Abstract
Distinctive histological variants of lung cancer are increasingly recognized to have specific genetic changes that impact tumor biology and response to therapy. In this study, we evaluated true papillary adenocarcinoma of the lung, proposed as a distinct diagnostic category with relatively poor response to therapy, to determine whether these tumors also have specific molecular alterations that would affect sensitivity to chemotherapy. Specifically, we measured protein levels of P53, ERCC1 and RRM1 by immunohistochemistry and evaluated the KEAP1 gene for mutations, correlating mutations of this gene with total and nuclear expression of the NRF2 transcription factor. We found high levels of P53 in 23 of the 55 specimens (41.8%), similar to the rate of P53 gene mutations observed in general for pulmonary adenocarcinoma, and levels of ERCC1 and RRM1 also showed distributions similar to those reported generally for NSCLC. However, KEAP1 alterations were observed at a significantly higher frequency in papillary adenocarcinoma tumors (60%) than what has been reported previously for NSCLC (3% to 19%). These mutations of KEAP1 were associated with increased nuclear accumulation of NRF2 in tumors, as expected for functional alterations. Thus, high rates of KEAP1 mutations and NRF2 overexpression in true papillary adenocarcinoma could be related to poor prognosis and chemotherapy resistance. Furthermore, this distinctive molecular characteristic supports the recognition of true papillary adenocarcinoma as a diagnostic entity.
Keywords: Pulmonary papillary adenocarcinoma, KEAP1 mutation, NRF2 expression, NSCLC
Introduction
Specific mutations in lung cancer appear to be relatively restricted to particular histologically defined phenotypes. For example, mutations of the RB gene are common in small cell cancers but rare in non small lung cell cancers (NSCLC), whereas mutations (or methylation) of P16 are characteristic in the non-small cell cancers [1]. Among NSCLCs, coding mutations of the KRAS and EGFR genes are relatively restricted to adenocarcinoma [2], with EGFR mutations being particular prevalent in adenocarcinoma tumors with the bronchioalveolar histological pattern of growth [3]. Thus, genomic data for lung cancer cannot be generalized across various histological types of the disease.
True papillary adenocarcinoma has been recently recognized as a distinctive histomorphologic subtype of lung adenocarcinoma by World Health Organization (WHO) classification [4], and the molecular characteristics of this subtype of lung cancer have not yet been defined. This variant appears to have a less favorable prognosis and poorer response to chemotherapeutic agents [5] than other forms of pulmonary adenocarcinoma, and thus recognition of this histological pattern of lung cancer could have clinical significance. However, many pathologists do not routinely recognize the morphological characteristics of papillary adenocarcinoma, and the actual incidence and significance of correctly classifying true papillary cancers is still not known.
While molecular characteristics of pulmonary papillary adenocarcinoma are undefined, several molecular alterations might be suspected based on associations with poor response of lung cancers to commonly-used agents. For example, aberrant p53 expression was found to be associated with resistance to cisplatin-based chemotherapy [6], and more recently, high expression levels of the excision repair cross-complementation 1 (ERCC1) and ribonucleotide reductase M1 (RRM1) genes have been associated with poor prognosis and poor response of non-small cell lung cancers (NSCLC) to cisplatin and gemcitabine [7, 8, 9]. In addition, the Kelch-like ECH-associated protein 1 (KEAP1) and the nuclear factor erythroid-2 related factor 2 (NRF2) signaling pathway have been identified to play important roles in the cellular response to oxidative stress, electrophiles and xenobiotics [10, 11, 12, 13], and KEAP1 mutations lead to constitutively active NRF2 and subsequent protection of cancer cells from chemotherapeutic drugs [14, 15] The frequency of these mutations in lung cancers is still uncertain; our previous study and others have shown that KEAP1 gene mutations occur in approximately 20% of NSCLCs (particularly in adenocarcinoma) [16, 17], while a more recent report found only one mutation among 31 cases of NSCLC [18]. Undoubtedly, these variations in reported rates of KEAP1 mutations are due, at least in part, to differences in lung cancer phenotypes evaluated in the various studies.
