
Animal experiment
Fawn-hooded hypertensive rat were from our own colony, derived from the original colony at Erasmus University Rotterdam (FHH/EUR) maintained by Dr. A. Provoost. FHH dams were supplied with molsidomine (Sigma-Aldrich, Zwijndrecht, Netherlands) in drinking water (120 mg/L) 2 weeks before to 4 weeks after birth. Control FHH mothers and their offspring received regular tap water. All offspring from 4 weeks of age received regular tap water and regular chow (Special Diets Services, Witham, Essex, England). Offspring were sacrificed at 2 days, 2 weeks, 36 weeks (males), and 42 weeks (females). The adult ages were chosen when renal injury in males and females was similar. Kidneys were isolated and snap-frozen (for microarray analysis), kept on ice (for Western blotting and polysome profiling), or fixed in formaldehyde (for immunohistochemistry). Note that although functional and morphological data from the adult rats have been published previously (Koeners et al., 2008b), all microarray data and all data pertaining to renal ribosomal proteins in adult kidneys is novel. Directly after weaning of the pups, the dams were placed in metabolic cages without food but with access to water with 2% glucose and 24-h urine was collected on antibiotic/antimycotic solution (Sigma-Aldrich) to prevent degradation of NO metabolites. NO metabolites were determined as described (Bongartz et al., 2010). Sentinel animals were housed under the same conditions and regularly monitored for infections by nematodes, pathogenic bacteria, and antibodies for rodent viral pathogens (International Council for Laboratory Animal Science, Nijmegen, Netherlands). The Utrecht University Board for studies in experimental animals approved the protocol.
Microarray
For an overview and extensive explanation of microarray data processing, please see Appendix. In short, a piece of snap-frozen kidney was put in 1 mL TRIzol (Invitrogen, Breda, Netherlands) containing 100–150 mg 1 mm glass beads (BioSpec Products, Bartlesville, OK, USA) and immediately homogenized in 30 s using a mini-beadbeater (BioSpec). The total RNA was isolated according to the manufacturer’s instructions. Total RNA was purified using NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany). Samples were then put on Illumina BeadChips (RatRef-12) by ServiceXS1 (Leiden, Netherlands). Kidneys from 2 days, 2 weeks, and adult FHH of both genders were used (at least n = 5/group). All samples were randomly placed on different arrays in order to minimize variation between BeadChips and between arrays.
After calculating the average intensity per probe, all arrays were Log2-transformed and Quantile normalized. The arrays were grouped and the average intensity was calculated. The significance of the differences in intensity between the groups was calculated using Cyber t-test. This final data containing normalized data, average intensity per group and statistical significance between groups were used in data evaluation. The data are submitted as MIAME-complaint to GEO2 under accession number GSE27725.
The number of genes differentially expressed in the molsidomine samples were counted per age in each gender. These were then compared in order to elucidate whether there were genes persistently affected by molsidomine. The 40 genes that were most differentially regulated (20 up and 20 down) by molsidomine were collected at each age for each gender.
All genes encoding for ribosomal proteins were collected. The differentially expressed genes encoding for ribosomal proteins were compared at each age. In order to determine whether the effect of molsidomine on ribosomal genes was stronger than on general gene expression profiles, the ratio of differentially regulated ribosomal genes to the whole ribosomal gene population was compared to the ratio of total differentially expressed genes with whole microarray data in a size test.
Western blot analysis
Fresh kidney samples were lysed on ice in lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100; all from Sigma-Aldrich) plus protease inhibitors (Santa Cruz Biotech, #29130) and subjected to centrifugation at 14K rpm at 4°C for 10 min. Protein content in the supernatants was quantified using Biorad Protein Assay. 6× Laemmli loading buffer was added to 50 μg samples which were then boiled for 5 min and loaded on a 10% SDS/PAGE gel. Transfers to PVDF membranes (Millipore, #IPVH00010) were done overnight at 15 V at 4°C, blocked in 5% milk/TBST solution for 1 h at RT, and subjected to blotting with αL36a (Abnova, #H00006173-M02) or α-actin (Santa Cruz Biotech, #1616) at dilutions of 1:200 in blocking buffer overnight at 4°C. Either α-mouse (L36a) or α-rabbit (actin) HRP-conjugated secondary antibodies (GE Healthcare, #NXA931 and NA934) were used at a dilution of 1:5000 in TBST for 20 min at RT. Blots were washed 3× in TBST for 10 min at RT. Detection was done with the ECL Advance Western Blot Detection Kit (GE Healthcare, #RPN2135). Quantifications were performed using a GS-800 densitometer (Biorad, Veenendaal, Netherlands) and Quantity One software (Biorad).
Polysome profiling
The kidneys from FHH pups from control dams or dams treated with molsidomine were collected at age 2 days and 2 weeks, maintained fresh on ice, and processed for polysome profiling on the same day. For polysome profiling of adult tissue frozen kidney tissue was used. Comparisons were only performed between treated and control rats of both genders at each age. Livers from 2-week-old pups were used to determine tissue-specificity.
All steps of this protocol were performed at 4°C or on ice. Gradients of 17–50% sucrose (11 ml) in gradient buffer (110 mM KAc, 20 mM MgAc2, and 10 mM HEPES pH 7.6) were prepared on the day prior to use. Kidneys were lysed in 500 μl polysome lysis buffer (gradient buffer containing 100 mM KCl, 10 mM MgCl2, 0.1% NP-40, 2 mM DTT, and 40 U/ml RNasin; Promega, Leiden, Netherlands) using a dounce homogenizer. The samples were centrifuged at 1200 g for 10 min to remove debris and loaded onto sucrose gradients. The gradients were ultracentrifuged for 2 h at 40,000 rpm in an SW41Ti rotor (Beckman-Coulter, USA). The gradients were displaced into a UA6 absorbance reader (Teledyne ISCO, USA) using a syringe pump (Brandel, USA) containing 60% sucrose. Absorbance was recorded at an OD of 254 nm. All chemicals came from Sigma-Aldrich unless stated otherwise.
Tissue NO content
Endogenous NO levels in kidney and liver tissues of FHH pups were assayed at 2 weeks by NO trapping with iron-dithiocarbamate (Fe-DETC) complexes, as previously described (Koeners et al., 2007; Van Faassen et al., 2008). Briefly, the yields of paramagnetic NO-Fe(II)-DETC complexes (mononitrosyl-iron complexes, MNIC) in tissues was quantified with EPR spectroscopy of intact frozen tissue sections at 77K. The tissues were reduced at room temperature with dithionite (50 mM for 15 min) to remove the overlapping EPR signal from paramagnetic Cu(II)-DETC complexes as commonly found in biological materials.
Immunohistochemistry
Kidneys were fixed overnight at room temperature in 4% formaldehyde. The tissue was embedded in paraffin and 4 μm sections were made on silanized glass slides. The slides were baked at 58°C overnight, deparaffinated, and rehydrated. Endogenous peroxidase was blocked using citric acid. For antigen retrieval, the sections were boiled for 20 min in citrate buffer (pH 6) and allowed to cool slowly. The sections were then blocked in 1% BSA (w/v) in PBS and incubated with rabbit-anti-pH3 (Santa Cruz Biotech, #1791) overnight at 4°C. The sections were then incubated in anti-rabbit Powervision PO (Immunologic, #DPVR110 HRP) for 30 min at RT and developed using DAB. Finally the sections were counterstained in hematoxylin, dehydrated, and enclosed in pertex. The quantification was performed as follows: At 20× magnification random fields were chosen, taking care not to include the edges of the tissue. The number of positive cells was counted in three fields of two sections per kidney. The average of these six counts was used for analysis.
Statistics
For statistics in microarray, please refer to the methodology. For other measurements the values are expressed as means ± SEM. Data were compared with unpaired t-test, one-way ANOVA, and two-way ANOVA where appropriate followed by post hoc test Student-Newman–Keuls. P < 0.05 is considered significant.
Results
Biometrical data
Biometrical data of FHH offspring and the number of rats studied are collected in Table 1. Note that adult kidney weight and tail-cuff blood pressure data, which were published previously (Koeners et al., 2008b), are included in the table for the sake of convenience. Molsidomine treatment decreased the kidney weight relative to body weight in 2-day-old females (P < 0.05) but not at older ages. However, in both male and female 2-week-old FHH rats relative kidney weight was unchanged. The kidneys of adult males exposed to perinatal molsidomine weighed less than controls, probably in association with reduced injury (Koeners et al., 2008b). Perinatal molsidomine decreased systolic blood pressure in adult FHH offspring (Koeners et al., 2008b). NO metabolites were determined in a 24-h collection of urine from FHH dams to substantiate the direct effects of molsidomine in their pups. Indeed, maternal urine NOx was increased by molsidomine (n = 4) vs. controls (n = 4) from 1.6 ± 0.1 to 2.6 ± 0.2 μmol/(100 g BW)/d (P < 0.01).
Table 1.
Biometrical data of control FHH and FHH during molsidomine (2 days and 2 weeks) or after perinatal molsidomine (adult).
| Males | Females | |||
|---|---|---|---|---|
| Controls | Molsidomine | Controls | Molsidomine | |
| 2 DAYS | ||||
| Number of pups/number of litters | 12/7 | 12/7 | 12/6 | 19/10 |
| RKW/BW (mg/g) | 4.9 ± 0.1 | 4.8 ± 0.1 | 5.3 ± 0.1 | 4.9 ± 0.1# |
| 2 WEEKS | ||||
| Number/litters | 15/8 | 17/10 | 13/8 | 18/10 |
| RKW/BW (mg/g) | 5.2 ± 0.1 | 5.3 ± 0.1 | 5.3 ± 0.1 | 5.5 ± 0.1 |
| ADULT | ||||
| Number/litters | 23/10 | 13/4 | 24/10 | 16/5 |
| RKW/BW (mg/g) | 3.8 ± 0.1 | 3.4 ± 0.03# | 4.4 ± 0.1 | 4.3 ± 0.1 |
| Systolic blood pressure (mmHg) | 158 ± 3 | 139 ± 4# | 145 ± 5 | 118 ± 5# |
#P < 0.05 vs. controls of same gender. Biometrical data in adults published previously (Koeners et al., 2008b) are included for the sake of convenience.
Tissue NO content
Although the NO-donor increased maternal NO metabolite excretion, the NO status of 2-week-old renal (and liver) tissue was unchanged as assayed by EPR spectroscopy of NO trapped with iron-dithiocarbamate complexes (Table 2). Unfortunately, the NO trapping procedure is not possible in 2-day-old pups.
