Abstract
The sister phyla dinoflagellates and apicomplexans inherited a drastically reduced mitochondrial genome (mitochondrial DNA, mtDNA) containing only three protein-coding (cob, cox1, and cox3) genes and two ribosomal RNA (rRNA) genes. In apicomplexans, single copies of these genes are encoded on the smallest known mtDNA chromosome (6 kb). In dinoflagellates, however, the genome has undergone further substantial modifications, including massive genome amplification and recombination resulting in multiple copies of each gene and gene fragments linked in numerous combinations. Furthermore, protein-encoding genes have lost standard stop codons, trans-splicing of messenger RNAs (mRNAs) is required to generate complete cox3 transcripts, and extensive RNA editing recodes most genes. From taxa investigated to date, it is unclear when many of these unusual dinoflagellate mtDNA characters evolved. To address this question, we investigated the mitochondrial genome and transcriptome character states of the deep branching dinoflagellate Hematodinium sp. Genomic data show that like later-branching dinoflagellates Hematodinium sp. also contains an inflated, heavily recombined genome of multicopy genes and gene fragments. Although stop codons are also lacking for cox1 and cob, cox3 still encodes a conventional stop codon. Extensive editing of mRNAs also occurs in Hematodinium sp. The mtDNA of basal dinoflagellate Hematodinium sp. indicates that much of the mtDNA modification in dinoflagellates occurred early in this lineage, including genome amplification and recombination, and decreased use of standard stop codons. Trans-splicing, on the other hand, occurred after Hematodinium sp. diverged. Only RNA editing presents a nonlinear pattern of evolution in dinoflagellates as this process occurs in Hematodinium sp. but is absent in some later-branching taxa indicating that this process was either lost in some lineages or developed more than once during the evolution of the highly unusual dinoflagellate mtDNA.
Keywords: mitochondrion, Apicomplexa, RNA editing, organelle genome
Introduction
Mitochondrial genome evolution has seemingly occurred in fits and starts across eukaryotic diversity, with some lineages displaying little change in this relict bacterial genome and others radically reorganizing it (Feagin 2000; Burger et al. 2003; Gray et al. 2004; Spencer and Gray 2010; Vlcek et al. 2010). Two sister phyla, the dinoflagellate algae and apicomplexan parasites, provide rich material for studying mitochondrial genome evolution as both demonstrate remarkable modification of this genome. Dinoflagellates include major ocean primary producers, essential symbionts of reef building corals, and a variety of parasites of vertebrates, invertebrates, and protistan hosts (Hackett et al. 2004; Stentiford and Shields 2005). Apicomplexans are almost exclusively parasitic and include such notorious human pathogens as the malaria causing Plasmodium spp. Despite great diversity of cell form and lifestyle shown by these two phyla, molecular phylogenies and the presence of common ultrastructural features firmly indicate their sister relationship (Cavalier-Smith 1991; Wolters 1991; Van de Peer and De Wachter 1997; Fast et al. 2002; Burki et al. 2007; Rodriguez-Ezpeleta et al. 2007; Gould et al. 2008).
The common ancestor of apicomplexans and dinoflagellates inherited a drastically reduced mitochondrial genome with only three protein-coding genes (cytochrome b, cob; cytochrome oxidase subunit 1, cox1; and cytochrome oxidase subunit 3, cox3) and two ribosomal RNAs (rRNAs), the small subunit (SSU) and large subunit (LSU) (Wilson and Williamson 1997; Nash et al. 2008; Waller and Jackson 2009). In both taxa, the rRNA genes are highly fragmented, with upwards of 20 genes encoding these fragments identified in the best characterized apicomplexan mitochondrial DNA (mtDNA), from Plasmodium falciparum (Feagin et al. 1997, and see Genbank accession M76611). In apicomplexans, single copies of protein-coding and rRNA genes are compactly encoded on the smallest known mitochondrial chromosome, at 6 kb (Wilson and Williamson 1997). In contrast, the mitochondrial genome (mtDNA) in dinoflagellates has undergone massive genome amplification and recombination, and this has generated multiple copies of each gene and gene fragments that are generally linked to one another in multiple different combinations (Norman and Gray 2001; Jackson et al. 2007; Nash et al. 2007; Slamovits et al. 2007; Kamikawa et al. 2009). Noncoding sequence has similarly undergone duplication and rearrangement, and in several dinoflagellate species contains inverted repeats with the potential to form secondary structures (Norman and Gray 2001; Jackson et al. 2007; Nash et al. 2007). This recombination and duplication has made assembly of genomic fragments impossible, and the ultimate size and overall organization of the dinoflagellate mtDNA remains unknown (Waller and Jackson 2009). Preliminary pulse field gel electrophoresis experiments suggest that the genome is spread over multiple linear chromosomes with an upper chromosome size limit of ∼30 kb (Nash et al. 2008).