In this investigation, we first evaluated levels of p53, ERCC1, and RRM1 using immunohistochemistry (IHC), then, evaluated the frequency and types of KEAP1 mutations using direct sequencing methods, and correlated KEAP1 mutation status with patterns of NRF2 expression in these tumors. These studies are intended to both identify mechanisms that might impact the clinical behavior of these cancers, and also to determine whether these cancer do have distinctive molecular characteristics that would support the recognition of this category of lung cancer.
Materials and Methods
Pulmonary papillary adenocarcinoma samples
We identified a total of 58 cases of true pulmonary papillary adenocarcinoma in the surgical pathology archives of The Johns Hopkins Hospital using the diagnostic criteria adapted by the WHO [1, 2], and a tissue microarray (TMA) was prepared using core samples (1 mm diameter, 3 cores/ case) of paraffin tissue blocks from individual cases. Adequately preserved DNA for sequencing was available from tissue samples from 20 of these cases. Use of human tissue and clinical information was reviewed and approved by the Institutional Review Board for Human Subjects.
Immunohistochemistry
For P53, a primary polyclonal antibody of goat anti-human C-terminal peptide (C-19, Santa Cruz Biotechnology, Santa Cruz, California) was used at 1:50 dilution. A primary rabbit anti-human polyclonal antibody raised against a C-terminal RRM1 peptide and a mouse anti-human monoclonal antibody raised against the full length ERCC1 protein (Abcam Inc., Cambridge, Massachusetts, United States) were both used at 1:100 dilutions. To stain for NRF2, a rabbit polyclonal antibody (C-20, Santa Cruz Biotechnology, Santa Cruz, California) was used at 1:250 dilution. For all antibodies, after washing, a secondary antibody conjugated with peroxidase was applied to detect and visualize the specific antigen-antibody complexes using LASB System-HRP assay kit (Dako, California). Both positive and negative controls were included in the procedures.
DNA Purification, amplification, and sequence analysis
Primers for PCR amplification and sequencing were designed in the area of exon 3 of KEAP1 [16] and synthesized by Invitrogen (Carlsbad, California, United States), and amplification of DNA from early passage A549 cells (American Type Culture Collection, Manassas, Virginia, USA) and from primary tumor samples was performed using Taq polymerase (Qiagen, California, United States) as previously described [16]. All potential genetic alterations were confirmed by bi-directional sequencing and testing a second, independent DNA sample from the tumor. All chromatograms alterations of KEAP1 sequences were validated by a repeat sequencing and re-analyzed by manual review.
Results
Characteristics of papillary adenocarcinomas
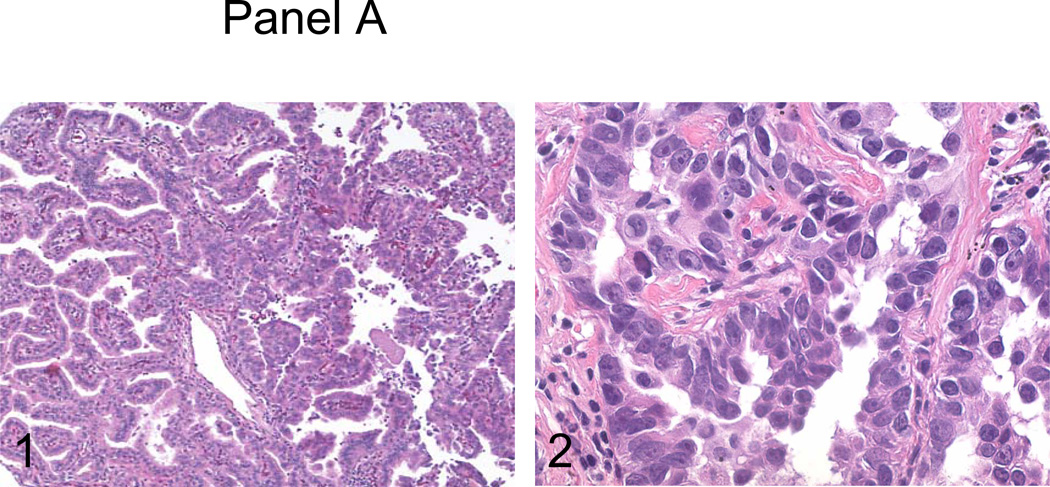
All cases selected for this study were characterized morphologically by numerous papillary structures with central vascular cores constituting at least 75% of the tumor volume (Figure 1, panel A1). The papillary structures were further complicated by tertiary branches and tufts, and consisted of individual tumor cells with high nuclear to cytoplasm ratio and markedly atypical nuclei (Figure 1, panel A2). Among patients with available clinical information, 46 of 49 (93.9%) were current or former smokers. Cases included 15 cases (27%) stage pT1, 28 cases (51%) stage pT2, two cases (4%) stage pT3, and 10 cases (18%) stage pT4.