Table 2.
Nitric oxide yields (pmol MNIC/mg tissue) determined by EPR in kidneys and liver in of 2-week-old control FHH and 2-week-old FHH offspring of dams treated with molsidomine.
| Males | Females | |||
|---|---|---|---|---|
| Controls | Molsidomine | Controls | Molsidomine | |
| 2 WEEKS | ||||
| Number/litters | 4/2 | 7/4 | 4/2 | 8/4 |
| NO yield in kidney (left and right) | 0.49 ± 0.03 | 0.53 ± 0.01 | 0.50 ± 0.04 | 0.49 ± 0.02 |
| NO yield in liver | 1.30 ± 0.08 | 1.39 ± 0.04 | 1.35 ± 0.08 | 1.32 ± 0.04 |
MNIC, mononitrosyl-iron complexes.
Microarray
Perinatal treatment with molsidomine significantly affected transcription of hundreds of genes at 2 days and at older ages (see Figure A1 in Appendix). The data also clearly shows that the transcriptional effect of molsidomine differs between ages. Few genes remained differentially expressed at all ages and those that did displayed bidirectional expression between ages.
The 40 most differentially expressed genes (20 induced and 20 reduced) were nearly all different between males and females at the same age. Several genes encoding for ribosomal proteins were present in the top 20 genes in males at all ages, however were less present in the top 20 of females (see Tables A1 in Appendix).
The present study is focused on the changes in ribosomal protein gene expression. Remarkably, these ribosomal protein genes in males were differentially induced by molsidomine at 2 days, then differentially reduced by molsidomine at both 2 weeks and in adults (see Figures 1 and 2; the collection of genes are shown in Table A2 in Appendix). These changes in ribosomal protein gene expression were significant at all ages (P < 0.05) and this effect was specific for ribosomal genes only in males (vs. all genes in the microarray; P < 0.001).
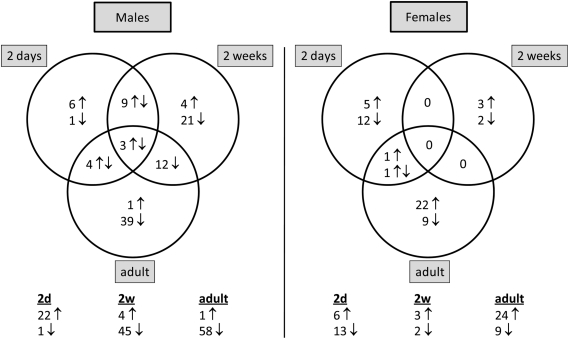
Figure 1.
Number of ribosomal protein genes that are differentially expressed by molsidomine at each age per gender. The distribution of these genes between ages per gender is shown in the Venn diagram. Below the diagram is noted the total number of ribosomal protein genes that are significantly differentially expressed in molsidomine vs. control FHH rats at each age.
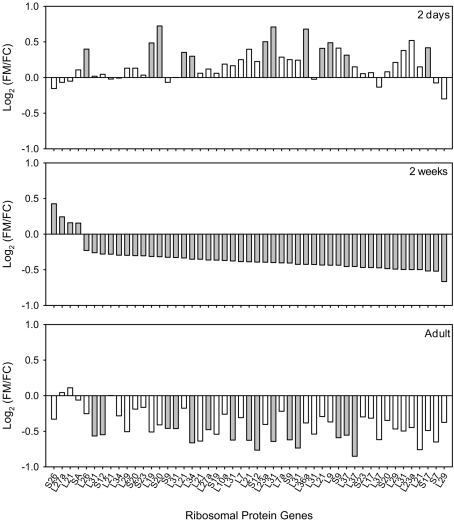
Figure 2.
Ribosomal protein genes in kidneys of FHH males differentially expressed by molsidomine at 2 weeks and whether these genes were also regulated at other ages. All genes that are significantly differentially expressed by molsidomine at 2 weeks are ranked (middle panel). Some of these genes were also regulated at 2 days (upper panel) and in adults (lower panel). The ratio of the genes is expressed as Log2(FM/FC), where FM and FC are the normalized intensities of gene expression in the molsidomine and control groups, respectively. Significant differential expression is indicated by a closed bar, non-significant differential expression by an open bar.
Protein expression of L36a
In order to determine if the changes in ribosomal protein gene expression suggested by the microarray analysis could be verified at the protein level, we analyzed the expression of ribosomal protein L36a at 2 days and 2 weeks. Table A2 in Appendix shows that genes coding for the ribosomal protein L36a are subject to some of the most significant up-regulation at 2 days and down-regulation at 2 weeks (and in adults) in molsidomine-treated FHH males. Note that more than one ribosomal protein L36a gene from different chromosomes is listed on Table 2. Western blot analysis of kidney samples of FHH rats demonstrated that ribosomal protein L36a protein tends to be down-regulated by molsidomine in 2 week males (Figure 3). Quantification of three independent experiments verified that this change in ribosomal protein L36a protein expression occurred only in molsidomine-treated males at 2 weeks (Figure 3). These results suggest that despite the increase in ribosomal protein gene expression seen in 2-day-old molsidomine-treated FHH males, this increases does not manifest as an increase in ribosomal protein levels (see also Figure 4). In contrast, the western blot results suggest that the decrease in ribosomal protein gene expression at 2 weeks in molsidomine-treated rats indeed does affect L36a proteins levels, as also supported by the profiles of Figure 4.
Figure 3.
Western Blot on ribosomal protein L36a. Western blot analysis was performed on kidneys lysates from 2 day and 2 week control and molsidomine FHH rats of both genders using an antibody against ribosomal protein L36a (upper panel). The measured band intensity of ribosomal protein L36a was normalized by α-actin of the same sample (ribosomal protein L36a/actin). The molsidomine effect on ribosomal protein L36a/actin is expressed relative to the age- and gender matched control, shown in graph by the dotted line (lower panel). The reduction in 2 week molsidomine males is borderline significant (P = 0.1). All 3 replicates are biological.
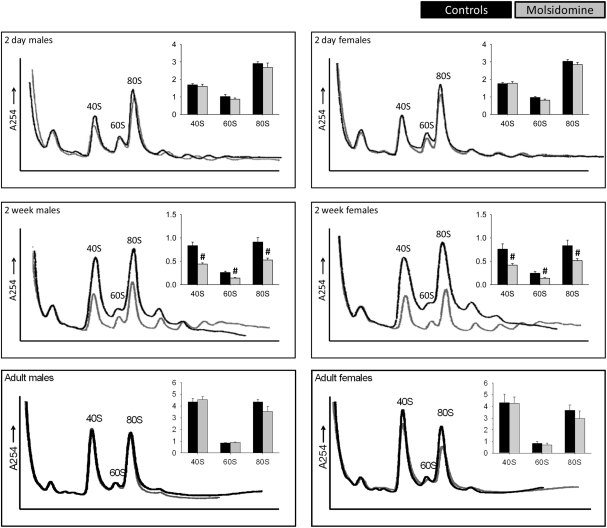
Figure 4.
Polysome profiles of kidneys. Kidneys from control 2 day old (males n = 5, females n = 6), 2 week old (n = 8 in both genders), and adult (n = 4 in both genders) and molsidomine-treated 2 day old (males n = 4, females n = 6), 2 week old (n = 9 in both genders) and adult (n = 5 in both genders) FHH rats were profiled to measure the total number of assembled ribosome structures. The peaks of 40S, 60S, and 80S were normalized against the left-most peak and the results shown in the corresponding histograms. #P < 0.01 vs. untreated of the same peak.
Ribosomal evaluation
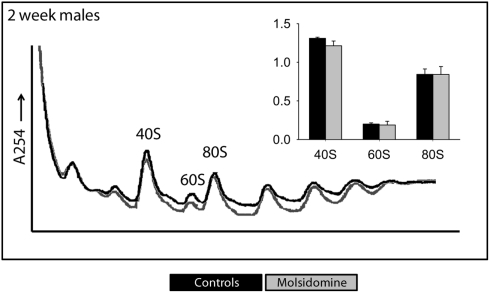
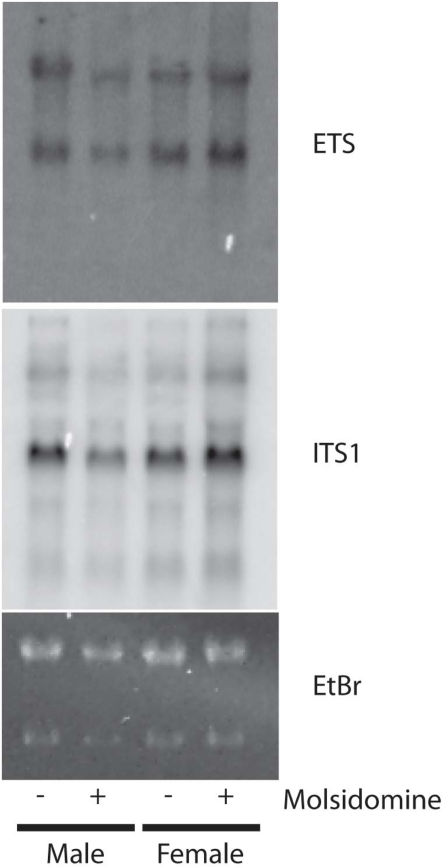
Polysome profiling was performed on kidneys and the effects on the peaks representing the small ribosomal subunit (40S), large ribosomal subunit (60S), and monosome (80S) were determined (Figure 4). Two days after birth and in adults, molsidomine had no effect on polysome profiles, but 2 weeks after birth all peaks were significantly reduced by molsidomine in both males and females (P < 0.01). This effect may be specific to the kidney, as no effect of molsidomine was observed in males at 2 weeks of age in liver polysome profiles (Figure 5). Northern blotting was performed on total RNA from 2 week FHH kidneys in order to determine rates of rRNA processing (Figure A2 in Appendix). However, no differences were observed in processing using probes binding to either the external transcribed spacer (ETS) or the internal transcribed spacer 1 (ITS1).
Figure 5.
Polysome profiles of livers. Livers from 2-week-old FHH males were profiled to measure the total number of assembled ribosome structures. The peaks of 40S, 60S, and 80S were normalized against the left-most peak and the results shown in the corresponding histograms. Black and gray lines are from the control and molsidomine groups, respectively.
