Abstract
The field of non-coding RNAs (ncRNAs) encompasses a wide array of RNA classes that are indispensible for the regulation of cellular activities. However, de-regulation of these ncRNAs can also play key roles in malignant transformation and cancer cell behavior. In this article we survey a select group of microRNAs and long ncRNAs that appear to contribute in keys ways to the development of acute and chronic leukemias, as well as contribute to their diagnosis, prognosis, and potentially, their treatment.
Keywords: non-coding RNA, AML, ALL, CLL, CML
Introduction
Non-coding RNAs (ncRNAs) are regarded as regulators of cell cycle progression, proliferation, and fate. There are numerous classes of ncRNAs, including very long ncRNAs, PIWI-associated ncRNAs, and small interfering RNAs. However few have been described with respect to hematopoiesis. Here we review the current understanding of the role of microRNAs (miRNA) and long non-coding RNAs (lncRNAs) in the pathogenesis of acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), chronic lymphocytic leukemia (CLL), and chronic myeloid leukemia (CML).
Regulatory microRNAs in Leukemia
MicroRNAs, negative regulators of gene expression, are initially transcribed as long primary miRNAs by RNA polymerase II, and subsequently processed by the nuclear ribonuclease Drosha and the cytoplasmic dsRNA-specific endonuclease Dicer. Mature miRNAs, averaging 22 nucleotides, associate with the RNA induced silencing complex (RISC) that aids in generating their repressive functions. The most efficient miRNA targeting is achieved by mature miRNAs binding to the 3′-untranslated regions (UTR) of target messenger RNAs (mRNA) leading to translational inhibition or mRNA cleavage (Zhang et al., 2007). miRNAs also bind within coding regions and 5′UTRs, through which additional regulation of mRNA translation can be mediated (Figure 1; Ørom et al., 2008; Tay et al., 2008). miRNAs have garnered much interest due to their role as post-transcriptional regulators of genes involved in numerous physiologic and developmental processes including cellular proliferation, cell cycle progression, and apoptosis (reviewed in Lee and Ambros, 2001; Cheng et al., 2005; Hayashita et al., 2005; Ivanovska et al., 2008). Altered miRNA expression has been implicated in the pathology of leukemia, where some miRNAs are proposed to function either as tumor suppressor genes (TS) or oncogenes (Table 1; Esquela-Kerscher and Slack, 2006).
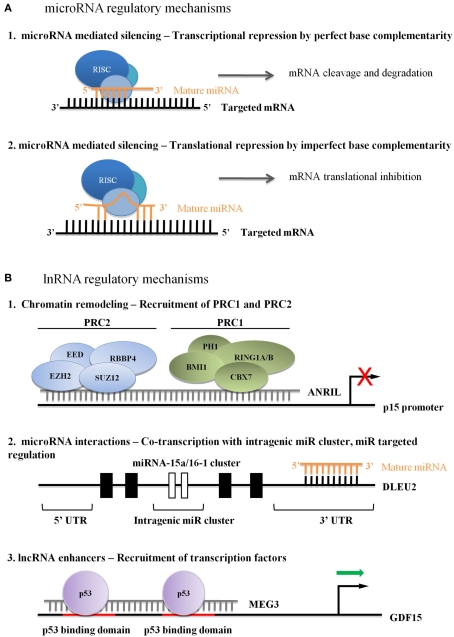
Figure 1.
(A) Regulatory microRNA mechanisms in leukemia. (A1) Perfect base complementarity between miRNA and target mRNA can lead to target mRNA degradation. (A2) Imperfect base complementarity between miRNA and target mRNA can lead to mRNA translational repression. (B) Currently hypothesized mechanisms of lncRNA regulation in leukemia. (B1) Recruitment of PcGs leads to stable repression, although the mechanism of lncRNA association with the target sequence is unknown. (B2) lncRNAs can influence miRNA cluster transcriptional activity, and can be post-transcriptionally regulated by miRNAs. (B3) Activator lncRNAs enhance transcriptional or binding affinity of transcription factors, although the mechanism of lncRNA association with the target sequence is unknown.
Table 1.
Characteristics of discussed ncRNAsa.
| ncRNA(s) | ncRNA class | Target | Disease association | Clinical relevance | Citation |
|---|---|---|---|---|---|
| miR-29b | miRNA | DNMT3a and DNMT3b, DNMT1 (indirect) | AML | Predictive response to therapy; potentially therapeutic | Garzon et al. (2009b) |
| miR-126/126* | miRNA | N/A | AML | Diagnostic: CBF | Li et al. (2008) |
| miR-224, -368, -382 | miRNA | N/A | AML | Diagnostic: t(15; 17) | Li et al. (2008) |
| miR-126, -126*, -224, -368, -382, -17-5p, -20a | miRNA | N/A | AML | Diagnostic: MLL-rearrangement | Li et al. (2008) |
| miR-21 | miRNA | PTEN | AML | Diagnostic: t(6; 11) vs. t(9; 11), prognostic | Garzon et al. (2008a) |
| miR-29b | miRNA | TCL1 | AML | Diagnostic: balanced 11q23 translocations; potentially therapeutic | Garzon et al. (2008a) |
| miR-155 | miRNA | Unknown in AML | AML | Diagnostic: FLT3–ITD mutations | Garzon et al. (2008a) |
| miR-199a, -191 | miRNA | N/A | AML | Prognostic | Garzon et al. (2008a) |
| miR-181 | miRNA | TLR and IL1-β | AML | Prognostic | Marcucci et al. (2008) |
| (miR-128a, -128b) vs. (miR-223, let-7b] | miRNA | N/A | AML vs. ALL | Diagnostic | Mi et al. (2007) |
| miR-18a, -532, -218, -625, 193a, -638, -550, -663 | miRNA | N/A | ALL | Prognostic | Zhang et al. (2009a) |
| miR-143 | miRNA | MLL–AF4 | ALL | Prognostic, potentially therapeutic | Dou et al. (2011) |
| miR-15a, -16-1 | miRNA | Bcl2 | CLL | Potentially therapeutic | Bandi et al. (2009b) |
| miR-29, -181 | miRNA | TCL1 | CLL | Prognostic, potentially therapeutic | Pekarsky et al. (2006) |
| miR-15a, -195, -221, -23b, -155, -223, -29a, -24, -29b, -146, -16, -16-2, -29c | miRNA | N/A | CLL | Diagnostic: expressed ZAP-70 and unmutated IgVH vs. no ZAP-70 and mutated IgVH | San Jose-Eneriz et al. (2009) |
| miR-7, -23a, -26a, -29a, -29c, -30b, -30c, -100, -126, -134, -141, -183, -196b, -199a, -224, -362, -422b, 520a, -191 | miRNA | N/A | CML | Predictive response to therapy | San Jose-Eneriz et al. (2009) |
| ANRIL, p15AS | Antisense | p15 | AML/ALL | Diagnostic, potentially prognostic | Yu et al. (2008); Iacobucci et al. (2011) |
| lincRNA-p21 | lincRNA | BCR-ABL | Potentially CML | Potentially therapeutic | Notari et al. (2006); Du et al. (2010) |
| MEG3 | lincRNA | p53, GDF15 | MDS/AML | Prognostic | Benetatos et al. (2009) |
| Combined T-UCR and miR profile | UCR | N/A | CLL/AML | Diagnostic: tumor type; prognostic | Calin et al. (2007) |
| Dleu2 | lincRNA | miR-15a/16 | CLL | Diagnostic, potentially therapeutic | Migliazza et al. (2001); Lerner et al. (2009) |
| HOTAIRM1 | Antisense | HOXA1, HOXA4, CD11b, CD18 | Hematopoietic regulator | Potentially therapeutic | Zhang et al. (2009b) |
| EGO | Antisense | MBP, EDN | Hematopoietic regulator | Potentially therapeutic | Wagner et al. (2007) |
| lincRNA-a7 | lncRNA | SCL/TAL1 | Hematopoietic regulator | Potentially therapeutic | Ørom et al. (2010) |
a Non-coding RNAs having different diagnostic or therapeutic roles in different leukemia subtypes are listed separately.