Figure 1.
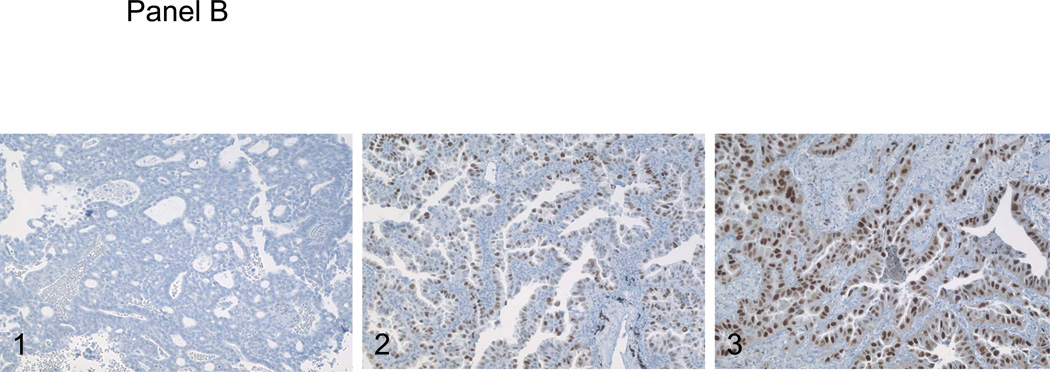
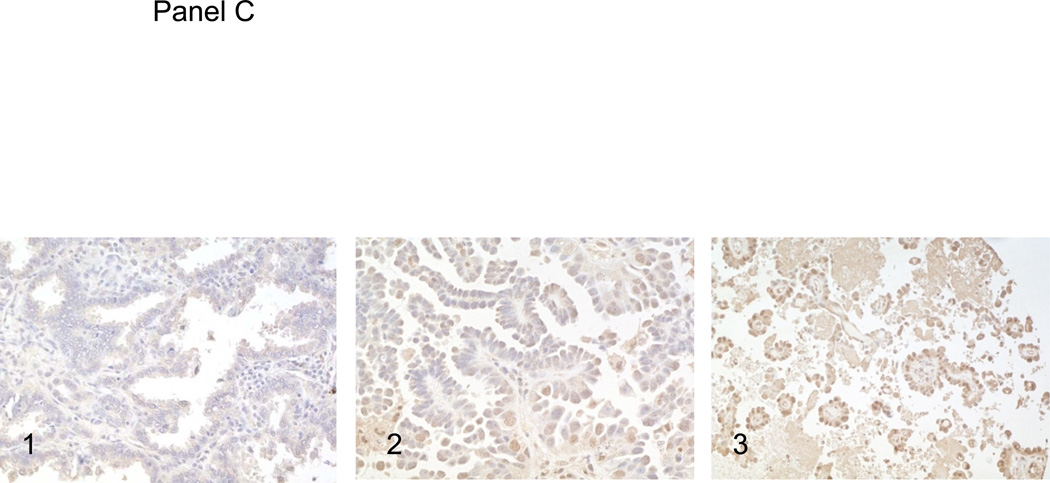
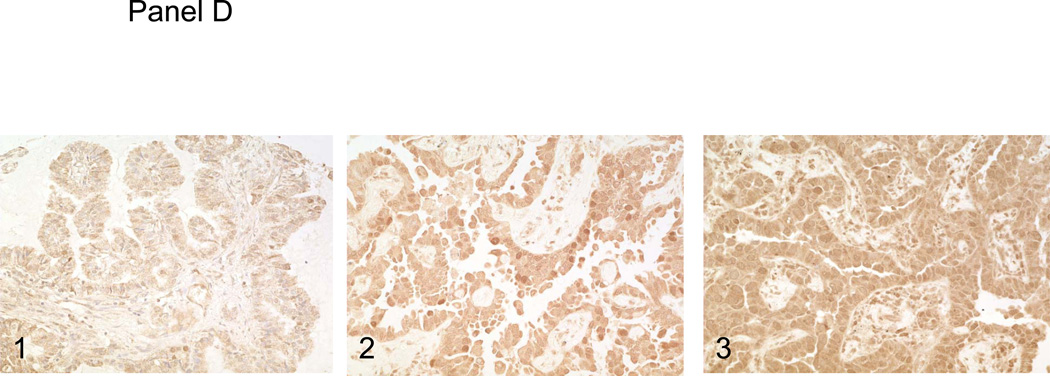
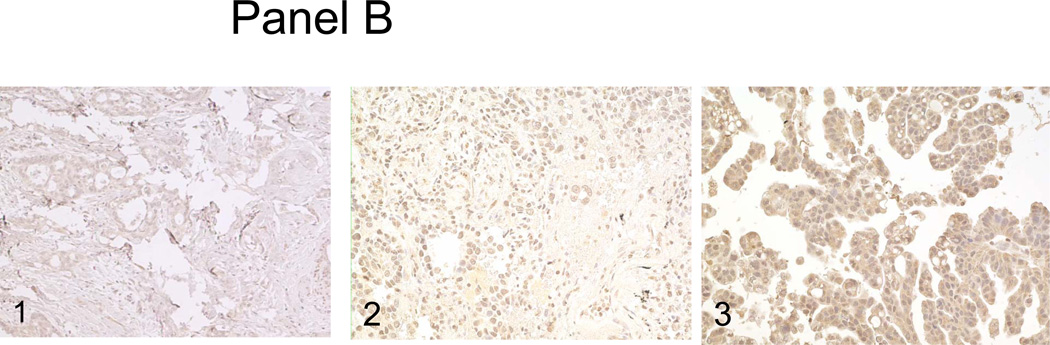
Panel A, the surgical resection specimen of pulmonary papillary adenocarcinoma. Panel B, C and D, immunohistochemical studies of P53, RRM1 and ERCC1. 1, background staining; 2, weakly staining; and 3, strongly staining.
Expression of P53, ERCC1, and RRM1 in papillary adenocarcinoma
Using immunohistochemistry to estimate the frequency of P53 mutations in pulmonary papillary adenocarcinoma, we observed high levels of P53 expression (2+ or 3+) in 23 of 55 (41.8%) of the cases of pulmonary papillary adenocarcinoma (Figure 1, panel B). This represents a frequency generally similar to the frequency of P53 mutations/overexpression reported previously for pulmonary adenocarcinoma in general. For RRM1 expression, 5/28 cases (17.6%) were negative, 15/28 cases (53.6%) stained weakly positive, and 8/28 cases (28.5%) stained strongly positive (Figure 1, panel C). For ERCC1, 5/28 cases (17.6%) of tumors were negative, 18/28 cases (64.2%) stained weakly positive, and 5/28 cases (17.9%) stained strongly positive (Figure 1, panel D). Although these data are only semi-quantitative, it is evident that the distributions of staining intensities for these two proteins are not significantly different than those reported for non-small cell cancer in general [5].