Immunohistochemistry
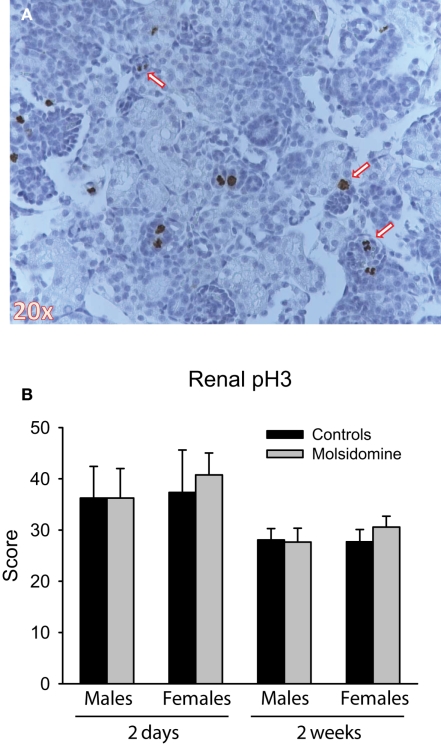
In order to determine if molsidomine resulted in a mitotic index change, we subjected kidneys from FHH rats to staining with a phospho-specific histone-3 (pH3) antibody (Figure 6). We saw no difference in the number of pH3-positive cells on molsidomine-treated FHH rat kidney slices compared to controls. As expected the number of pH3-positive cells decreased as the age of the rats increased. No pH3-positive cells were observed in adults (data not shown).
Figure 6.
Phospho-histone H3 (pH3) immunohistochemistry. Histology was performed on the kidneys from 2 day, 2 week and adult control and molsidomine FHH rats of both genders for pH3. An example of positive staining for pH3 in 2 day female molsidomine-treated FHH rat is shown in (A). Several arrows indicate cells in mitosis staining positive for pH3. The number of positive cells at 2 days and 2 weeks is shown in (B). Note that data in adults is not shown as all stained sections were completely negative for pH3.
Discussion
Nitric oxide donors are known to inhibit proliferation of mesangial and other glomerular cells in vitro (Rupprecht et al., 2000). Little is known about the effects of NO-donors on early growth and nephrogenesis. Recently we observed that administration of NO-donors during early development ameliorates the long-term phenotype in the FHH rat model of progressive hypertension-linked renal injury (Koeners et al., 2008b). Assuming that the direct effects of the NO-donor in early development were related to an as of yet undefined aspect of renal development, the present study focused on a global analysis of ribosomal proteins as a key step in the post-transcriptional regulation of protein synthesis.
The prominent presence of ribosomal protein genes, especially in FHH males, on the lists of the most differentially expressed genes as a result of perinatal NO administration led us to examine how these changes in gene expression affected the protein levels of an individual ribosomal protein as well as the structures of mature, assembled ribosomes. Interestingly, when we measured the protein level of one of the most significantly differentially expressed gene in FHH males, ribosomal protein L36a, we found that only at 2 weeks of age did the change in gene expression correlate with a change in protein expression. This is in contrast to the ribosomal protein L36a protein levels at 2 days, which show no increase despite a substantial increase in ribosomal protein L36a gene expression at that age in males. This discrepancy may be due to the ribosome biogenesis machinery being saturated at 2 days of age and unable to incorporate higher levels of ribosomal proteins. The tight regulation of this biogenesis may likely be degrading excess ribosomal proteins at the protein level or blocking translation of ribosomal proteins at the mRNA level, although at present our data cannot distinguish between these two possibilities. No change in ribosomal protein L36a protein expression was observed in FHH females at 2 weeks of age, but this is not surprising given that no significant gene expression change of ribosomal protein L36a is seen in these animals. However, given the following data, it is likely that there is a reduction of one or more key ribosomal proteins at the protein level in FHH females at 2 weeks. Conceivably by the time the FHH rat has reached adulthood the kidney cells have adjusted the half-lives and/or degradation rates of certain ribosomal proteins in order to reach the normal number of mature ribosome structures.
The most significant finding of this study was that at 2 weeks after birth, i.e., at the end of nephrogenesis (Marquez et al., 2002), perinatal NO administration resulted in a global reduction of ribosome structures in both male and female FHH rats. All of the peaks representing major ribosome structures were found to be substantially decreased in molsidomine-treated FHH rats at 2 weeks of age. This global reduction in NO-treated females at 2 weeks was surprising because only two ribosomal protein genes, coding for ribosomal protein L16 and ribosomal protein L21, were significantly reduced. Ribosomal protein L16 gene expression was not reduced by molsidomine in FHH males at 2 weeks, but strikingly 6 out of the 45 genes that were significantly reduced coded for ribosomal protein L21. This suggests that ribosomal protein L21 may be a key ribosomal protein in the biogenesis of the 60S large subunit and may also play an unappreciated role in the biogenesis of the 40S subunit. Additionally, ribosomal protein L21 appears to be important in the development of craniofacial organs (Xie et al., 2009) and a missense mutation in L21 leads to hereditary hypotrichosis simplex in humans (Zhou et al., 2011). Our findings suggest that ribosomal protein L21 may have an as yet unrecognized role in the development of blood pressure control mechanisms of the kidney.
To our knowledge, this is the one of the most striking examples of a drug-induced decrease in ribosome biogenesis in an animal model to date that is not accompanied by deleterious effects. For example, rapamycin, a powerful inhibitor of the mTOR pathway that directly regulates ribosome biogenesis, when injected into rats had only a slight effect on polysome profiles of liver tissue (Reiter et al., 2004). However, doses of rapamycin as low as 1 ng/mL have been shown to negatively affect cell function and contribute to cell death, for instance in rodent islet cells (Bell et al., 2003; Tanemura et al., 2009). Moreover, many vertebrate and invertebrate models of deficiency of a single ribosomal protein due to gene deletion, knockdown, or missense mutations often show severe phenotypes (Caldarola et al., 2009), and models of biallelic loss of every ribosomal protein gene (with the one exception of ribosomal protein L22; Anderson et al., 2007) results in lethality. In contrast, our data suggest that a decrease of ribosome biogenesis during a critical period of nephrogenesis results in permanent physiologic changes to the kidney, which in turn ameliorate the hypertension phenotype late in life. Interestingly, the effects of exogenous NO on ribosomal biogenesis did not affect the liver, underlining the crucial role of NO in the developing cardiovascular system (Bustamante et al., 1996). An obvious change linked to a decrease in ribosome biogenesis would be a reduction in proliferation. However, immunohistochemistry did not reveal a change in proliferation. Note that although SIN-1, the active metabolite of molsidomine, can generate peroxynitrite in vitro, it appears to function solely as a NO-donor at in vivo oxygen concentrations (Singh et al., 1999). Indeed, recently we supplemented molsidomine to rescue cardiac function in rats with cardiorenal failure and found no increase in 3-nitrotyrosine in heart, kidney, or liver (Bongartz et al., 2010).
The mechanism of action of NO administered perinatally to FHH rats on ribosomal protein gene expression and the subsequent reduction of ribosome structures remains unclear. Indeed, direct measurement of whole kidney NO content at 2 weeks failed to show any effect of maternal NO-donor treatment, suggesting that the decrease in ribosome biogenesis at 2 weeks is programmed by an earlier event. Possibly maternal molsidomine intake results in increased placental transfer of NO adducts and increased fetal renal NO content. Alternatively, indirect effects on the placental circulation could play a role. Indeed, although previous studies (Cai et al., 2000) suggest a link between NO, rRNA synthesis and proliferation, we were unable to establish any differences in pre-rRNA levels in our model that would support direct effects of exogenous NO on rRNA production (Figure A2 in Appendix). This suggests that the effect of NO at 2 weeks more likely lies within epigenetically programmed transcriptional changes of ribosomal protein genes, as demonstrated by the microarray results. Note that in a previous study we did find an increase in offspring kidney NO content when spontaneously hypertensive rat (SHR) dams were treated with citrulline (Koeners et al., 2007). Subsequently siblings of these SHR had a lower blood pressure than control SHR. The results of the present study suggest that mechanisms underlying the antihypertensive effects of perinatal administration of the NO-donor molsidomine in rats with genetic hypertension could be quite different.
It has been previously established that perinatal NO administration alleviates the hypertension phenotype in FHH rats (Koeners et al., 2008b). The present study suggests a potential mechanism underlying this phenotype alleviation. At this stage we have no direct proof that the remarkable ribosomal changes we observe provide the causal mechanism for the beneficial effects of maternal NO on the blood pressure and renal function of the offspring. However, our data demonstrate a novel possibility that long-term amelioration of hypertension by NO in gestation may induce epigenetic changes that affect the postnatal transcription of ribosomal protein genes and a reduction of ribosome structures during a critical period of nephrogenesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Expert assistance: Paula P. Martens (UMC Utrecht), Jeroen Korving, and Harry Begthel (Hubrecht Institute). This study was financially supported by the Dutch Kidney Foundation (NSN C06.2168) and Royal Netherlands Academy of Arts and Sciences (KNAW).
Appendix
Methodology microarray
After the hybridization of the samples to Illumina BeadChips RatRef-12 the raw data set containing all probes of each BeadArray was received. Each sample consisted of approximately 1 million beads, interrogating about 23K unique probes encoding genes (n = 22523), controls, housekeeping genes, and negative controls (n = 825). Each gene is probed with at least 30 beads. Before averaging the intensities of beads per probe, the outliers were removed, using the same approach to removing outliers as in BeadStudio. The removal of outliers was based on median ± 3× median absolute deviation (MAD) and all beads with intensities outside the area of median ± 3× MAD were removed. Outliers were frequent for each probe. In our hands close to 90% of the probes had outliers. Note that the outliers in “empty” beads, the negative controls (NC), were not removed, as they were considered to be background noise of the beads themselves. After removal of outliers (average 3 beads) the number of beads per probe averaged 39 beads.
After the test, the intensities of beads per probe are averaged for further data processing. Before the normalization procedure, the significance of a call from a gene was detected by applying a detection score that is dependent on the distribution of the intensities and the average intensity of the gene and NC (regardless of the number of NC). In this framework we performed the appropriate Student’s t-test between the beads of a gene and the average intensity of all NC on the same array including testing (in)equality between two population variances in order to enhance reliability of the t-test. All probes with a significant call above the negative background were considered biologically active in the respective sample.
The software (T4Illumina) was written to process the raw data by averaging the intensities and determining the call of a gene, including determining the (un)equality of variance and many more. This software is available at request.
The significance of the presence of a gene in a group of samples (Figure A1) is outside the scope of the present study and will not be handled in this document. Note that all genes presented in the present study are significantly biologically active.