miRNAs in AML
Acute myeloid leukemia is characterized by the abnormal proliferation and accumulation of immature myeloblasts in the bone marrow and blood. Diagnosis is based on the presence of malignant blasts in the bone marrow as well as distinctive translocations and gene mutations. AML represents a group of heterogeneous disorders with striking differences in survival based on cytogenetics, prognostic gene mutations, and age and has an average long-term overall survival (OS) rate of approximately 25–60% depending on the above characteristics in patients over age 55 (Heerema-McKenney and Arber, 2009; http://seer.cancer.gov, accessed September 24, 2011).
Golub et al. (1999) first demonstrated that expression profiling of protein-encoding genes could distinguish AML from ALL, pioneering the way for expression-based diagnostic approaches. The advent of miRNA expression profiling further distinguished AML from ALL and has been used to explore possible associations of miRNAs with regard to diagnosis, prognosis, and response to treatment (Calin and Croce, 2006; Mi et al., 2007). Garzon et al. (2008b) reported that the expression of miR-29 family member miR-29b is often down-regulated in primary AML with respect to bone marrow CD34+ progenitors. miR-29b directly targets the 3′UTR of DNA methyltransferases DNMT3a and DNMT3b and indirectly targets DNMT1 via negative regulation of SP1, a transactivator of DNMT1. Down-regulation of the methyltransferases caused by forced expression of miR-29b leads to altered DNA methylation and subsequent repression of TS genes such as p15INK4b and ESR1 often observed in myeloid leukemogenesis (Garzon et al., 2009b). Restoration of miR-29b increases sensitivity to the hypomethylating agent, decitabine, in patients with AML over age 60 (Blum et al., 2010). Forced expression of miR-29b in AML cell lines and primary AML samples reduces cell growth and induces apoptosis, indicative of its TS activity in acute leukemia. Additional support was provided by the dramatic reduction of tumors in a xenograft leukemia model in response to miR-29b over-expression. Following miR-29b expression, gene expression analysis of the AML cell line K562 showed that miR-29b, along with miR-29a, target pathways involved with cell cycle, proliferation, and apoptosis (Garzon et al., 2009a). Together these data suggest that synthetic miR-29b oligonucleotides could potentially serve as a therapeutic approach in AML.
Just as numerous groups have identified mRNA signatures distinguishing AML subclasses, several groups have associated miRNA signatures with cytogenetic abnormalities and predictors of outcome in AML (Bullinger et al., 2004; Valk et al., 2004). Li et al. reported that miRNA expression signatures accurately discriminate between AMLs with the common translocations t(15; 17), t(8; 21), and inv(16), all of which represent favorable prognosis rearrangements that result in the disruption of core-binding factors (CBF). Expression of two (miR-126/126*), three (miR-224, -368, and -382), and seven (miR-126, -126*, -224, -368, -382, -17-5p, and -20a) miRNAs distinguishes CBF t(8; 21) and inv16, t(15; 17) and MLL-rearrangement AMLs, respectively, from one another (Li et al., 2008). Up-regulation of miR-21 distinguishes AMLs with t(6; 11) from those with t(9; 11), while down-modulation of miR-29 correlates with balanced 11q23 translocations. miR-155 shows increased expression in patients with AML characterized by FLT3–ITD mutations (Garzon et al., 2008a).
Expression patterns of miRNAs are also associated with prognosis. Marcucci et al. (2008) have associated a worse prognosis in patients with t(6; 11) who display increased expression of miR-21, an inhibitor of the TS PTEN. This group has also shown that patients with AML that express elevated miR-199a and -191 have a significantly lower OS and event-free survival (Garzon et al., 2008a). The miR-181 family has been shown to contribute to an aggressive AML phenotype through mechanisms associated with the activation of pathways controlled by toll-like receptors and interleukin-1β (Marcucci et al., 2008).
miRNAs in ALL
Acute lymphoblastic leukemia represents a heterogeneous group of disorders that result in the malignant transformation of lymphoblasts at various stages of differentiation. Although more common in children, ALL also occurs in adults, with a shift from childhood-prevalent to adulthood-prevalent subtypes during adolescence. A diagnosis of ALL occurs when blood and bone marrow samples show a large number of abnormal lymphocyte blasts. Treatment options are based on ALL subtype and prognostic factors such as age.