Mutations of KEAP1 in papillary adenocarcinoma
To determine the frequency of mutations in KEAP1 in pulmonary papillary adenocarcinoma, we amplified and sequenced exon 3 of the KEAP1 gene in DNA derived from the 20 tumor samples with suitable frozen tissue. In all, 12 of the 20 cases (60%) showed mutations of KEAP1, which is significantly more frequent than the 18.5% of cases (10 of 54 cases) with KEAP1 mutations that we observed in non-selected cases of pulmonary non-small cell carcinomas (p<0.05) and strikingly more frequent than the one of 31 cases more recently reported.
Representative electropherograms are shown in Figure 2, panel A. overall, four of the 12 mutations (cases P4, P5, P6 and P8) were characterized by GCC replacing TTA at the codon corresponding to the 367th amino acid in the Kelch domain of KEAP1, which leads to premature termination and truncated KEAP1 protein (Figure 2, panel A1). Three tumors (P17, P18, and P19) showed mutations that replace tyrosine with phyenylalanine in the 334th amino acid, resulting in a frameshift in the Kelch domain of KEAP1 (Figure 2, panel A2). Three tumors (P2, P10 and P14) showed insertion of a stop-codon at the position of the 346th amino acid, leading to premature termination and truncated KEAP1 protein (Figure 2, panel A3). Two tumors (P3 and P15) showed an insertion of A and G, that results in premature termination and truncated KEAP1 protein (Figure 2, panel A4). Finally, three tumors (P5, P6 and P8) showed amino acid substitution of glycine at 333th to cysteine in the Kelch domains of KEAP1. Interestingly, 9 of the 12 mutations would result in premature termination and truncated KEAP1 protein, and 3 of the 12 tumors had more than one mutation affecting the KEAP1 gene.
Figure 2.
Panel A, electropherograms of KEAP1 mutations. 1, the replacement of GCC to TTA at 367th amino acid in the Kelch domain., 2, the replacement of 334th amino acid of tyrosine with phyenylalanine, 3, the insertion of a stop-codon at 346th amino acid, 4, the insertion of A and G. Panel B, immunohistochemical studies of NRF2. 1, the normal lung parenchyma. 2, the tumor cells with wild-type KEAP. 3, the tumor cells with KEAP1 mutations
Abnormal expression and distribution of NRF2 protein in papillary adenocarcinoma
To evaluate expression and cellular localization of NRF2 protein in pulmonary papillary adenocarcinoma tumors, we performed immunohistochemistry using a polyclonal anti-NRF2 antibody. The overall expression of NRF2 protein for 55 pulmonary papillary adenocarcinoma tumors is summarized in Table 1. Normal lung parenchyma shows weak, and predominantly cytoplasmic, staining of NRF2 protein (Figure 2, panel B1), and eight tumor tissues with wild-type KEAP1 also demonstrated predominantly cytoplasmic staining of NRF2 (Figure 2, panel B2). By contrast, all 12 tumor tissues harboring KEAP1 mutations showed an increased level of NRF2 staining both in the nucleus and cytoplasm (Figure 2, panel B3), suggesting that there is increased nuclear accumulation and activation of NRF2 in tumor cells in the setting of KEAP1 mutation.
Table 1.
Summary of NRF2 and P53 expressions in the pulmonary papillary adenocarcinomas.
| Stage (cases) | Score | P53 stain Case numbers |
NRF2 stain Case numbers |
|---|---|---|---|
| pT1 (n=15) | |||
| 0 | 6 | 5 | |
| 1 | 4 | 3 | |
| 2 | 5 | 7 | |
| pT2 (n=28) | |||
| 0 | 8 | 3 | |
| 1 | 10 | 6 | |
| 2 | 10 | 9 | |
| pT3 (n=2) | |||
| 0 | 1 | 0 | |
| 1 | 1 | 1 | |
| 2 | 0 | 1 | |
| pT4 (n=10) | |||
| 0 | 0 | 0 | |
| 1 | 2 | 3 | |
| 2 | 8 | 7 |
Note: Staining was scored semi-quantitatively using a three tier system: 0, undetectable; 1+, weakly positive; 2+, moderately to intensely positive.