The averaged intensities, after removal of outliers, of the probes from all BeadArrays are Log2-transformed and quantile normalized. Next the arrays were grouped accordingly and the average intensity per group was calculated. Finally the significance of the differences in intensities between the groups was calculated using Cyber t-test. Cyber-T is a statistics program designed specifically for microarray data (http://cybert.ics.uci.edu/). The normalization, averaging, and statistics procedures can all be performed by the software Flex- Array (http://gqinnovationcenter.com/services/bioinformatics/ flexarray/index.aspx?l=e).
Northern blot
Total RNA was isolated from kidneys using Trizol using the procedure as recommended by the supplier (Invitrogen). Five microgram of RNA was run on a formaldehyde 1% agarose gel in MOPS buffer and DEPC-treated water for 4 h at 50 V. The gel was soaked in 50 mM NaOH for 15 min followed by 5 min in DEPC-treated water followed by 30 min in 10× SSC buffer. The RNA was transferred to a positively charged nylon membrane (GE Healthcare) overnight by capillary action and bound to the membrane by UV-crosslinking at 120 mJ. The blots were hybridized with DNA probes overnight in ExpressHyb Hybridization Buffer (Clontech) at 65°C. ETS and ITS1 DNA probes were made using the following primers: ETS FW: 5′-GTCTCGGTACGGGTGTGTC-3′ and REV 5′-TTTTTCCCCTTCCTCCTTTC-3′; ITS1 FW 5′-GGCCTGTGTGAGTGTTCCTC-3′ and REV 5′-TCAAGGGAAGA GCGAGAAAA-3′. Probes were labeled with 32P-αCTP (Perkin-Elmer) using a random primer DNA labeling system (Invitrogen). Following hybridization, blots were washed twice for 30 min with 0.1% SDS/0.2× SSC at 65°C. Blots were exposed on storage phosphorimaging screens (Molecular Dynamics) overnight and scanned using a Typhoon Scanner (GE Healthcare).
Table A1.
Collection of the top 20 strongest up- and down-regulated genes at all ages in each gender.
| Entrez gene ID | Symbol | Definition | Gene expression | ||
|---|---|---|---|---|---|
| 2 Days | 2 Weeks | Adult | |||
| A: TOP 40 OF MALE FHH OF 2 DAYS OLD (20 INDUCED AND 20 REDUCED) | |||||
| 366411 | RGD1560729 | Ribosomal protein S24 | 0.820 | −0.217 | −0.354 |
| 500817 | RGD1563124 | 40S ribosomal protein S20 | 0.723 | −0.317 | −0.409 |
| 287029 | RGD1562055 | Ribosomal protein L31 | 0.709 | −0.399 | −0.642 |
| 501876 | RGD1563431 | Large subunit ribosomal protein L36a | 0.680 | −0.424 | −0.383 |
| 499560 | LOC499560 | LRRG00135 | 0.657 | 0.049 | 0.185 |
| 289384 | RGD1560186 | Ribosomal protein L37 | 0.632 | −0.192 | −0.844 |
| 500451 | RGD1562259 | 40S ribosomal protein S20 | 0.534 | −0.114 | −0.452 |
| 362896 | Stat6 | Signal transducer and activator of transcription 6 (predicted), transcript variant 1 | 0.526 | −0.221 | −0.058 |
| 501562 | LOC501562 | ORF2 consensus sequence encoding endonuclease and reverse transcriptase minus RNaseH | 0.514 | 0.009 | 0.181 |
| 171069 | Usmg5 | Upregulated during skeletal muscle growth 5 | 0.508 | −0.372 | −0.651 |
| 367923 | LOC367923 | 60S ribosomal protein L23a | 0.503 | −0.394 | −0.403 |
| 362894 | RGD1310066 | mKIAA1002 protein | 0.493 | 0.214 | 0.146 |
| 314434 | RGD1559566 | 60S ribosomal protein L9 | 0.488 | −0.436 | −0.369 |
| 291075 | Peci | Peroxisomal delta3, delta2-enoyl-coenzyme A isomerase | 0.487 | −0.242 | 0.313 |
| 680294 | LOC680294 | Ribosomal protein L19 | 0.485 | −0.312 | −0.510 |
| 315338 | Hoxc10 | Homeo box C10 | 0.480 | −0.137 | −0.223 |
| 500438 | LOC500438 | ORF2 consensus sequence encoding endonuclease and reverse transcriptase minus RNaseH | 0.478 | −0.083 | 0.026 |
| 500923 | RGD1565798 | Tumor protein, translationally controlled 1 | 0.478 | 0.030 | −0.106 |
| 301434 | Clk1 | CDC-like kinase 1 | 0.474 | 0.081 | 0.282 |
| 502887 | RGD1563551 | Ribosomal protein L31 | 0.470 | −0.397 | −0.523 |
| 287422 | Per1 | Period homolog 1 (Drosophila) | −0.401 | 0.432 | 0.824 |
| 304063 | Ets2 mapped | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | −0.415 | 0.104 | −0.062 |
| 25458 | Gss | Glutathione synthetase | −0.415 | 0.134 | 0.320 |
| 116546 | Ralb | v-ral simian leukemia viral oncogene homolog B (ras related) | −0.417 | −0.007 | 0.090 |
| 25122 | Scnn1a | Sodium channel, non-voltage-gated, type I, alpha | −0.417 | 0.218 | 0.337 |
| 64017 | Enpep | Glutamyl aminopeptidase | −0.422 | 0.102 | 0.787 |
| 29144 | Canx | Calnexin | −0.431 | 0.298 | 0.689 |
| 24244 | Calm3 | Calmodulin 3 | −0.443 | 0.037 | 0.067 |
| 500300 | LOC500300 | Hypothetical protein MGC6835 | −0.446 | 0.090 | 1.036 |
| 290551 | Chdh | Choline dehydrogenase | −0.456 | 0.256 | −0.028 |
| 291081 | RGD1309427 | Tubulin, beta-like | −0.462 | 0.018 | −0.344 |
| 29517 | Sgk | Serum/glucocorticoid regulated kinase | −0.466 | 0.661 | 0.862 |
| 294313 | Mtch1 | Mitochondrial carrier homolog 1 (C. elegans) | −0.503 | −0.072 | −0.138 |
| 498290 | RGD1561353 | OTTHUMP00000044730 | −0.506 | 0.040 | 0.267 |
| 25365 | Actg2 | Actin, gamma 2, smooth muscle, enteric | −0.560 | 0.018 | 0.079 |
| 117273 | Rhoa | ras homolog gene family, member A | −0.572 | −0.047 | −0.171 |
| 501211 | LOC501211 | LOC501211 | −0.608 | 0.074 | −0.596 |
| 287571 | Taf15 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor | −0.712 | 0.255 | 0.049 |
| 25279 | Cyp24a1 | Cytochrome P450, subfamily 24 | −0.849 | 0.342 | −0.634 |
| 25633 | Dnase1 | Deoxyribonuclease I | −0.861 | 0.298 | 0.233 |
| B: TOP 40 OF MALE FHH OF 2 WEEKS OLD (20 INDUCED AND 20 REDUCED) | |||||
| 498789 | RGD1564865 | 20-alpha-hydroxysteroid dehydrogenase | 0.056 | 0.685 | −0.110 |
| 29517 | Sgk | Serum/glucocorticoid regulated kinase | −0.466 | 0.661 | 0.862 |
| 81676 | Hnmt | Histamine N-methyltransferase | 0.028 | 0.625 | 1.057 |
| 25526 | Ptgds | Prostaglandin D2 synthase (brain) | −0.089 | 0.586 | −0.559 |
| 362188 | RGD1311652 | MGC52019 protein | 0.149 | 0.584 | 0.291 |
| 83810 | Trpv1 | Transient receptor potential cation channel, subfamily V, member 1 | 0.022 | 0.583 | −0.373 |
| 246234 | Slc34a3 | Solute carrier family 34 (sodium phosphate), member 3 | −0.149 | 0.559 | −0.108 |
| 171072 | Sult1c2 | Sulfotransferase family, cytosolic, 1C, member 2 | 0.326 | 0.513 | 0.662 |
| 57300 | Aadac | Arylacetamide deacetylase (esterase) | −0.051 | 0.508 | 0.040 |
| 25256 | Fmo1 | Flavin containing monooxygenase 1 | 0.092 | 0.506 | 0.044 |
| 363227 | Obfc2a | Oligonucleotide/oligosaccharide-binding fold containing 2A | 0.145 | 0.452 | −0.222 |
| 79428 | Luzp1 | Leucine zipper protein 1 | −0.086 | 0.443 | 0.119 |
| 287422 | Per1 | Period homolog 1 | −0.401 | 0.432 | 0.824 |
| 362216 | Mrps26 | Mitochondrial ribosomal protein S26 | −0.154 | 0.427 | −0.330 |
| 311826 | Rexo4 | REX4, RNA exonuclease 4 homolog (S. cerevisiae) | −0.083 | 0.421 | 0.125 |
| 299944 | RGD1304605 | Hypothetical LOC299944 | 0.126 | 0.406 | 0.477 |
| 303926 | Igsf11 | Immunoglobulin superfamily, member 11 | 0.187 | 0.403 | 0.059 |
| 293343 | MGC94288 | 4632419K20Rik protein | −0.167 | 0.401 | 0.274 |
| 24267 | Comt | Catechol-O-methyltransferase | −0.156 | 0.397 | −0.180 |
| 81641 | Anpep | Alanyl (membrane) aminopeptidase | 0.007 | 0.395 | 0.083 |
| 364108 | RGD1562073 | Ribosomal protein S17 | 0.418 | −0.517 | −0.490 |
| 29258 | Rps7 | Ribosomal protein S7 | −0.076 | −0.520 | −0.650 |
| 60414 | Rhd | Rh blood group, D antigen | 0.076 | −0.521 | 0.066 |