Mi et al. (2007) have shown that expression signatures of as few as two miRNAs can discriminate ALL from AML; miR-128a and -128b were significantly higher in ALL, whereas miR-223 and let-7b were expressed at significantly higher levels in AML. This study also demonstrated that lineage discriminating miRNAs could differentiate ALL samples from AML samples even when both leukemias displayed the same translocation/fusion events such as t(11; 19)(q23; p13.3)/MLL–ENL (Mi et al., 2007). Zhang et al. identified an 8-miRNA-expression profile (miR-18a, -532, -218, -625, -193a, -638, -550, and -663) that differentiates good from poor steroid response in pediatric ALL. miRNA expression signatures were also able to identify relapse and non-relapse pediatric cases (Zhang et al., 2009a). miR-143 has been shown to negatively regulate the MLL–AF4 fusion protein, a product of the translocation t(4; 11)(q21; q23), which confers poorer prognosis compared to other MLL arrangements. Restoration of miR-143 in MLL–AF4-positive cells induced apoptosis by reducing MLL–AF4 fusion protein levels, suggesting that forced expression of miR-128 could serve as a therapeutic agent in MLL–AF4 ALL (Dou et al., 2011).
miRNAs in CLL
Chronic lymphocytic leukemia, the most common leukemia found in adults, results from immature, malignant resting B cell lymphocytes overexpressing the anti-apoptotic B cell lymphoma 2 (Bcl2) protein and accumulating in the bone marrow and the blood (Cimmino et al., 2005). Of those diagnosed with CLL, most are over the age of 50. CLL exhibits clinical heterogeneity as marked by the observation that some patients present with aggressive leukemia requiring immediate treatment while others require no intervention for many years (Li et al., 2011).
The first demonstration of miRNA association with pathogenesis was discovered in CLL (Calin et al., 2002). The Croce group showed that miR-15a and -16 reside in the fragile chromosomal band 13q14, which is deleted in more than half of CLL cases, as well as some prostate tumor and retinoblastoma samples (Chen et al., 2001; Kivelä et al., 2003). They went on to show that miR-15a and -16 negatively regulate cell growth and cell cycle progression (Calin et al., 2002). Additionally, over-expression of these miRNAs has been shown to repress Bcl2 expression and induce apoptosis in a leukemic cell line model, indicating a potential therapeutic role for miR-15a and -16 in the treatment of Bcl2-overexpressing tumors (Bandi et al., 2009a).
Two markers of aggressive CLL include up-regulated 70-kDa zeta-associated protein (ZAP-70) and unmutated immunoglobulin variable genes (IgVH). When these factors are expressed at high levels they are associated with high levels of TCL1, an oncogene that co-activates the anti-apoptotic oncoprotein AKT and aids in regulatory pathways involved in cell survival and death. miR-29 and -181 have been shown to negatively regulate TCL1. Clinical CLL samples show miR-29 and -181 expression is inversely related to TCL1 expression, suggesting that these miRNAs could potentially serve as prognostic markers of CLL progression and as therapeutic agents in aggressive forms of TCL1-overexpressing CLL (Pekarsky et al., 2006). Calin et al. (2005) demonstrated that 13 miRNAs (miR-15a, -195, -221, -23b, -155, -223, -29a, -24, -29b, -146, -16, -16-2, and -29c) could discriminate between patients expressing ZAP-70 and unmutated IgVH and those not expressing ZAP-70 and mutated IgVH, thereby distinguishing aggressive from indolent CLL.
miRNAs in CML
Chronic myeloid leukemia is a malignant clonal stem cell disorder characterized by an increase of mature granulocytes in the bone marrow and blood. It often expresses the constitutively active BCR-ABL tyrosine kinase formed by a translocation that links Abl1 on chromosomal band 9q34 to a portion of BCR on chromosomal band 22q11. CML is often suspected on the basis an extremely elevated white blood cell count with maturation of white blood cells and few or no leukemic blasts. Treatment and prognosis depend primarily on the phase of CML, i.e., whether in chronic, accelerated, or blast crisis. The development of the BCR-ABL1 kinase inhibitor, imatinib mesylate, as well as other Bcr–Abl inhibitors, has significantly improved treatment and outcome of patients with CML (Kantarjian et al., 2002). Jose-Eneriz et al. (2009) identified 19 miRNAs (18 up-regulated: miR-7, -23a, -26a, -29a, -29c, -30b, -30c, -100, -126, -134, -141, -183, -196b, -199a, -224, -362, -422b, -520a, and 1 down-regulated: miR-191) that are differentially expressed between imatinib resistant and responder samples.
Long Non-Coding RNAs in Leukemia
Long non-coding RNAs are ncRNAs greater than 200 nucleotides long, transcribed by RNA polymerase II or III, and can account for nearly 60% of all non-ribosomal and non-mitochondrial RNA in human cells (Kapranov et al., 2010). LncRNAs are involved in transcriptional silencing, chromatin remodeling, and gene activation (reviewed in Huarte and Rinn, 2010; Gibb et al., 2011) and originate from many chromosomal environments, including antisense to protein-coding genes, intergenic DNA (termed “lincRNAs” for long intergenic non-coding RNAs’), and from ultraconserved regions (Carninci et al., 2005). Excluding the latter, only a few lncRNA primary sequences are evolutionarily conserved (Guttman et al., 2009; Baker, 2011). In contrast to miRNAs, several cases have shown that conservation of secondary structure is more important to preserving ncRNA function than nucleotide sequence (Yap et al., 2010). Although lncRNAs in leukemia are not as extensively characterized as they are in some other tumor types, recent progress has identified several high profile targets that could alter how lncRNAs are viewed in terms of leukemia development, classification, and therapeutic targeting.
Suppressors of Tumor Suppressors are Oncogenes
The INK4A–ARF–INK4B locus is deregulated in up to 40% of human cancers (Sherr, 1998; Kim and Sharpless, 2006). In addition to p15INK4b, p14ARF, and p16INK4a, TS genes interact closely with p53 and Rb to regulate cell cycle progression (Bandi et al., 2009a). Pasmant et al. identified a polyadenylated lncRNA that was transcribed antisense to p15INK4b. The full-length transcript, ANRIL (antisense ncRNAs in the INK4 locus) has several isoforms (Pasmant et al., 2007). The p15AS variant, isolated from two AML cell lines, was significantly up-regulated in 11 of 16 AML and ALL primary samples (Yu et al., 2008). The authors demonstrated that p15AS was responsible for Dicer-independent silencing of p15INK4b by altering H3K9me2 and H3K4me2 levels at both endogenous and exogenous p15INK4b promoters. EZH2 and SUZ12 were required for stable silencing of p15INK4b, even after p15AS repression, indicating the likely recruitment of the polycomb repressive complex 2 (PRC2). Other reports demonstrated ANRIL-CBX7 interacts with either di- or tri-methylated H3K27, implying that ANRIL-mediated silencing works through both PRC1 and PRC2 complexes (Figure 1; Group et al., 2005; Kotake et al., 2011; Margueron and Reinberg, 2011). A study by Iacobucci et al. (2011) involving acute leukemia and normal peripheral blood cells showed a statistically significant association between an ANRIL nucleotide polymorphism and ALL phenotype, demonstrating the importance of regulated ANRIL expression in primary leukemia.