We then explored the possibility of an association between mutations of KEAP1 and p53. Among the 8 tumor tissues with wild-type KEAP1, 3 cases (37.5%) demonstrated high expression of P53 protein. In contrast, 8 of 12 (67%) cases with KEAP1 mutations showed high expression of P53 protein, consistent with mutations of the P53 gene. Thus, occurrences of mutations of P53 and KEAP1 are positively correlated (difference between groups significant by chi-square test, p=0.003).
Discussion
We evaluated protective pathways in pulmonary papillary adenocarcinoma, a histomorphologically distinctive subtype of lung adenocarcinoma. We found that the frequency of high expression levels of P53, ERCC1, and RRM1 are similar in cases of pulmonary papillary to those observed previously in other types of pulmonary adenocarcinoma. However, we found a remarkably high KEAP1 mutation frequency (12/20 cases, 60%) in pulmonary papillary adenocarcinoma, which is significantly greater than our previously reported rates of 18% for pulmonary non-small cell carcinoma in general and 28% for adenocarcinoma [16], and strikingly higher than the frequency of approximately 3% recently reported for NSCLC [18]. As expected, KEAP1 mutations were associated with increased nuclear expression of the NRF2 transcription factor in these papillary cancers, but some cases without KEAP1 mutations also displayed increased NRF2 expression and nuclear localization. These high rates of KEAP1 mutations and NRF2 overexpression in true papillary adenocarcinoma of the lung could be related to the reported poor prognosis and chemotherapy resistance of these cancers, and we propose that this distinctively high frequency of KEAP1 mutations supports the recognition of true papillary adenocarcinoma as a diagnostic entity.
The KEAP1 alterations found in pulmonary papillary adenocarcinoma tumors included insertions, deletions, and frameshifts, with the most frequent type of mutation in pulmonary papillary adenocarcinoma (involving 58.3% (7/12) of the mutations) consisting of a single base insertion that would result in a premature stop codon and thus a truncated KEAP1 protein. Remarkably, 25% (3/20) of the cases with mutations were found to have more than one mutation affecting the KEAP1 gene. Taken together with our previous data and others, our present study indicates that KEAP1 gene alterations are frequent in pulmonary adenocarcinomas, particularly in papillary adenocarcinomas.
We also found that cancer cells of pulmonary papillary adenocarcinoma demonstrate an increased accumulation of NRF2 by IHC staining, and in tumors with KEAP1 mutations, both nuclear and cytoplasmic accumulations of NRF2 are markedly increased. These data suggest that frequent mutations of KEAP1 in pulmonary papillary adenocarcinoma could significantly reduce the sensitivity of these cancers to chemotherapy. Increased NRF2 staining was also demonstrated in several cases of pulmonary papillary adenocarcinoma with wild-type KEAP1, suggesting that there are other mechanisms that could contribute to stabilization and induction of NRF2. Possible mechanisms for this include: 1) increased levels of the nuclear oncoprotein prothymosin, which regulates the intranuclear dissociation of KEAP1-NRF2 complex [19]; 2) somatic mutations in Nrf2 gene which impair its binding with Keap1 [19,20]; 3) alternative splicing of KEAP1, resulting in a nonfunctional KEAP1 protein in cancer cells [21]; 4) methylation of CpG-rich regions of KEAP1 promoter, resulting in decreased expression of this protein [22]; and 5) phosphorylation of NRF2 by protein kinase(s) associated with the MAPK/ERK signaling cascade, leading to stabilization of NRF2 [23]. Evidently, mechanisms for regulation of KEAP1-NRF2 pathway are complex, and further studies are needed to understand exaggerated NRF2 responses in cancer cells even in the absence of KEAP1 mutations.