| 494500 | Yc2 | Glutathione S-transferase Yc2 subunit | 0.390 | −0.550 | −0.078 |
| 25748 | Alas2 | Aminolevulinic acid synthase 2, erythroid | 0.021 | −0.569 | 0.078 |
| 24615 | S100a4 | S100 calcium-binding protein A4 | −0.090 | −0.573 | −0.730 |
| 300024 | LOC300024 | Ly6-B antigen gene | −0.202 | −0.575 | −0.118 |
| 500988 | RGD1564560 | RCK | −0.111 | −0.623 | 0.905 |
| 500019 | RGD1564980 | 60S ribosomal protein L29 (P23) | −0.300 | −0.666 | −0.375 |
| 25250 | Cox8h | Cytochrome c oxidase subunit VIII-H (heart/muscle) | −0.037 | −0.689 | 0.061 |
| 54249 | Cfd | Complement factor D (adipsin) | 0.001 | −0.702 | −0.297 |
| 361619 | MGC72973 | Beta-glo | −0.232 | −0.747 | −0.638 |
| 291541 | Cidea | Cell death-inducing DNA fragmentation factor, alpha subunit-like effector A | −0.234 | −0.797 | 0.132 |
| 25357 | Thrsp | Thyroid hormone responsive protein | 0.132 | −0.829 | 0.021 |
| 293522 | Eraf | Erythroid associated factor | −0.189 | −0.834 | −0.170 |
| 54232 | Ca3 | Carbonic anhydrase 3 | −0.249 | −0.867 | −0.830 |
| 25475 | Lgals5 | Lectin, galactose binding, soluble 5 | −0.218 | −0.918 | −0.225 |
| 81639 | Alox15 | Arachidonate 15-lipoxygenase | −0.010 | −1.005 | 0.035 |
| 79451 | Fabp4 | Fatty acid binding protein 4, adipocyte | −0.189 | −1.067 | −0.061 |
| 24860 | Ucp1 | Uncoupling protein 1 (mitochondrial, proton carrier) | −0.083 | −1.330 | 0.041 |
| C: TOP 40 OF ADULT MALE FHH (20 INDUCED AND 20 REDUCED) | |||||
| 117556 | Sv2b | Synaptic vesicle glycoprotein 2b | 0.046 | −0.471 | 1.766 |
| 313089 | Slc7a13 | Solute carrier family 7 (cationic amino acid transporter, y + system) member 13 | 0.150 | 0.029 | 1.419 |
| 498211 | RGD1560523 | S-adenosylmethionine synthetase gamma form (methionine adenosyltransferase) | −0.037 | 0.124 | 1.251 |
| 50672 | Ednrb | Endothelin receptor type B | 0.223 | −0.396 | 1.238 |
| 117560 | Klf9 | Kruppel-like factor 9 | −0.138 | −0.059 | 1.220 |
| 501085 | LOC501085 | Sulfotransferase K1 | −0.087 | 0.290 | 1.181 |
| 497772 | LOC497772 | Hypothetical gene supported by NM_030827 | 0.183 | 0.259 | 1.178 |
| 501610 | LOC501610 | Gag-Pol polyprotein | 0.023 | 0.099 | 1.123 |
| 316256 | Tnfrsf21 | Tumor necrosis factor receptor superfamily, member 21 | −0.013 | 0.252 | 1.112 |
| 367201 | RGD1562392 | Sulfotransferase K1 (rSULT1C2) | 0.007 | 0.136 | 1.109 |
| 500621 | LOC500621 | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase | −0.238 | 0.034 | 1.104 |
| 313729 | Errfi1 | ERBB receptor feedback inhibitor 1 | 0.002 | 0.084 | 1.091 |
| 310395 | LOC310395 | Nocturnin (CCR4 protein homolog) | −0.232 | 0.105 | 1.074 |
| 361272 | Dhtkd1 | Dehydrogenase E1 and transketolase domain-containing 1 | 0.066 | 0.126 | 1.060 |
| 81676 | Hnmt | Histamine N-methyltransferase | 0.028 | 0.625 | 1.057 |
| 364773 | LOC364773 | Liver regeneration-related protein LRRG07 | −0.370 | −0.192 | 1.043 |
| 500300 | LOC500300 | Hypothetical protein MGC6835 | −0.446 | 0.090 | 1.036 |
| 24861 | Ugt1a1 | UDP glycosyltransferase 1 family, polypeptide A1 | 0.246 | 0.091 | 1.027 |
| 171361 | Eef1a1 | Eukaryotic translation elongation factor 1 alpha 1 | 0.101 | −0.260 | 1.007 |
| 301276 | RGD1564912 | Mut protein | −0.090 | 0.083 | 0.999 |
| 500180 | LOC500180 | IG kappa-chain V–V region K2 precursor | 0.002 | 0.024 | −0.796 |
| 502302 | LOC502302 | 40S ribosomal protein S19 | 0.002 | −0.034 | −0.796 |
| 29191 | Tac2 | Tachykinin 2 | −0.008 | 0.014 | −0.805 |
| 362506 | Ccl19 | Chemokine (C–C motif) ligand 19 | 0.043 | −0.170 | −0.812 |
| 363074 | RGD1309779 | ENSANGP00000021391 | −0.061 | −0.042 | −0.815 |
| 499300 | Ptprcap | Protein tyrosine phosphatase, receptor type, C polypeptide-associated protein | 0.009 | −0.007 | −0.817 |
| 361537 | Tyrobp | Tyro protein tyrosine kinase binding protein | 0.030 | −0.129 | −0.824 |
| 54232 | Ca3 | Carbonic anhydrase 3 | −0.249 | −0.867 | −0.830 |
| 299269 | isg12(b) | Putative ISG12(b) protein | −0.072 | −0.169 | −0.833 |
| 292654 | RGD1564549 | Hypothetical protein FLJ20512 | −0.173 | 0.064 | −0.840 |
| 289384 | RGD1560186 | Ribosomal protein L37 | 0.632 | −0.192 | −0.844 |
| 360918 | Pf4 | Platelet factor 4 | 0.106 | −0.299 | −0.845 |
| 498744 | RGD1561310 | Ribosomal protein L37 | 0.151 | −0.454 | −0.851 |
| 367077 | RGD1562835 | 40S ribosomal protein S26 | 0.170 | −0.348 | −0.852 |
| 24440 | Hbb | Hemoglobin beta chain complex | −0.237 | −0.127 | −0.857 |
| 25632 | Hba-a2 | Hemoglobin alpha, adult chain 2 | −0.105 | −0.016 | −0.860 |
| 299848 | RGD1565117 | 40S ribosomal protein S26 | 0.029 | −0.217 | −0.882 |
| 499131 | LOC499131 | NADH:ubiquinone oxidoreductase B15 subunit | −0.075 | −0.156 | −0.909 |
| 140608 | Atp5i | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit e | −0.139 | −0.008 | −0.971 |
| 692000 | LOC692000 | Dolichol-phosphate mannosyltransferase subunit 3 | −0.190 | 0.095 | −1.014 |
| D: TOP 40 OF FEMALE FHH OF 2 DAYS OLD (20 INDUCED AND 20 REDUCED) | |||||
| 499638 | LOC499638 | LRRGT00057 | 1.238 | 0.031 | 0.087 |
| 361117 | LOC361117 | LRRGT00149 | 1.206 | −0.117 | −0.136 |
| 500285 | LOC500285 | LRRGT00176 | 1.198 | −0.039 | 0.029 |
| 498105 | LOC498105 | LRRGT00176 | 1.186 | 0.049 | −0.044 |
| 498076 | LOC498076 | RIKEN cDNA 2410116I05 | 1.124 | 0.041 | 0.103 |
| 500586 | LOC500586 | LRRGT00057 | 1.110 | −0.046 | −0.062 |
| 498245 | LOC498245 | LRRGT00176 | 1.105 | 0.037 | −0.039 |
| 313278 | RGD1561090 | Protein tyrosine phosphatase, receptor type, D | 0.978 | −0.065 | 0.104 |
| 362803 | LOC362803 | Putative RNA binding protein 1 | 0.888 | 0.045 | −0.109 |
| 500380 | LOC500380 | LRRGT00008 | 0.885 | 0.117 | −0.258 |
| 498623 | LOC498623 | LRRGT00176 | 0.844 | 0.044 | −0.102 |
| 501156 | LOC501156 | LRRGT00176 | 0.793 | 0.074 | −0.293 |
| 500988 | RGD1564560 | RCK | 0.791 | −0.432 | −0.473 |
| 498979 | LOC498979 | LRRGT00194 | 0.763 | 0.035 | −0.147 |
| 25365 | Actg2 | Actin, gamma 2, smooth muscle, enteric | 0.752 | −0.435 | −0.126 |
| 309902 | Cxxc6 | CXXC finger 6 | 0.738 | 0.107 | −0.008 |
| 500343 | LOC500343 | LRRGT00176 | 0.734 | 0.110 | −0.138 |
| 315604 | Ddx6 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | 0.597 | −0.305 | −0.006 |
| 498217 | LOC498217 | LRRG00135 | 0.595 | 0.063 | −0.015 |
| 308831 | Odz4 | Odd Oz/ten-m homolog 4 (Drosophila) | 0.595 | −0.052 | 0.140 |
| 24832 | Thy1 | Thymus cell antigen 1, theta | −0.336 | −0.161 | 0.319 |
| 366942 | RGD1565809 | Ribosomal protein L15 | −0.345 | −0.070 | 0.010 |
| 362850 | Angptl4 | Angiopoietin-like 4 | −0.352 | −0.311 | −0.221 |
| 25475 | Lgals5 | Lectin, galactose binding, soluble 5 | −0.356 | −0.162 | 0.207 |
| 64473 | Bpnt1 | Bisphosphate 3′-nucleotidase 1 | −0.359 | −0.134 | 0.009 |
| 338475 | Nrep | Neuronal regeneration-related protein | −0.370 | −0.154 | 0.300 |
| 294282 | Rps18 | Ribosomal protein S18 | −0.377 | −0.016 | 0.021 |
| 140582 | Ddit4l | DNA-damage-inducible transcript 4-like | −0.389 | −0.186 | −0.055 |
| 360941 | LOC360941 | ORF7 | −0.420 | 0.033 | 0.076 |
| 117273 | Rhoa | ras homolog gene family, member A | −0.423 | −0.239 | −0.189 |
| 29222 | Ptma | Prothymosin alpha | −0.426 | −0.167 | 0.207 |
| 315745 | Fem1b | Feminization 1 homolog b (C. elegans) | −0.448 | −0.074 | 0.167 |
| 124440 | Rpl 41 | Ribosomal protein L41 | −0.455 | 0.034 | 0.067 |
