Abstract
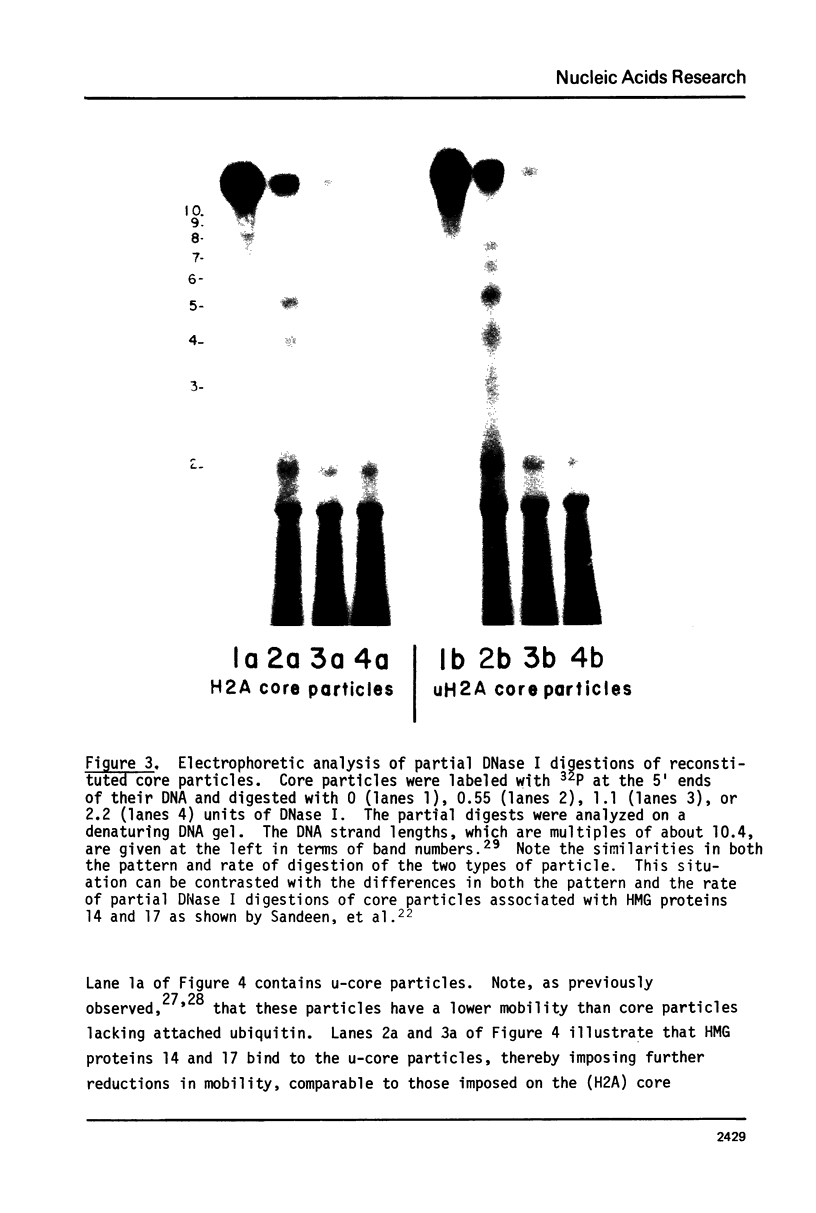
We have purified uH2A (A24) and reconstituted it, in place of H2A, into high molecular weight nucleohistone containing the core histones and DNA. uH2A-containing core particles were then prepared by nuclease digestion and studies on these particles were carried out. We show that two uH2A molecules can be accommodated within a core particle. We also show that the presence of two uH2A molecules in a core particle does not alter significantly either the pattern or the rate of DNase I digestion as compared to both reconstituted and native core particles. Finally, we show that HMG proteins 14 and 17 can bind to uH2A-containing core particles. We conclude that uH2A has little influence on structure at the level of individual nucleosomes.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright S. C., Wiseman J. M., Lange R. A., Garrard W. T. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980 Apr 25;255(8):3673–3684. [PubMed] [Google Scholar]
- Ballal N. R., Kang Y. J., Olson M. O., Busch H. Changes in nucleolar proteins and their phosphorylation patterns during liver regeneration. J Biol Chem. 1975 Aug 10;250(15):5921–5925. [PubMed] [Google Scholar]
- Bonner W. M., Pollard H. B. The presence of F3-F2a1 dimers and F1 oligomers in chromatin. Biochem Biophys Res Commun. 1975 May 5;64(1):282–288. doi: 10.1016/0006-291x(75)90250-8. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Gazit B., Cedar H. Nuclease sensitivity of active chromatin. Nucleic Acids Res. 1980 Nov 25;8(22):5143–5155. doi: 10.1093/nar/8.22.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri C. P., Gorovsky M. A. DNase I sensitivity of ribosomal genes in isolated nucleosome core particles. Nucleic Acids Res. 1980 Jan 11;8(1):197–214. doi: 10.1093/nar/8.1.197-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I. L., Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci U S A. 1977 Mar;74(3):864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I. L., French M. F., Musso R., Busch H. Presence of protein A24 in rat liver nucleosomes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5492–5495. doi: 10.1073/pnas.74.12.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I. L., Wilson G., Ballal N. R., Busch H. Chromatin conjugate protein A24 is cleaved and ubiquitin is lost during chicken erythropoiesis. J Biol Chem. 1980 Nov 25;255(22):10555–10558. [PubMed] [Google Scholar]
- Hunt L. T., Dayhoff M. O. Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem Biophys Res Commun. 1977 Jan 24;74(2):650–655. doi: 10.1016/0006-291x(77)90352-7. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Johns E. W. A method for the selective extraction of histone fractions f2(a)1 and f2(a)2 from calf thymus deoxyribonucleoprotein at pH7. Biochem J. 1967 Nov;105(2):611–614. doi: 10.1042/bj1050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. High-resolution fractionation of nucleosomes: minor particles, "whiskers," and separation of mononucleosomes containing and lacking A24 semihistone. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3244–3248. doi: 10.1073/pnas.77.6.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Precise location of DNase I cutting sites in the nucleosome core determined by high resolution gel electrophoresis. Nucleic Acids Res. 1979 Jan;6(1):41–56. doi: 10.1093/nar/6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Mardian J. K., Paton A. E., Bunick G. J., Olins D. E. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980 Sep 26;209(4464):1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., True R., Burch J. B., Kunkel G. Semihistone protein A24 replaces H2A as an integral component of the nucleosome histone core. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1030–1034. doi: 10.1073/pnas.76.3.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Matsui S. I., Seon B. K., Sandberg A. A. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Penefsky H. S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1977 May 10;252(9):2891–2899. [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Todd R. D., Garrard W. T. Two-dimensional electrophoretic analysis of polynucleosomes. J Biol Chem. 1977 Jul 10;252(13):4729–4738. [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- West M. H., Bonner W. M. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980 Jul 8;19(14):3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]