In addition to genome expansion, dinoflagellates have radically modified the presentation and expression of their mitochondrial genes. Standard AUG start codons for transcripts of the three mtDNA protein-coding genes are missing (Jackson et al. 2007; Nash et al. 2007; Slamovits et al. 2007; Waller and Jackson 2009), and dinoflagellates have taken the additional and highly unusual step of abandoning standard terminator codons. Transcripts for cox1 and cob both lack known stop codons (or any apparent alternative), whereas in cox3 transcripts, a standard UAA stop codon is generated but only through oligoadenylation following an in-frame U nucleotide. In the dinoflagellate Karlodinium micrum (synonym: Karlodinium veneficum Ballantine, see Bergholtz et al. 2006) transcripts of cox3 also require messenger RNA (mRNA) trans-splicing, a process that joins two cox3 precursor transcripts which are encoded separately in the genome to form a complete coding sequence (Jackson et al. 2007; Waller and Jackson 2009); cox3 is also trans-spliced in all other later-branching dinoflagellates that have been investigated (Jackson CJ, Waller RF, unpublished data). Furthermore, extensive substitutional RNA editing (up to 6% of nucleotides) of mtDNA transcripts is seen in most dinoflagellates and has been detected in ∼25 species so far (Lin et al. 2002, 2008; Zhang and Lin 2005; Jackson et al. 2007). With the exception of alternative start codons, all of these characters are absent from apicomplexans.
Altogether, it is evident that mitochondrial genome evolution has taken a remarkably accelerated route within dinoflagellate radiation. From the relative simplicity of the apicomplexan state, dinoflagellates have dramatically expanded the genome size, gene copy number, and complexity of processes necessary to express these genes, albeit with no apparent increase in the function of this organelle genome. Insights into how and when this process of genome elaboration occurred require examination of taxa that diverged throughout dinoflagellate radiation. Most of the available data for dinoflagellate mtDNAs come from the later-branching dinophycean species (those possessing a typical dinokaryon Adl et al. 2005), and the unusual character states observed seem to be common to all these (Norman and Gray 2001; Jackson et al. 2007; Kamikawa et al. 2007, 2009; Nash et al. 2007). Some insights into the mtDNA of a lower branching taxon are available from the dinoflagellate Oxyrrhis marina, where additional novel character states are observed (Slamovits et al. 2007). In this study, we have performed an extensive characterization of the mtDNA from the basal dinoflagellate Hematodinium sp., to further investigate mtDNA character states in early dinoflagellate evolution. Hematodinium sp. is a parasitic dinoflagellate, belonging to the Syndiniales, and infects the hemolymph of crustaceans. It has a number of life stages, including several aflagellate feeding trophont and sporont stages and two motile transmissive dinospore life stages both with characteristic dinoflagellate transverse and longitudinal flagella (Appleton and Vickerman 1998). We have generated extensive genomic and transcriptomic data from the Hematodinium sp. mitochondrial genome, which together indicate that the Hematodinium sp. mtDNA shares many of the derived character states seen in later-branching dinoflagellates, while also exhibiting some notable exceptions which may reflect ancestral dinoflagellate states.
Materials and Methods
Cell Culture, Nucleic Acid Extraction, and mtDNA Cloning
Hematodinium sp. ex Nephrops norvegicus was cultured in the dark at 10 °C in Nephrops saline media, supplemented with 5% fetal calf serum and penicillin, streptomycin, and gentamycin (Appleton and Vickerman 1998). Karlodinium micrum was cultured in Guillard's f2 media at 16 °C on a 12-h light/12-h dark cycle. Cells were harvested by centrifugation (10 min, 2,600 g), and genomic DNA was extracted using Plant DNAzol (Invitrogen) according to the manufacturer's instructions. For K. micrum, total RNA was extracted using Trizol (Invitrogen). Polymerase chain reaction (PCR) was used to amplify Hematodinium sp. mtDNA fragments using oligonucleotides (20–24 nt) designed from mitochondrial genes identified in a cDNA library (see below) using BLAST. PCR products were cloned into pGEM-T Easy vector (Promega, Madison, Wisconsin) and fully sequenced. Analysis of DNA sequences was performed using the software package Sequencher 4.7 (Gene Codes Corporation, Ann Arbor, Michigan, USA) and Geneious Pro 5.1 (Biomatters). Protein alignments were performed using Geneious Pro 5.1. Genbank accessions: HE610721–HE610773.