| 314397 | Rin3 | Ras and Rab interactor 3 | −0.497 | −0.156 | −0.188 |
| 89813 | Pdk4 | Pyruvate dehydrogenase kinase, isoenzyme 4 | −0.498 | −0.357 | 0.533 |
| 287571 | Taf15 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor | −0.503 | −0.197 | 0.113 |
| 308568 | RGD1309326 | RIKEN cDNA 2410002F23 | −0.539 | −0.154 | −0.089 |
| 246771 | Slc25a25 | Solute carrier family 25 (mitochondrial carrier, phosphate carrier) member 25 | −0.575 | −0.393 | −0.111 |
| 25633 | Dnase1 | Deoxyribonuclease I | −0.588 | −0.250 | −0.274 |
| 360739 | Ranbp1 | RAN binding protein 1 | −0.597 | −0.306 | 0.453 |
| E: TOP 40 OF FEMALE FHH OF 2 WEEKS OLD (20 INDUCED AND 20 REDUCED) | |||||
| 24567 | Mt1a | Metallothionein 1a | −0.154 | 0.424 | −0.075 |
| 64522 | Slc5a2 | Solute carrier family 5 (sodium/glucose cotransporter) member 2 | −0.110 | 0.419 | −0.362 |
| 362334 | Tmem140 | Transmembrane protein 140 | 0.205 | 0.359 | −0.333 |
| 313430 | Fam151a | Family with sequence similarity 151, member A | −0.061 | 0.325 | −0.358 |
| 360895 | Rrp15 | Ribosomal RNA processing 15 homolog (S. cerevisiae) | 0.067 | 0.324 | −0.001 |
| 362334 | Tmem140 | Transmembrane protein 140 | 0.207 | 0.284 | 0.051 |
| 289419 | Nuak2 | NUAK family, SNF1-like kinase, 2 | 0.037 | 0.278 | −0.257 |
| 307098 | Net1 | Neuroepithelial cell transforming gene 1 | −0.058 | 0.278 | −0.049 |
| 24484 | Igfbp3 | Insulin-like growth factor binding protein 3 | 0.267 | 0.270 | −0.206 |
| 303002 | RGD1308952 | mKIAA0665 protein | 0.052 | 0.263 | −0.163 |
| 85385 | Shc1 | src homology 2 domain-containing transforming protein C1 | 0.094 | 0.261 | 0.123 |
| 300659 | Rnf26 | Ring finger protein 26 | −0.166 | 0.253 | 0.085 |
| 404871 | Olr557 | Olfactory receptor 557 (predicted) | 0.023 | 0.251 | −0.213 |
| 360228 | LOC360228 | WDNM1 homolog | 0.257 | 0.248 | −0.243 |
| 116565 | Lrpap1 | Low density lipoprotein receptor-related protein associated protein 1 | 0.036 | 0.248 | −0.180 |
| 25619 | Plau | Plasminogen activator, urokinase | 0.225 | 0.242 | −0.071 |
| 361367 | Amfr | Autocrine motility factor receptor | 0.043 | 0.239 | −0.135 |
| 171179 | Keg1 | Kidney expressed gene 1 | 0.014 | 0.238 | −0.115 |
| 310810 | Sdfr2 | Stromal cell derived factor receptor 2 | 0.211 | 0.232 | 0.027 |
| 299027 | Eif2s3x | Eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked | −0.021 | 0.228 | 0.096 |
| 24856 | Ttr | Transthyretin | 0.115 | −0.331 | 0.062 |
| 309809 | RGD1310495 | KIAA1919 protein | −0.028 | −0.336 | −0.184 |
| 311676 | Tmepai | Transmembrane, prostate androgen induced RNA | 0.033 | −0.340 | 0.199 |
| 89813 | Pdk4 | Pyruvate dehydrogenase kinase, isoenzyme 4 | −0.498 | −0.357 | 0.533 |
| 246781 | Ptpn7 | Protein tyrosine phosphatase, non-receptor type 7 | −0.008 | −0.364 | 0.101 |
| 501194 | LOC501194 | Hypothetical protein D330021B20 | 0.249 | −0.365 | −0.110 |
| 291555 | Atp8b1 | ATPase, class I, type 8B, member 1 | 0.091 | −0.392 | −0.097 |
| 246771 | Slc25a25 | Solute carrier family 25 (mitochondrial carrier, phosphate carrier) member 25 | −0.575 | −0.393 | −0.111 |
| 303132 | Aff4 | AF4/FMR2 family, member 4 | −0.014 | −0.399 | 0.057 |
| 295660 | RGD1564400 | Eukaryotic translation initiation factor 5 | −0.057 | −0.407 | −0.231 |
| 361296 | Rnf125 | Ring finger protein 125 | 0.270 | −0.407 | −0.134 |
| 64353 | Pdlim5 | PDZ and LIM domain 5 | −0.063 | −0.411 | −0.297 |
| 25365 | Actg2 | Actin, gamma 2, smooth muscle, enteric | 0.752 | −0.435 | −0.126 |
| 498859 | LOC498859 | LOC498859 | −0.002 | −0.449 | −0.111 |
| 315190 | Upk3a | Uroplakin 3A | 0.317 | −0.459 | −0.297 |
| 501530 | LOC501530 | LOC501530 | 0.442 | −0.487 | 0.438 |
| 290704 | LOC290704 | Palladin | 0.371 | −0.516 | −0.324 |
| 94197 | Rab14 | RAB14, member RAS oncogene family | 0.122 | −0.565 | −0.194 |
| 25373 | Ahsg | Alpha-2-HS-glycoprotein | 0.031 | −0.711 | 0.092 |
| 498989 | LOC498989 | Ab2-143 | 0.646 | −0.728 | −0.269 |
| F: TOP 40 OF ADULT FEMALE FHH (20 INDUCED AND 20 REDUCED) | |||||
| 361734 | Ms4a4a | Membrane-spanning 4-domains, subfamily A, member 4 | −0.013 | 0.006 | 1.155 |
| 117518 | Ccl17 | Chemokine (C–C motif) ligand 17 | −0.091 | −0.034 | 1.052 |
| 406161 | C4-2 | Complement component 4, gene 2 | 0.048 | 0.197 | 0.885 |
| 304349 | RGD1559588 | Cell surface receptor FDFACT | 0.010 | −0.105 | 0.877 |
| 500172 | LOC500172 | Immunoglobulin kappa-chain | 0.067 | −0.083 | 0.871 |
| 29517 | Sgk | Serum/glucocorticoid regulated kinase | 0.142 | 0.027 | 0.861 |
| 498982 | RGD1560020 | Myb proto-oncogene protein | 0.143 | 0.078 | 0.801 |
| 298906 | Pqlc3 | PQ loop repeat containing 3 | −0.044 | −0.012 | 0.795 |
| 116676 | Aldh1a2 | Aldehyde dehydrogenase family 1, subfamily A2 | 0.216 | 0.134 | 0.790 |
| 64195 | Mgl1 | Macrophage galactose N-acetyl-galactosamine specific lectin 1 | −0.077 | −0.030 | 0.769 |
| 29168 | Ubd | Ubiquitin D | −0.115 | 0.087 | 0.754 |
| 313438 | Dock11 | Dedicator of cytokinesis 11 | 0.024 | −0.113 | 0.727 |
| 681872 | LOC681872 | Interleukin 19 | 0.038 | −0.033 | 0.726 |
| 246143 | Nradd | Neurotrophin receptor associated death domain | −0.032 | 0.041 | 0.726 |
| 84032 | Col3a1 | Collagen, type III, alpha 1 | 0.057 | −0.029 | 0.716 |
| 24366 | Fgb | Fibrinogen, B beta polypeptide | 0.022 | 0.195 | 0.709 |
| 502902 | RGD1565140 | Clecsf12 protein | −0.066 | −0.009 | 0.692 |
| 501405 | LOC501405 | GTPase activating protein testicular GAP1 | −0.015 | −0.013 | 0.691 |
| 24251 | Cd53 | CD53 antigen | 0.035 | −0.049 | 0.681 |
| 298975 | Scin | Scinderin | 0.095 | 0.095 | 0.679 |
| 309527 | Ch25h | Cholesterol 25−hydroxylase | 0.055 | −0.014 | −0.713 |
| 286926 | Tfpi2 | Tissue factor pathway inhibitor 2 | 0.019 | −0.005 | −0.725 |
| 293538 | RGD1565366 | Hmx2 protein | 0.256 | −0.049 | −0.737 |
| 24517 | Junb | Jun-B oncogene | 0.116 | −0.039 | −0.745 |
| 24451 | Hmox1 | Heme oxygenase (decycling) 1 | 0.155 | 0.248 | −0.771 |
| 292868 | Klks3 | Kallikrein, submaxillary gland S3 | −0.113 | −0.039 | −0.771 |
| 363483 | Dmrtc1c | DMRT-like family C1c | 0.096 | 0.076 | −0.808 |
| 24498 | Il6 | Interleukin 6 | 0.055 | −0.099 | −0.826 |
| 78965 | Csf1 | Colony stimulating factor 1 (macrophage) | 0.131 | 0.064 | −0.837 |
| 24296 | Cyp1a1 | Cytochrome P450, family 1, subfamily a, polypeptide 1 | 0.102 | −0.062 | −0.857 |
| 24508 | Irf1 | Interferon regulatory factor 1 | −0.214 | 0.033 | −0.894 |
| 25542 | Ccl3 | Chemokine (C–C motif) ligand 3 | 0.102 | 0.064 | −0.923 |
| 25464 | Icam1 | Intercellular adhesion molecule 1 | −0.177 | −0.058 | −0.965 |
| 25361 | Vcam1 | Vascular cell adhesion molecule 1 | 0.133 | −0.038 | −1.099 |
| 362993 | Rnd1 | Rho family GTPase 1 | 0.054 | −0.031 | −1.161 |
| 24770 | Ccl2 | Chemokine (C–C motif) ligand 2 | −0.007 | −0.083 | −1.232 |
| 116637 | Ccl4 | Chemokine (C–C motif) ligand 4 | 0.067 | 0.029 | −1.322 |
| 114105 | Cxcl2 | Chemokine (C–X–C motif) ligand 2 | −0.091 | 0.069 | −1.504 |
| 25651 | Selp | Selectin, platelet | −0.078 | −0.032 | −1.531 |
| 81503 | Cxcl1 | Chemokine (C–X–C motif) ligand 1 | −0.075 | −0.010 | −2.156 |
The difference in gene expression between molsidomine-treated and control FHH was noted as Log2(FM/FC), where FM and FC are the normalized intensities. Red and blue numbers indicate gene expression differentially induced and reduced by molsidomine, respectively.
Table A2.