Acting downstream of ANRIL, lincRNA-p21 is a 3.1-kb transcript induced by p53 expression that represses cellular proliferation by both p53-dependent and independent mechanisms (Huarte et al., 2010). LincRNA-p21 contains two canonical p53 binding sites in a promoter distinct from its closest neighbor, CDNK1A. Independent knock-down of either p53 or lincRNA-p21 produces significantly overlapping gene set enrichments (p < 10−200), implying at least a partial overlap of apoptosis-induction mechanisms. Interestingly, in p53−/− cell lines, lincRNA-p21 cannot direct proper localization and function of the pre-mRNA binding protein hnRNP-K (Huarte et al., 2010). Although these functional studies were done in mouse embryonic fibroblasts and human lung carcinoma cell lines, two reports demonstrate that BCR-ABL stimulates hnRNP-K expression and stability, subsequently promoting tumor progression (Notari et al., 2006; Du et al., 2010). Although the activity of lincRNA-p21 in acute or chronic leukemia is currently unknown, these data suggest that further work in CML is warranted.
p53-Interacting Tumor Suppressor lncRNAs
A second p53-regulating lincRNA, MEG3, was highlighted in human malignancies when investigators reported high expression levels in normal gonadotrophs and severely reduced expression in tumor-derived gonadotroph cells (Zhang et al., 2003). Forcing re-expression of MEG3 in HCT-116 cells leads to p53 accumulation and inhibition of cellular proliferation, indicative of a high-level regulator of p53-dependent TS activities (Zhang et al., 2003; Zhou et al., 2007). Although MEG3 isoforms can contain several small open reading frames, they are not required for p53-mediated cellular activities (Zhou et al., 2007; Zhang et al., 2010a). Instead, MEG3 secondary structure is critical to maintaining function, including down-regulation of MDM2 expression and enhanced p53 binding to a specific subset of gene promoters, including GDF15 (Figure 1; Zhang et al., 2010a). Of interest, MEG3 can suppress cell growth in p53null cells, indicating p53-independent activities as well (Zhou et al., 2007).
Expression of the MEG3-DLK1 locus is tightly regulated by two differentially methylated regions (DMRs), which are hypermethylated in a subset of solid tumors and suppress MEG3 expression (Kagami et al., 2010; Astuti et al., 2005). Benetatos et al. (2009) examined a cohort of 85 patients with either myelodysplastic syndrome (MDS) or AML. They found that 48% (20/42) of patients with AML displayed aberrant hypermethylation of the MEG3 promoter, which significantly correlated with decreased OS (HR 1.98, p = 0.04). In MDS, 35% (15/43) of patients displayed aberrant hypermethylation, a result that trended toward decreased survival but was not quite significant (HR 2.15, p = 0.072; Benetatos et al., 2009). An independent assessment of 40 AML samples by a second group confirmed aberrant methylation in the MEG3-associated DMRs in AML samples, but not in normal controls (Khoury et al., 2010). Importantly, neither study showed MEG3 hypermethylation to be associated with karyotype or disease subtype (Benetatos et al., 2009). Although additional studies will be needed to determine the functional role of MEG3 in leukemia, the importance of its interactions with MDM2, p53, and GDF15, as well as p16 in pituitary carcinomas, will likely demonstrate a role for MEG3 in leukemia (Zhang et al., 2010b).
ncRNA–ncRNA Regulation: Transcribed Ultraconserved Regions and Host Genes
Within the human genome, 481 transcribed genomic segments have been identified that are 100% conserved between orthologous regions in the human, mouse, and rat (Bejerano et al., 2004; Calin et al., 2007). Of these “transcribed ultraconserved regions” (T-UCRs), 39% are contained within intergenic sequences and 43% are intronic; the remainder are exonic or exon-overlapping (Mestdagh et al., 2010). Like miRNAs, T-UCR locations are closely associated with genomic fragile sites and ubiquitously expressed T-UCRs are frequently associated with cancer-associated genomic regions (CAGRs; Calin et al., 2004, 2007). Microarray analysis has shown that T-UCRs differentially expressed in human malignancies are highly likely to be associated with CAGRs of that tumor type (Calin et al., 2007).
Transcribed ultraconserved regions expression has been used to predict disease outcome in both CLL and neuroblastoma samples (Calin et al., 2007). In a survey of 133 cancers and 22 normal tissues, a profile of 19 T-UCRs (8 up- and 11 down-regulated) could differentiate between normal, CLL, colorectal, and hepatocarcinoma samples (Calin et al., 2007). This study also showed that the expression of five T-UCRs (three intronic and two intergenic) could divide a CLL cohort into two prognostic groups, previously defined by low (favorable) vs. high (poor) ZAP-70 expression (Calin et al., 2007). Expression of these diagnostic T-UCRs negatively correlated with the previously defined CLL miRNA signature, suggesting a mechanism for miRNA regulation of these T-UCRs (Calin et al., 2005). Of the diagnostic T-UCR profile, three (uc.160, uc.346A, and uc.348) contain miRNAs recognition sites, including targeting by miR-155:: miR-24:: and miR-29 (Calin et al., 2005). Repressive activity of miR-155 was confirmed in an in vitro assay, while the negative correlations were observed between miR-155?uc.346A and miR-24?uc pairings; 160 were validated in the aforementioned diagnostic cohort (Calin et al., 2007). miR-155 over-expression was identified in CLL, while in AML its up-regulation was associated with expanded bone marrow granulocyte and monocyte proliferation (reviewed in Faraoni et al., 2009) and miR-24 loss-of-function has been linked to methotrexate resistance in HCT-116 cells (Mishra et al., 2009). Interestingly, miR-29a has also been shown to regulate MEG3 expression in hepatocarcinoma cell lines (Braconi et al., 2011). Although it is currently unknown whether miRNAs repress T-UCRs, or whether the altered expression of the T-UCRs affects the primary target of the leukemia-associated miRNAs, it is clear that careful regulation of these two ncRNA species is critical to understanding the disease state and for developing ncRNA-associated classification or ncRNA-directed targeted therapy. In addition to miRNA regulation, differential methylation of T-UCR-associated CpG islands (CGI) may also control expression. In a study of 15 leukemia cell lines and 64 primary leukemia samples, differential methylation was seen at three of these CGI (∼60% in cell lines and approximately 18% in primary samples), although the relevance of CGI-mediated hypermethylation to leukemia etiology remains unclear (Lujambio et al., 2010). A much larger cohort will need to be investigated before diagnostic implications can be verified.