Generation of cDNA Library
DNA-free total RNA was extracted from in vitro cultured Hematodinium sp. trophonts using the RNAqueous mini kit (Ambion), following the manufacturer's protocol. Approximately 40–100 ng of total RNA was used to generate ∼10 μg of cDNA, using the SMARTer PCR cDNA synthesis kit (Clontech). Amplified cDNA was sequenced using the 454 Titanium chemistry on a GS-FLX platform (Roche) by Genome Quebec (Montreal, Canada). Reads representing mtDNA transcripts were detected using BLAST within the software package Geneious Pro 5.1 (Biomatters).
Northern Blot Analysis
Hybridization probe templates for cob, cox1, and cox3 were generated using PCR from full-length cDNA for each gene cloned into pGEM-T Easy vector. The cob (932 nt), cox1 (1,118 nt), and cox3 (681 nt) probes corresponded to positions 99-1030, 248-1365, and 48-728 of respective full-length cDNAs for each gene encompassing ∼80% of the open reading frame for each respective gene. Probe templates for Hematodinium sp. and K. micrum rRNA Northerns were generated by PCR using genomic DNA. Hematodinium sp. probes for LSUA (119 nt), LSUE (181 nt), LSUF (95 nt), RNA8 (91 nt), SSUB (76 nt), SSUA1 (70 nt), and SSUA2 (63 nt) correspond to positions 37-155, 1-181, 15-109, 1-91, 1-76, 27-96, and 1-63 of respective cDNAs for each gene. Karlodinium micrum probes for RNA10 (95 nt), LSUG (110 nt), RNA7 (76 nt), and RNA8 (87 nt) correspond to positions 1-95, 1-97, 1-76, and 1-87 of respective cDNAs for each gene; LSUE and RNA2 probes were full-length. PCR fragments were purified from gels and random hexamer-based probes were constructed using the Prime-a-gene labeling system (Promega) and 32P labeled dATP, according to the manufacturer's instructions.
For cob, cox1, and cox3 Northerns, total RNA was extracted using Trizol (Invitrogen), separated on a 1% denaturing agarose/formaldehyde gel (5 μg per lane), transferred to Hybond N+ membrane (GE Healthcare) via Northern blotting according to the manufacturer's instructions, and RNA was cross-linked by UV irradiation. For rRNA fragment Northerns, equivalent total RNA was separated on 5% polyacrylamide/urea gels. Probe hybridization was performed overnight at 65 °C in modified Church's buffer (51 g Na2HPO4.2H20, 16.8 g anhydrous NaH2PO4, 4 ml of 0.5 M ethylenediaminetetraacetic acid, and 70 g sodium dodecyl sulphate per liter). Following hybridization, membranes were washed twice for 5 min each in 4 × saline sodium citrate (SSC) + 0.5% SDS then with the following series: 2 × SCC + 0.5% SDS; 4 × SSC + 0.5% SDS; 2 × SCC + 0.5% SDS; 4 × SSC + 0.5% SDS. All wash steps were carried out for 1 h at 65 °C. Membranes were visualized using X-ray film.
Circular Reverse Transcriptase PCR
Total Hematodinium sp. RNA was ligated head to tail using an RNA ligase (Promega) according to the manufacturer's instructions. First strand cDNA synthesis across the ligated mRNA ends was performed for cox1, cob, and cox3 using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions and using a gene specific primer designed for cDNA sequences (described above). Subsequently, PCR was performed with cDNA as template, using primers directed outward toward the gene termini. PCR products were ligated into the pGEM T-easy vector (Promega), cloned, and fully sequenced.
Results
In this investigation, genomic sequences from the Hematodinium sp. mitochondrial genome were generated by multiple PCR amplifications between mitochondrial genes. Transcriptome data were primarily generated by 454 deep sequencing of Hematodinium sp. cDNAs as well as individual RT-PCRs. Mitochondrial sequences from the transcriptome were further used to inform PCR primer design for the genomic study.