In summary, our finding of frequent KEAP1 mutations in pulmonary papillary adenocarcinoma and the association between loss of functional KEAP1 and increased NRF2 activity suggest important roles for the KEAP1-NRF2 pathway in the regulation of antioxidants, detoxification enzymes, and drug transporters activity in this subset of pulmonary adenocarcinoma. The relatively disproportionate frequency of KEAP1 mutations in pulmonary papillary adenocarcinoma supports the recognition of these cancers as a unique subset of lung adenocarcinoma.
Acknowledgement
QKL is the recipient of the Research Seed Grant Award from the American Society of Cytopathology. EG, SB and AS are supported by the National Cancer Institute (P50 CA058184) and the Flight Attendant Medical Research Institute, and SB and AS are also supported by RO1 CA140492. Authors also acknowledge Ellen Tully’s technical support. This research is proved by the Institutional Review Board.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Wikenheiser-Brokamp KA. Retinoblastoma regulatory pathway in lung cancer. Curr Mol Med. 2006;6:783–793. doi: 10.2174/1566524010606070783. Review. [DOI] [PubMed] [Google Scholar]
- 2.Harris TJR, McCormick F. The molecular pathology of cancer. Nat Rev Clin Oncol. 2010;7:251–265. doi: 10.1038/nrclinonc.2010.41. [DOI] [PubMed] [Google Scholar]
- 3.Okada A, Shimmyo T, Hashimoto T, Kobayashi Y, Miyagi Y, Ishikawa Y, Nakagawa K, Hayashi J, Tsuchiya E. Predictive advantage of a cell type classification for pulmonary adenocarcinoma coupled with data for p53, K-ras and EGFR alterations. Cancer Sci. 2010;101:1745–1753. doi: 10.1111/j.1349-7006.2010.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colby TV, Noguchi M, Henschke C, Vazquez MF, Geisinger K, Yokose T, Ohori P, Rami-Porta R, Franks T, Shimosato Y, Matsuno Y, Khoor A, Westra WH, et al. Adenocarcinoma. In: Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. World Health Organization classification of tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon: IACR Press; 2003. pp. 35–44. [Google Scholar]
- 5.Silver SA, Askin FS. True papillary carcinoma of the lung: a distinct cliniclpathologic entity. Am J Surg Pathol. 1997;21:43–51. doi: 10.1097/00000478-199701000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Rusch V, Klimstra D, Venkatraman E, Oliver J, Martini N, Gralla R, Kris M, Dmitrovsky E. Aberrant p53 expression predicts clinical resistance to cisplatin-based chemotherapy in locally advanced non-small cell lung cancer. Cancer Res. 1995;55:5038–5042. [PubMed] [Google Scholar]
- 7.Bepler G, Kusmartseva I, Sharma S, Gautam A, Cantor A, Sharma A, Simon G. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non–small-cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 9.Ceppi P, Volante M, Novello S, Rapa I, Danenberg KD, Danenberg PV, Cambieri A, Selvaggi G, Saviozzi S, Calogero R, Papotti M, Scagliotti GV. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 11.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, Gabrielson G, Feinstein E, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1–2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda H, Tanaka T, Takahama U. Cisplatin generates superoxide anion by interaction with DNA in a cell-free system. Biochem Biophys Res Commun. 1994;203:1175–1180. doi: 10.1006/bbrc.1994.2306. [DOI] [PubMed] [Google Scholar]
- 18.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, Minna JD, Stewart DJ, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki H, Nonaka M, Fujii Y, Yamakawa Y, Fukai I, Kiriyama M, Sasaki M. Expression of the prothymosin-a gene as a prognostic factor in lung cancer. Surg Today. 2001;31:936–938. doi: 10.1007/s005950170040. [DOI] [PubMed] [Google Scholar]
- 20.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, Eom HS, Yoo NJ, Lee SH. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220:446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, Kong AN. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem. 2000;275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]