Long non-coding RNAs targeting by miRNAs is only one example of ncRNA-ncRNA regulation. Deleted in leukemia 2 (Dleu2) is an lncRNA transcribed from 13q14 (Liu et al., 1997; Migliazza et al., 2001). The Myc-repressed Dleu2 transcript is a host gene for the miR-15a/miR-16-1 cluster, providing the primary transcript from which miR-15a and miR-16-1 are processed (Figure 1; Klein et al., 2010). While de-regulation of these two miRNAs has long been associated with CLL, deletion-mapping studies have demonstrated that the Dleu2 transcript is frequently disrupted in CLL cell line, and increased expression of this gene leads to reduced proliferation and clonogenicity (Lerner et al., 2009). Although further analysis is needed in order to validate these observations, Dleu2 serves as an important example of the intricate co-regulation of lncRNA and miRNAs.
Hematopoietic Regulator lncRNAs as Potential Oncogenic Genes
Newly discovered lncRNAs are being characterized at a rapid pace. In hematopoiesis, the antisense lincRNA, HOTAIRM1, has recently been identified as an essential regulator of myeloid cell differentiation. This lincRNA is transcribed from within the HOXA cluster and regulates HOXA1, HOXA4, CD11b, and CD18 during retinoic acid-induced differentiation of an acute promyelocytic leukemia cell line (Zhang et al., 2009b). EGO is expressed during eosinophil development and is essential for major basic protein and eosinophil-derived neurotoxin mRNA expression (Wagner et al., 2007). Ørom et al. (2010) identified several lncRNA activators, one of which strongly induced expression of SCL/TAL1. Although these lncRNAs have not yet been associated with hematopoietic malignancies, such critical regulators of cell fate are likely to be identified as potent regulators of tumorigenicity.
Conclusion
It has become evident in recent years that the de-regulation of miRNAs and lncRNAs plays a critical role in malignant transformation, tumor cell behavior and, in particular, hematologic malignancies. These ncRNAs could prove to be increasingly useful in the development of much needed novel diagnostic, prognostic, and therapeutic strategies for acute and chronic leukemias.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jason E. Farrar for his critical evaluation of this manuscript. This work was in part supported by grants from the United States Department of Defense, the NIH (CA120535 and a TARGET initiative through the Children’s Oncology Group and CureSearch) as well as from the Lyles Parachini Fund, the Michael Corb Fund, and the Michael Garil Leukemia Survivors Fund and Program. Dr. Arceci is supported in part through the endowed King Fahd Professorship in Pediatric Oncology at Johns Hopkins.
References
- Astuti D., Latif F., Wagner K., Gentle D., Cooper W. N., Catchpoole D., Grundy R., Ferguson-Smith A. C., Maher E. R. (2005). Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br. J. Cancer 92, 1574–1580 10.1038/sj.bjc.6602478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. (2011). Long noncoding RNAs: the search for function. Nat. Methods. 8, 379–383 10.1038/nmeth0411-275 [DOI] [Google Scholar]
- Bandi N., Zbinden S., Gugger M., Arnold M., Kocher V., Hasan L., Kappeler A., Brunner T., Vassella E. (2009a). miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 69, 5553–5559 10.1158/0008-5472.CAN-08-4277 [DOI] [PubMed] [Google Scholar]
- Bandi N., Zbinden S., Gugger M., Arnold M., Kocher V., Hasan L., Kappeler A., Brunner T., Vassella E. (2009b). miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non‚ Äí Small cell lung cancer. Cancer Res. 69, 5553–5559 10.1158/0008-5472.CAN-08-4277 [DOI] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W. J., Mattick J. S., Haussler D. (2004). Ultraconserved elements in the human genome. Science 304, 1321–1325 10.1126/science.1098119 [DOI] [PubMed] [Google Scholar]
- Benetatos L., Hatzimichael E., Dasoula A., Dranitsaris G., Tsiara S., Syrrou M., Georgiou I., Bourantas K. L. (2009). CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk. Res. 34, 148–153 10.1016/j.leukres.2009.06.019 [DOI] [PubMed] [Google Scholar]
- Blum W., Garzon R., Klisovic R. B., Schwind S., Walker A., Geyer S., Liu S., Havelange V., Becker H., Schaaf L., Mickle J., Devine H., Kefauver C., Devine S. M., Chan K. K., Heerema N. A., Bloomfield C. D., Grever M. R., Byrd J. C., Villalona-Calero M., Croce C. M., Marcucci G. (2010). Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc. Natl. Acad. Sci. U.S.A. 107, 7473–7478 10.1073/pnas.1002650107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C., Kogure T., Valeri N., Huang N., Nuovo G., Costinean S., Negrini M., Miotto E., Croce C. M., Patel T. (2011). MicroRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30, 4750–4756 10.1038/onc.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullinger L., DöHner K., Bair E., FröHling S., Schlenk R. F., Tibshirani R., DöHner H., Pollack J. R. (2004). Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 350, 1605–1616 10.1056/NEJMoa031046 [DOI] [PubMed] [Google Scholar]
- Calin G. A., Croce C. M. (2006). MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 66, 7390–7394 10.1158/0008-5472.CAN-06-0800 [DOI] [PubMed] [Google Scholar]
- Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C. M. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S. E., Iorio M. V., Visone R., Sever N. I., Fabbri M., Iuliano R., Palumbo T., Pichiorri F., Roldo C., Garzon R., Sevignani C., Rassenti L., Alder H., Volinia S., Liu C.-G., Kipps T. J., Negrini M., Croce C. M. (2005). A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353, 1793–1801 10.1056/NEJMoa050995 [DOI] [PubMed] [Google Scholar]
- Calin G. A., Liu C.-G., Ferracin M., Hyslop T., Spizzo R., Sevignani C., Fabbri M., Cimmino A., Lee E. J., Wojcik S. E., Shimizu M., Tili E., Rossi S., Taccioli C., Pichiorri F., Liu X., Zupo S., Herlea V., Gramantieri L., Lanza G., Alder H., Rassenti L., Volinia S., Schmittgen Thomasâ d., Kipps T. J., Negrini M., Croce C. M. (2007). Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 10.1016/j.ccr.2007.07.027 [DOI] [PubMed] [Google Scholar]
- Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kodzius R., Shimokawa K., Bajic V. B., Brenner S. E., Batalov S., Forrest A. R. R., Zavolan M., Davis M. J., Wilming L. G., Aidinis V., Allen J. E., Ambesi-Impiombato A., Apweiler R., Aturaliya R. N., Bailey T. L., Bansal M., Baxter L., Beisel K. W., Bersano T., Bono H., Chalk A. M., Chiu K. P., Choudhary V., Christoffels A., Clutterbuck D. R., Crowe M. L., Dalla E., Dalrymple B. P., De Bono B., Gatta G. D., Di Bernardo D., Down T., Engstrom P., Fagiolini M., Faulkner G., Fletcher C. F., Fukushima T., Furuno M., Futaki S., Gariboldi M., Georgii-Hemming P., Gingeras T. R., Gojobori T., Green R. E., Gustincich S., Harbers M., Hayashi Y., Hensch T. K., Hirokawa N., Hill D., Huminiecki L., Iacono M., Ikeo K., Iwama A., Ishikawa T., Jakt M., Kanapin A., Katoh M., Kawasawa Y., Kelso J., Kitamura H., Kitano H., Kollias G., Krishnan S. P. T., Kruger A., Kummerfeld S. K., Kurochkin I. V., Lareau L. F., Lazarevic D., Lipovich L., Liu J., Liuni S., Mcwilliam S., Babu M. M., Madera M., Marchionni L., Matsuda H., Matsuzawa S., Miki H., Mignone F., Miyake S., Morris K., Mottagui-Tabar S., Mulder N., Nakano N., Nakauchi H., Ng P., Nilsson R., Nishiguchi S., Nishikawa S., Nori F., Ohara O., Okazaki Y., Orlando V., Pang K. C., Pavan W. J., Pavesi G., Pesole G., Petrovsky N., Piazza S., Reed J., Reid J. F., Ring B. Z., Ringwald M., Rost B., Ruan Y., Salzberg S. L., Sandelin A., Schneider C., Schönbach C., Sekiguchi K., Semple C. A., Seno S., Sessa L., Sheng Y., Shibata Y., Shimada H., Shimada K., Silva D., Sinclair B., Sperling S., Stupka E., Sugiura K., Sultana R., Takenaka Y., Taki K., Tammoja K., Tan S. L., Tang S., Taylor M. S., Tegner J., Teichmann S. A., Ueda H. R., van Nimwegen E., Verardo R., Wei C. L., Yagi K., Yamanishi H., Zabarovsky E., Zhu S., Zimmer A., Hide W., Bult C., Grimmond S. M., Teasdale R. D., Liu E. T., Brusic V., Quackenbush J., Wahlestedt C., Mattick J. S., Hume D. A., Kai C., Sasaki D., Tomaru Y., Fukuda S., Kanamori-Katayama M., Suzuki M., Aoki J., Arakawa T., Iida J., Imamura K., Itoh M., Kato T., Kawaji H., Kawagashira N., Kawashima T., Kojima M., Kondo S., Konno H., Nakano K., Ninomiya N., Nishio T., Okada M., Plessy C., Shibata K., Shiraki T., Suzuki S., Tagami M., Waki K., Watahiki A., Okamura-Oho Y., Suzuki H., Kawai J., Hayashizaki Y., FANTOM Consortium, and RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Chen C., Frierson H. F., Jr., Haggerty P. F., Theodorescu D., Gregory C. W., Dong J.-T. (2001). An 800-kb region of deletion at 13q14 in human prostate and other carcinomas. Genomics 77, 135–144 10.1006/geno.2001.6633 [DOI] [PubMed] [Google Scholar]
- Cheng A. M., Byrom M. W., Shelton J., Ford L. P. (2005). Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33, 1290–1297 10.1093/nar/gki844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., Rassenti L., Alder H., Volinia S., Liu C.-G., Kipps T. J., Negrini M., Croce C. M. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949 10.1073/pnas.0506654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou L., Zheng D., Li J., Li Y., Gao L., Wang L., Yu L. (2011). Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene. 10.1038/onc.2011.248 [DOI] [PubMed] [Google Scholar]
- Du Q., Wang L., Zhu H., Zhang S., Xu L., Zheng W., Liu X. (2010). The role of heterogeneous nuclear ribonucleoprotein K in the progression of chronic myeloid leukemia. Med. Oncol. 27, 673–679 10.1007/s12032-009-9373-y [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F. J. (2006). Oncomirs [mdash] microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- Faraoni I., Antonetti F. R., Cardone J., Bonmassar E. (2009). miR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta 1792, 497–505 [DOI] [PubMed] [Google Scholar]
- Garzon R., Garofalo M., Martelli M. P., Briesewitz R., Wang L., Fernandez-Cymering C., Volinia S., Liu C. G., Schnittger S., Haferlach T., Liso A., Diverio D., Mancini M., Meloni G., Foa R., Martelli M. F., Mecucci C., Croce C. M., Falini B. (2008a). Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. U.S.A. 105, 3945–3950 10.1073/pnas.0800135105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Volinia S., Liu C.-G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T., Flomenberg N., Marcucci G., Calin G. A., Kornblau S. M., Kantarjian H., Bloomfield C. D., Andreeff M., Croce C. M. (2008b). MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111, 3183–3189 10.1182/blood-2007-07-098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Heaphy C. E. A., Havelange V., Fabbri M., Volinia S., Tsao T., Zanesi N., Kornblau S. M., Marcucci G., Calin G. A., Andreeff M., Croce C. M. (2009a). MicroRNA 29b functions in acute myeloid leukemia. Blood 114, 5331–5341 10.1182/blood-2009-03-211938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C. E. A., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., Havelange V., Volinia S., Blum W., Rush L. J., Perrotti D., Andreeff M., Bloomfield C. D., Byrd J. C., Chan K., Wu L.-C., Croce C. M., Marcucci G. (2009b). MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 113, 6411–6418 10.1182/blood-2008-07-170589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb E. A., Brown C. J., Lam W. L. (2011). The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 10, 38. 10.1186/1476-4598-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub T. R., Slonim D. K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J. P., Coller H., Loh M. L., Downing J. R., Caligiuri M. A., Bloomfield C. D., Lander E. S. (1999). Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537 10.1126/science.286.5439.531 [DOI] [PubMed] [Google Scholar]