Ribosomal protein genes in kidneys of FHH males differentially expressed by molsidomine (FM, 2 days and 2 weeks) or after perinatal molsidomine (FM, adult) vs. controls (FC).
| Entrez gene ID | Ribosome | Chromosome | Gene expression | ||
|---|---|---|---|---|---|
| 2 Days | 2 Weeks | Adult | |||
| A: 2 DAYS | |||||
| 366411 | S24 | 5 | 0.820 | −0.217 | −0.354 |
| 500817 | S20 | 7 | 0.723 | −0.317 | −0.409 |
| 287029 | L31 | 10 | 0.709 | −0.399 | −0.642 |
| 501876 | L36a | 13 | 0.680 | −0.424 | −0.383 |
| 289384 | L37 | 13 | 0.632 | −0.192 | −0.844 |
| 500451 | S20 | 5 | 0.534 | −0.114 | −0.452 |
| 367923 | L23a | 3 | 0.503 | −0.394 | −0.403 |
| 314434 | L9 | X | 0.488 | −0.436 | −0.369 |
| 680294 | L19 | – | 0.485 | −0.312 | −0.510 |
| 502887 | L31 | 4 | 0.470 | −0.397 | −0.523 |
| 500322 | S7 | 4 | 0.462 | −0.192 | −0.411 |
| 500510 | L21 | 5 | 0.437 | −0.044 | −0.432 |
| 498078 | L7a | 11 | 0.427 | −0.331 | −0.354 |
| 364108 | S17 | 14 | 0.418 | −0.517 | −0.490 |
| 294700 | L21 | 2 | 0.409 | −0.434 | −0.293 |
| 287417 | L26 | 10 | 0.399 | −0.228 | −0.253 |
| 295439 | L21 | 2 | 0.353 | −0.334 | −0.175 |
| 314733 | S19 | 7 | 0.329 | −0.330 | −0.638 |
| 289715 | L37 | 14 | 0.314 | −0.454 | −0.554 |
| 364059 | L34 | 13 | 0.299 | −0.352 | −0.662 |
| 365800 | L36a | 4 | 0.280 | −0.014 | −0.131 |
| 498828 | L10 | 18 | 0.276 | −0.241 | −0.306 |
| 364825 | L36 | 18 | −0.199 | 0.117 | 0.126 |
| B: 2 WEEKS | |||||
| 362216 | S26 | 3 | −0.154 | 0.427 | −0.330 |
| 308719 | L27a | 1 | −0.068 | 0.243 | 0.044 |
| 503211 | L21 | 8 | −0.051 | 0.161 | 0.111 |
| 367030 | SA | 8 | 0.106 | 0.155 | −0.061 |
| 287417 | L26 | 10 | 0.399 | −0.228 | −0.253 |
| 298126 | L31 | 5 | 0.018 | −0.261 | −0.566 |
| 313283 | S12 | 5 | 0.045 | −0.279 | −0.548 |
| 287996 | L21 | 11 | −0.022 | −0.281 | 0.002 |
| 296870 | L34 | X | −0.008 | −0.295 | −0.283 |
| 364828 | L29 | 18 | 0.132 | −0.297 | −0.506 |
| 500559 | S20 | 5 | 0.132 | −0.302 | −0.188 |
| 498360 | S23 | 14 | 0.033 | −0.304 | −0.164 |
| 680294 | L19 | – | 0.485 | −0.312 | −0.510 |
| 500817 | S20 | 7 | 0.723 | −0.317 | −0.409 |
| 367102 | S9 | 8 | −0.068 | −0.325 | −0.458 |
| 299935 | L31 | 7 | 0.001 | −0.328 | −0.461 |
| 295439 | L21 | 2 | 0.353 | −0.334 | −0.175 |
| 364059 | L34 | 13 | 0.299 | −0.352 | −0.662 |
| 300731 | L21 | 8 | 0.061 | −0.352 | −0.638 |
| 499133 | L27a | 1 | 0.119 | −0.363 | −0.477 |
| 687298 | S19 | 10 | 0.059 | −0.365 | −0.541 |
| 366656 | L10a | 6 | 0.188 | −0.370 | −0.260 |
| 502854 | L31 | 4 | 0.166 | −0.376 | −0.624 |
| 297755 | L7 | 5 | 0.252 | −0.386 | −0.308 |
| 294781 | L21 | 2 | 0.397 | −0.388 | −0.627 |
| 65139 | S12 | 1 | 0.225 | −0.393 | −0.765 |
| 367923 | L23a | 3 | 0.503 | −0.394 | −0.403 |
| 287029 | L31 | 10 | 0.709 | −0.399 | −0.642 |
| 501604 | L7a | X | 0.287 | −0.402 | −0.218 |
| 81772 | S9 | 1 | 0.252 | −0.406 | −0.620 |
| 289401 | L31 | 13 | 0.243 | −0.424 | −0.735 |
| 501876 | L36a | 13 | 0.680 | −0.424 | −0.383 |
| 299740 | L31 | 7 | −0.024 | −0.426 | −0.539 |
| 294700 | L21 | 2 | 0.409 | −0.434 | −0.293 |
| 314434 | L9 | X | 0.488 | −0.436 | −0.369 |
| 300278 | S9 | X | 0.414 | −0.436 | −0.590 |
| 289715 | L37 | 14 | 0.314 | −0.454 | −0.554 |
| 498744 | L37 | 17 | 0.151 | −0.454 | −0.851 |
| 501058 | S23 | 8 | 0.054 | −0.467 | −0.297 |
| 363418 | L17 | 12 | 0.068 | −0.469 | −0.316 |
| 81770 | L37 | 2 | −0.136 | −0.472 | −0.618 |
| 500817 | S20 | 7 | 0.080 | −0.484 | −0.347 |
| 363861 | L29 | 12 | 0.212 | −0.490 | −0.467 |
| 366887 | L31 | 7 | 0.379 | −0.497 | −0.497 |
| 686564 | L23a | 14 | 0.520 | −0.497 | −0.446 |
| 364139 | L21 | 14 | 0.150 | −0.497 | −0.758 |
| 364108 | S17 | 14 | 0.418 | −0.517 | −0.490 |
| 29258 | S7 | 5 | −0.076 | −0.520 | −0.650 |
| 500019 | L29 | 4 | −0.300 | −0.666 | −0.375 |
| C: 36 WEEKS | |||||
| 360710 | L7a | 11 | 0.034 | 0.157 | 0.218 |
| 365300 | L7 | 1 | −0.007 | 0.004 | −0.213 |
| 294282 | S18 | 20 | −0.094 | −0.010 | −0.337 |
| 498837 | S27a | 18 | 0.196 | −0.043 | −0.342 |
| 315521 | L32 | 8 | 0.339 | 0.010 | −0.363 |
| 367250 | L7a | 9 | 0.021 | 0.060 | −0.371 |
| 365560 | L36a | 20 | 0.030 | −0.097 | −0.384 |
| 500451 | S20 | 5 | 0.534 | −0.114 | −0.452 |
| 367102 | S9 | 8 | −0.068 | −0.325 | −0.458 |
| 299935 | L31 | 7 | 0.001 | −0.328 | −0.461 |
| 499133 | L27a | 1 | 0.119 | −0.363 | −0.477 |
| 500559 | S20 | 5 | 0.220 | −0.339 | −0.490 |
| 309408 | S12 | 1 | −0.079 | −0.134 | −0.495 |
| 498954 | S16 | 19 | 0.099 | −0.171 | −0.499 |
| 302497 | L10a | X | 0.334 | −0.262 | −0.499 |
| 25347 | L39 | X | 0.014 | −0.145 | −0.512 |
| 302898 | L1 | 10 | 0.034 | 0.076 | −0.515 |
| 302528 | L37a | X | 0.006 | −0.041 | −0.520 |
| 502887 | L31 | 4 | 0.470 | −0.397 | −0.523 |
| 499752 | L7a | 3 | 0.115 | −0.093 | −0.527 |
| 124323 | S23 | 2 | 0.163 | −0.260 | −0.529 |
| 293754 | L16 | 1 | −0.278 | 0.116 | −0.548 |
| 313283 | S12 | 5 | 0.045 | −0.279 | −0.548 |
| 289715 | L37 | 14 | 0.314 | −0.454 | −0.554 |
| 315642 | L27a | 8 | −0.162 | −0.126 | −0.560 |
| 298495 | L35a | 5 | 0.296 | −0.332 | −0.565 |
| 298126 | L31 | 5 | 0.018 | −0.261 | −0.566 |
| 366689 | L21 | 6 | −0.128 | −0.317 | −0.575 |
| 81768 | L22 | 5 | 0.002 | −0.148 | −0.580 |
| 300278 | S9 | X | 0.414 | −0.436 | −0.590 |
| 292539 | L17 | 1 | 0.237 | −0.246 | −0.592 |
| 498363 | P2 | 14 | 0.481 | −0.130 | −0.597 |
| 299041 | P1 | X | 0.041 | −0.157 | −0.601 |
| 57809 | L35a | 1 | −0.112 | −0.210 | −0.614 |
| 81772 | S9 | 1 | 0.252 | −0.406 | −0.620 |
| 502854 | L31 | 4 | 0.166 | −0.376 | −0.624 |
| 294781 | L21 | 2 | 0.397 | −0.388 | −0.627 |
| 314054 | S10 | 6 | −0.053 | −0.170 | −0.631 |
| 314733 | S19 | 7 | 0.329 | −0.330 | −0.638 |
| 287029 | L31 | 10 | 0.709 | −0.399 | −0.642 |
| 498555 | P2 | 15 | 0.002 | −0.072 | −0.645 |
| 503110 | S19 | 7 | 0.008 | −0.047 | −0.651 |
| 64360 | L23 | 1 | −0.247 | 0.029 | −0.654 |
| 297459 | S25 | 4 | −0.247 | 0.160 | −0.659 |
| 314248 | S17 | 6 | 0.000 | −0.064 | −0.661 |
| 364059 | L34 | 13 | 0.299 | −0.352 | −0.662 |
| 171061 | L17 | 1 | 0.219 | −0.170 | −0.663 |
| 366485 | – | 5 | −0.275 | −0.376 | −0.665 |
| 500714 | L6 | 6 | 0.023 | −0.218 | −0.686 |
| 289401 | L31 | 13 | 0.243 | −0.424 | −0.735 |
| 27139 | S26 | 7 | 0.205 | −0.240 | −0.760 |
| 65139 | S12 | 1 | 0.225 | −0.393 | −0.765 |
| 498523 | L23a | 15 | −0.134 | −0.253 | −0.766 |
| 289932 | S7 | 15 | 0.098 | −0.295 | −0.785 |
| 502302 | S19 | 1 | 0.002 | −0.034 | −0.796 |
| 289384 | L37 | 13 | 0.632 | −0.192 | −0.844 |
| 498744 | L37 | 17 | 0.151 | −0.454 | −0.851 |
| 367077 | S26 | 8 | 0.170 | −0.348 | −0.852 |
| 299848 | S26 | 7 | 0.029 | −0.217 | −0.882 |
All genes that are significantly differentially expressed by molsidomine are arranged per age [(A) is newborn males, i.e., 2 days, (B) is 2-week-old males, and (C) is adult males, i.e., 36 weeks]. The difference in gene expression between molsidomine-treated and control FHH was noted as Log2(FM/FC), where FM and FC are the normalized intensities. Red and blue numbers indicate gene expression differentially induced and reduced by molsidomine, respectively. The symbols and the chromosomal locations are also shown. Ratios indicated in bold and red are increased differential expression and ratios indicated in bold and blue are decreased differential expression. Note that some genes in (A) may reappear in (B,C) and some genes in Table A2B may reappear in Table A2C.