- Group R. G. E. R., Genome Science G., The F. C., Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C. C., Suzuki M., Kawai J., Suzuki H., Carninci P., Hayashizaki Y., Wells C., Frith M., Ravasi T., Pang K. C., Hallinan J., Mattick J., Hume D. A., Lipovich L., Batalov S., Engström P. G., Mizuno Y., Faghihi M. A., Sandelin A., Chalk A. M., Mottagui-Tabar S., Liang Z., Lenhard B., Wahlestedt C. (2005). Antisense Transcription in the mammalian transcriptome. Science 309, 1564–1566 10.1126/science.1112009 [DOI] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., Huarte M., Zuk O., Carey B. W., Cassady J. P., Cabili M. N., Jaenisch R., Mikkelsen T. S., Jacks T., Hacohen N., Bernstein B. E., Kellis M., Regev A., Rinn J. L., Lander E. S. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. (2005). A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65, 9628–9632 10.1158/0008-5472.CAN-05-2352 [DOI] [PubMed] [Google Scholar]
- Heerema-McKenney A., Arber D. A. (2009). Acute myeloid leukemia. Hematol. Oncol. Clin. North Am. 23, 633–654 10.1016/j.hoc.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Huarte M., Guttman M., Feldser D., Garber M., Koziol M. J., Kenzelmann-Broz D., Khalil A. M., Zuk O., Amit I., Rabani M., Attardi L. D., Regev A., Lander E. S., Jacks T., Rinn J. L. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M., Rinn J. L. (2010). Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 19, R152–R161 10.1093/hmg/ddq353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I., Sazzini M., Garagnani P., Ferrari A., Boattini A., Lonetti A., Papayannidis C., Mantovani V., Marasco E., Ottaviani E., Soverini S., Girelli D., Luiselli D., Vignetti M., Baccarani M., Martinelli G. (2011). A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia. Leuk. Res. 35, 1052–1059 10.1016/j.leukres.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Ivanovska I., Ball A. S., Diaz R. L., Magnus J. F., Kibukawa M., Schelter J. M., Kobayashi S. V., Lim L., Burchard J., Jackson A. L., Linsley P. S., Cleary M. A. (2008). MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 28, 2167–2174 10.1128/MCB.01977-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose-Eneriz S. E., Roman-Gomez J., Jimenez-Velasco A., Garate L., Martin V., Cordeu L., Vilas-Zornoza A., Rodriguez-Otero P., Calasanz M. J., Prosper F., Agirre X. (2009). MicroRNA expression profiling in imatinib-resistant chronic myeloid leukemia patients without clinically significant ABL1-mutations. Mol. Cancer 8, 69. 10.1186/1476-4598-8-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M., O’sullivan M. J., Green A. J., Watabe Y., Arisaka O., Masawa N., Matsuoka K., Fukami M., Matsubara K., Kato F., Ferguson-Smith A. C., Ogata T. (2010). The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 6, e1000992. 10.1371/journal.pgen.1000992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H., Sawyers C., Hochhaus A., Guilhot F., Schiffer C., Gambacorti-Passerini C., Niederwieser D., Resta D., Capdeville R., Zoellner U., Talpaz M., Druker B. (2002). Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N. Engl. J. Med. 346, 645–652 10.1056/NEJMoa011573 [DOI] [PubMed] [Google Scholar]
- Kapranov P., St Laurent G., Raz T., Ozsolak F., Reynolds C. P., Sorensen P. H., Reaman G., Milos P., Arceci R. J., Thompson J. F., Triche T. J. (2010). The majority of total nuclear-encoded non-ribosomal RNA in a human cell is “dark matter” un-annotated RNA. BMC Biol. 8, 149. 10.1186/1741-7007-8-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury H., Suarez-Saiz F., Wu S., Minden M. D. (2010). An upstream insulator regulates DLK1 imprinting in AML. Blood 115, 2260–2263 10.1182/blood-2009-03-212746 [DOI] [PubMed] [Google Scholar]
- Kim W. Y., Sharpless N. E. (2006). The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275 10.1016/j.cell.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Kivelä T., Tuppurainen K., Riikonen P., Vapalahti M. (2003). Retinoblastoma associated with chromosomal 13q14 deletion mosaicism. Ophthalmology 110, 1983–1988 10.1016/S0161-6420(03)00484-6 [DOI] [PubMed] [Google Scholar]
- Klein U., Lia M., Crespo M., Siegel R., Shen Q., Mo T., Ambesi-Impiombato A., Califano A., Migliazza A., Bhagat G., Dalla-Favera R. (2010). The DLEU2/miR-15a/16-1 Cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17, 28–40 10.1016/j.ccr.2009.11.019 [DOI] [PubMed] [Google Scholar]
- Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. (2011). Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 30, 1956–1962 10.1038/onc.2010.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Ambros V. (2001). An Extensive Class of Small RNAs in Caenorhabditis elegans. Science 294, 862–864 10.1126/science.1065329 [DOI] [PubMed] [Google Scholar]
- Lerner M., Harada M., Lovén J., Castro J., Davis Z., Oscier D., Henriksson M., Sangfelt O., Grandér D., Corcoran M. M. (2009). DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp. Cell Res. 315, 2941–2952 10.1016/j.yexcr.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Li S., Moffett H. F., Lu J., Werner L., Zhang H., Ritz J., Neuberg D., Wucherpfennig K. W., Brown J. R., Novina C. D. (2011). MicroRNA expression profiling identifies activated B cell status in chronic lymphocytic leukemia cells. PLoS ONE 6, e16956. 10.1371/journal.pone.0016956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lu J., Sun M., Mi S., Zhang H., Luo R. T., Chen P., Wang Y., Yan M., Qian Z., Neilly M. B., Jin J., Zhang Y., Bohlander S. K., Zhang D.-E., Larson R. A., Le Beau M. M., Thirman M. J., Golub T. R., Rowley J. D., Chen J. (2008). Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc. Natl. Acad. Sci. U.S.A. 105, 15535–15540 10.1073/pnas.0710905105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Corcoran M., Rasool O., Ivanova G., Ibbotson R., Grander D., Iyengar A., Baranova A., Kashuba V., Merup M., Wu X., Gardiner A., Mullenbach R., Poltaraus A., Hultstrom A. L., Juliusson G., Chapman R., Tiller M., Cotter F., Gahrton G., Yankovsky N., Zabarovsky E., Einhorn S., Oscier D. (1997). Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene 15, 2463–2473 10.1038/sj.onc.1201643 [DOI] [PubMed] [Google Scholar]