Figure A1.
Total number of genes differentially expressed by molsidomine at each age per gender. Hundreds of genes were significantly affected by molsidomine at each age in both genders. The distribution of the genes over all ages is shown in a Venn diagram. The up and down arrows indicate genes that are differentially induced or reduced by molsidomine vs. control, respectively. Below the diagram are the total numbers of genes that are differentially expressed by molsidomine vs. control.
Figure A2.
Northern Blots. Northern blotting was performed on kidneys from 2 week FHH in order to determine rates of pre-rRNA processing. ETS: external transcribed spacer. ITS1: internal transcribed spacer 1. Ethidium bromide (EtBr) is shown as loading control. Note the slight decrease of molsidomine-treated males is due to loading (seen by EtBr) and not due to processing defects of pre-rRNA.
Footnotes
Abbreviations
FHH, fawn-hooded hypertensive rat; NO, nitric oxide.
References
- Anderson S. J., Lauritsen J. P., Hartman M. G., Foushee A. M., Lefebvre J. M., Shinton S. A., Gerhardt B., Hardy R. R., Oravecz T., Wiest D. L. (2007). Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity 26, 759–772 10.1016/j.immuni.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Bell E., Cao X., Moibi J. A., Greene S. R., Young R., Trucco M., Gao Z., Matschinsky F. M., Deng S., Markman J. F., Naji A., Wolf B. A. (2003). Rapamycin has a deleterious effect on MIN-6 cells and rat and human islets. Diabetes 52, 2731–2739 10.2337/diabetes.52.11.2731 [DOI] [PubMed] [Google Scholar]
- Bongartz L. G., Braam B., Verhaar M. C., Cramer M. J., Goldschmeding R., Gaillard C. A., Steendijk P., Doevendans P. A., Joles J. A. (2010). The nitric oxide donor molsidomine rescues cardiac function in rats with chronic kidney disease and cardiac dysfunction. Am. J. Physiol. Heart Circ. Physiol. 299, H2037–H2045 10.1152/ajpheart.00400.2010 [DOI] [PubMed] [Google Scholar]
- Bustamante S. A., Pang Y., Romero S., Pierce M. R., Voelker C. A., Thompson J. H., Sandoval M., Liu X., Miller M. J. (1996). Inducible nitric oxide synthase and the regulation of central vessel caliber in the fetal rat. Circulation 94, 1948–1953 [DOI] [PubMed] [Google Scholar]
- Cai C. Q., Guo H., Schroeder R. A., Punzalan C., Kuo P. C. (2000). Nitric oxide-dependent ribosomal RNA cleavage is associated with inhibition of ribosomal peptidyl transferase activity in ANA-1 murine macrophages. J. Immunol. 165, 3978–3984 [DOI] [PubMed] [Google Scholar]
- Caldarola S., De Stefano M. C., Amaldi F., Loreni F. (2009). Synthesis and function of ribosomal proteins – fading models and new perspectives. FEBS J. 276, 3199–3210 10.1111/j.1742-4658.2009.07036.x [DOI] [PubMed] [Google Scholar]
- Chen J. K., Chen J., Neilson E. G., Harris R. C. (2005). Role of mammalian target of rapamycin signaling in compensatory renal hypertrophy. J. Am. Soc. Nephrol. 16, 1384–1391 10.1681/ASN.2004100894 [DOI] [PubMed] [Google Scholar]
- Cowley A. W., Jr. (2008). Renal medullary oxidative stress, pressure-natriuresis, and hypertension. Hypertension 52, 777–786 10.1161/HYPERTENSIONAHA.107.092858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley S. D., Gurley S. B., Oliverio M. I., Pazmino A. K., Griffiths R., Flannery P. J., Spurney R. F., Kim H. S., Smithies O., Le T. H., Coffman T. M. (2005). Distinct roles for the kidney and systemic tissues in blood pressure regulation by the renin-angiotensin system. J. Clin. Invest. 115, 1092–1099 10.1172/JCI200523378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feelisch M. (1998). The use of nitric oxide donors in pharmacological studies. Naunyn Schmiedebergs Arch. Pharmacol. 358, 113–122 10.1007/PL00005231 [DOI] [PubMed] [Google Scholar]
- Garvin J. L., Herrera M., Ortiz P. A. (2011). Regulation of renal NaCl transport by nitric oxide, endothelin, and ATP: clinical implications. Annu. Rev. Physiol. 73, 359–376 10.1146/annurev-physiol-012110-142247 [DOI] [PubMed] [Google Scholar]
- Kasinath B. S., Mariappan M. M., Sataranatarajan K., Lee M. J., Feliers D. (2006). mRNA translation: unexplored territory in renal science. J. Am. Soc. Nephrol. 17, 3281–3292 10.1681/ASN.2006050488 [DOI] [PubMed] [Google Scholar]
- Kim Y. M., Son K., Hong S. J., Green A., Chen J. J., Tzeng E., Hierholzer C., Billiar T. R. (1998). Inhibition of protein synthesis by nitric oxide correlates with cytostatic activity: nitric oxide induces phosphorylation of initiation factor eIF-2 alpha. Mol. Med. 4, 179–190 [PMC free article] [PubMed] [Google Scholar]
- Koeners M. P., Braam B., Joles J. A. (2008a). Blood pressure follows the kidney: perinatal influences on hereditary hypertension. Organogenesis 4, 153–157 10.4161/org.4.3.6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeners M. P., Braam B., Van Der Giezen D. M., Goldschmeding R., Joles J. A. (2008b). A perinatal nitric oxide donor increases renal vascular resistance and ameliorates hypertension and glomerular injury in adult fawn-hooded hypertensive rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1847–R1855 10.1152/ajpregu.00073.2008 [DOI] [PubMed] [Google Scholar]
- Koeners M. P., Van Faassen E. E., Wesseling S., De Sain-Van Der Velden M., Koomans H. A., Braam B., Joles J. A. (2007). Maternal supplementation with citrulline increases renal nitric oxide in young spontaneously hypertensive rats and has long-term antihypertensive effects. Hypertension 50, 1077–1084 10.1161/HYPERTENSIONAHA.107.095794 [DOI] [PubMed] [Google Scholar]
- Marquez M. G., Cabrera I., Serrano D. J., Sterin-Speziale N. (2002). Cell proliferation and morphometric changes in the rat kidney during postnatal development. Anat. Embryol. 205, 431–440 10.1007/s00429-002-0262-9 [DOI] [PubMed] [Google Scholar]
- McMillen I. C., Robinson J. S. (2005). Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol. Rev. 85, 571–633 10.1152/physrev.00053.2003 [DOI] [PubMed] [Google Scholar]
- McStay B., Grummt I. (2008). The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24, 131–157 10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- Reiter A. K., Anthony T. G., Anthony J. C., Jefferson L. S., Kimball S. R. (2004). The mTOR signaling pathway mediates control of ribosomal protein mRNA translation in rat liver. Int. J. Biochem. Cell Biol. 36, 2169–2179 10.1016/j.biocel.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Rupprecht H. D., Akagi Y., Keil A., Hofer G. (2000). Nitric oxide inhibits growth of glomerular mesangial cells: role of the transcription factor EGR-1. Kidney Int. 57, 70–82 10.1046/j.1523-1755.2000.00828.x [DOI] [PubMed] [Google Scholar]
- Sanij E., Hannan R. D. (2009). The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 4, 374–382 10.4161/epi.4.6.9449 [DOI] [PubMed] [Google Scholar]
- Singh R. J., Hogg N., Joseph J., Konorev E., Kalyanaraman B. (1999). The peroxynitrite generator, SIN-1, becomes a nitric oxide donor in the presence of electron acceptors. Arch. Biochem. Biophys. 361, 331–339 10.1006/abbi.1998.1007 [DOI] [PubMed] [Google Scholar]
- Smallegange C., Hale T. M., Bushfield T. L., Adams M. A. (2004). Persistent lowering of pressure by transplanting kidneys from adult spontaneously hypertensive rats treated with brief antihypertensive therapy. Hypertension 44, 89–94 10.1161/01.HYP.0000129539.88028.e6 [DOI] [PubMed] [Google Scholar]
- Tanemura M., Saga A., Kawamoto K., Machida T., Deguchi T., Nishida T., Sawa Y., Doki Y., Mori M., Ito T. (2009). Rapamycin induces autophagy in islets: relevance in islet transplantation. Transplant. Proc. 41, 334–338 10.1016/j.transproceed.2008.10.030 [DOI] [PubMed] [Google Scholar]
- Van Dokkum R. P., Jacob H. J., Provoost A. P. (1998). Genetic differences define severity of renal damage after L-NAME-induced hypertension in rats. J. Am. Soc. Nephrol. 9, 363–371 [DOI] [PubMed] [Google Scholar]
- Van Faassen E. E., Koeners M. P., Joles J. A., Vanin A. F. (2008). Detection of basal NO production in rat tissues using iron-dithiocarbamate complexes. Nitric Oxide 18, 279–286 10.1016/j.niox.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Wilcox C. S. (2005). Oxidative stress and nitric oxide deficiency in the kidney: a critical link to hypertension? Am. J. Physiol. Regul. Integr. Comp. Physiol. 289, R913–R935 10.1152/ajpregu.00250.2005 [DOI] [PubMed] [Google Scholar]
- Xie M., Kobayashi I., Kiyoshima T., Nagata K., Ookuma Y., Fujiwara H., Sakai H. (2009). In situ expression of ribosomal protein L21 in developing tooth germ of the mouse lower first molar. J. Mol. Histol. 40, 361–367 10.1007/s10735-009-9249-7 [DOI] [PubMed] [Google Scholar]
- Zhou C., Zang D., Jin Y., Wu H., Liu Z., Du J., Zhang J. (2011). Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Hum. Mutat. 32, 710–714 10.1002/humu.21503 [DOI] [PubMed] [Google Scholar]