- Lujambio A., Portela A., Liz J., Melo S. A., Rossi S., Spizzo R., Croce C. M., Calin G. A., Esteller M. (2010). CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 29, 6390–6401 10.1038/onc.2010.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Radmacher M. D., Maharry K., MróZek K., Ruppert A. S., Paschka P., Vukosavljevic T., Whitman S. P., Baldus C. D., Langer C., Liu C.-G., Carroll A. J., Powell B. L., Garzon R., Croce C. M., Kolitz J. E., Caligiuri M. A., Larson R. A., Bloomfield C. D. (2008). MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 358, 1919–1928 10.1056/NEJMoa074256 [DOI] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. (2011). The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P., Fredlund E., Pattyn F., Rihani A., Van Maerken T., Vermeulen J., Kumps C., Menten B., De Preter K., Schramm A., Schulte J., Noguera R., Schleiermacher G., Janoueix-Lerosey I., Laureys G., Powel R., Nittner D., Marine J. C., Ringner M., Speleman F., Vandesompele J. (2010). An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 29, 3583–3592 10.1038/onc.2010.106 [DOI] [PubMed] [Google Scholar]
- Mi S., Lu J., Sun M., Li Z., Zhang H., Neilly M. B., Wang Y., Qian Z., Jin J., Zhang Y., Bohlander S. K., Le Beau M. M., Larson R. A., Golub T. R., Rowley J. D., Chen J. (2007). MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 104, 19971–19976 10.1073/pnas.0709313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliazza A., Bosch F., Komatsu H., Cayanis E., Martinotti S., Toniato E., Guccione E., Qu X., Chien M., Murty V. V. V., Gaidano G., Inghirami G., Zhang P., Fischer S., Kalachikov S. M., Russo J., Edelman I., Efstratiadis A., Dalla-Favera R. (2001). Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood 97, 2098–2104 10.1182/blood.V97.7.2098 [DOI] [PubMed] [Google Scholar]
- Mishra P. J., Song B., Mishra P. J., Wang Y., Humeniuk R., Banerjee D., Merlino G., Ju J., Bertino J. R. (2009). MiR-24 Tumor suppressor activity is regulated independent of p53 and through a target site polymorphism. PLoS ONE 4, e8445. 10.1371/journal.pone.0008445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notari M., Neviani P., Santhanam R., Blaser B. W., Chang J.-S., Galietta A., Willis A. E., Roy D. C., Caligiuri M. A., Marcucci G., Perrotti D. (2006). A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood 107, 2507–2516 10.1182/blood-2005-09-3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U. A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., Guigo R., Shiekhattar R. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U. A., Nielsen F. C., Lund A. H. (2008). MicroRNA-10a binds the 52UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Pasmant E., Laurendeau I., Héron D., Vidaud M., Vidaud D., Bièche I. (2007). Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 67, 3963–3969 10.1158/0008-5472.CAN-06-2004 [DOI] [PubMed] [Google Scholar]
- Pekarsky Y., Santanam U., Cimmino A., Palamarchuk A., Efanov A., Maximov V., Volinia S., Alder H., Liu C.-G., Rassenti L., Calin G. A., Hagan J. P., Kipps T., Croce C. M. (2006). Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 66, 11590–11593 10.1158/0008-5472.CAN-06-3613 [DOI] [PubMed] [Google Scholar]
- San Jose-Eneriz E., Roman-Gomez J., Jimenez-Velasco A., Garate L., Martin V., Cordeu L., Vilas-Zornoza A., Rodriguez-Otero P., Calasanz M. J., Prosper F., Agirre X. (2009). MicroRNA expression profiling in imatinib-resistant chronic myeloid leukemia patients without clinically significant ABL1-mutations. Mol. Cancer 8, 69. 10.1186/1476-4598-8-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C. J. (1998). Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12, 2984–2991 10.1101/gad.12.19.2984 [DOI] [PubMed] [Google Scholar]
- Tay Y., Zhang J., Thomson A. M., Lim B., Rigoutsos I. (2008). MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128 10.1038/nature07299 [DOI] [PubMed] [Google Scholar]
- Valk P. J. M., Verhaak R. G. W., Beijen M. A., Erpelinck C. A. J., Van Doorn-Khosrovani S. B. V. W., Boer J. M., Beverloo H. B., Moorhouse M. J., Van Der Spek P. J., Lã Wenberg B., Delwel R. (2004). Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 350, 1617–1628 10.1056/NEJMoa040465 [DOI] [PubMed] [Google Scholar]
- Wagner L. A., Christensen C. J., Dunn D. M., Spangrude G. J., Georgelas A., Kelley L., Esplin M. S., Weiss R. B., Gleich G. J. (2007). EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood 109, 5191–5198 10.1182/blood-2006-07-036657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K. L., Li S., Muñoz-Cabello A. M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M. J., Zhou M.-M. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674 10.1016/j.molcel.2010.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A. P., Cui H. (2008). Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206 10.1038/nature06468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007). microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302, 1–12 10.1016/j.ydbio.2006.08.028 [DOI] [PubMed] [Google Scholar]
- Zhang H., Luo X.-Q., Zhang P., Huang L.-B., Zheng Y.-S., Wu J., Zhou H., Qu L.-H., Xu L., Chen Y.-Q. (2009a). MicroRNA patterns associated with clinical prognostic parameters and cns relapse prediction in pediatric acute leukemia. PLoS ONE 4, e7826. 10.1371/journal.pone.0007826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lian Z., Padden C., Gerstein M. B., Rozowsky J., Snyder M., Gingeras T. R., Kapranov P., Weissman S. M., Newburger P. E. (2009b). A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113, 2526–2534 10.1182/blood-2008-04-154138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Rice K., Wang Y., Chen W., Zhong Y., Nakayama Y., Zhou Y., Klibanski A. (2010a). Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 151, 939–947 10.1210/en.2009-1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhou Y., Klibanski A. (2010b). Isolation and characterization of novel pituitary tumor related genes: a cDNA representational difference approach. Mol. Cell. Endocrinol. 326, 40–47 10.1016/j.mce.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhou Y., Mehta K. R., Danila D. C., Scolavino S., Johnson S. R., Klibanski A. (2003). A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 88, 5119–5126 10.1210/jc.2003-030222 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D. L., Gejman R., Ansell P. J., Zhao J., Weng C., Klibanski A. (2007). activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742 10.1074/jbc.M611329200 [DOI] [PubMed] [Google Scholar]