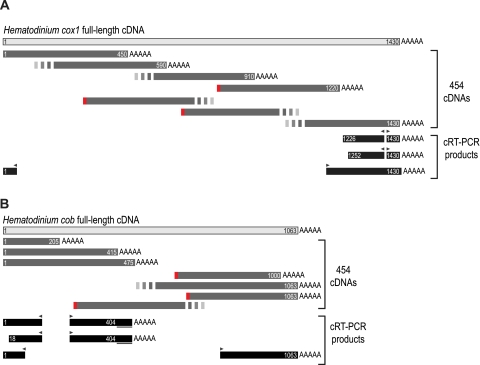
PCR Sampling of the Hematodinium sp. mtDNA
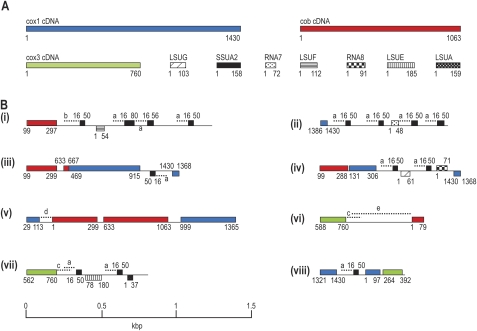
From characterized dinoflagellate and apicomplexan mtDNAs, only three protein-coding genes, cob, cox1, and cox3, are known, and these sequences were used for priming sites to PCR amplify Hematodinium sp. mtDNA. Haphazard primer combinations were used to test for various possible patterns of gene linkage and to generate further mtDNA sequence between these coding regions. Multiple PCR products resulted from these amplifications, and a selection of 26 mtDNA fragments were cloned and fully sequenced (see fig. 1 for representative fragments and supplementary fig. 1, Supplementary Material online for additional sequences). These amplicons totaled ∼17 kb of sequence and revealed many copies of gene fragments corresponding to the three protein genes (cob, cox1, and cox3) as well as coding sequence for several of the fragmented LSU and SSU rRNAs known from apicomplexans and other dinoflagellates (LSUE, LSUG, LSUA, LSUF, RNA7, RNA8, SSUA, and SSUB). Individual genes were represented multiple times (e.g., 16 cob, 15 cox1, and 29 SSUA sequences) and in numerous different genomic contexts. Most mtDNA amplicons contained internal gene fragments in addition to those used for priming (up to seven gene fragments represented per amplicon, fig. 1 fragment iv), revealing relatively tight packing of these gene fragments. For the protein-coding genes, 10 different fragmentation points were observed for cox1, 10 for cob, and two for cox3. There was no evidence in the mtDNA amplicons of additional known mitochondrial genes other than those five present in apicomplexans and other dinoflagellates mtDNAs (for the three protein and the two fragmented rRNAs). To search for novel mitochondrial transcripts encoded by these mtDNA elements, BLAST searches were made against the Hematodinium sp. transcriptome (described below). This test found no evidence of further genes or transcriptional elements.
FIG. 1.—
Schematic of Hematodinium sp. cDNAs (A) and eight mtDNA fragments generated by PCR (B). Gene sequences in (A) correspond to the longest cDNA data generated for each gene. Mitochondrial sequences are drawn to scale, with coding sequence in (B) on either the forward or the reverse strand indicated above or below the line, respectively. All mtDNA fragments represent single PCR amplicons. Colored blocks indicate protein-coding genes, textured black boxes indicate rRNA genes. cDNA lengths (in nucleotides) are indicated in (A), and corresponding nucleotide matches in PCR fragments are indicated in (B). Duplicated intergenic sequences (>99% identity) between PCR fragments are indicated by dashed lines and matching lowercase letters. Some lowercase letters have corresponding matches in supplementary figure 1 (Supplementary Material online).
PCR Amplification of Full-Length Genomic Sequence for Cob, Cox1, and Cox3
Although many different gene fragments were recovered using the above strategy, this sampling method did not generate any complete coding sequences for the known dinoflagellate mitochondrial genes. To test for intact genes, PCR was used to amplify full coding sequences, and for all three protein-encoding genes, complete genes were found in Hematodinium sp. genomic DNA. This included cox3, which is not found in intact form in most other dinoflagellates (O. marina, which apparently encodes a full-length genomic cox3 copy as part of a cob–cox3 fusion, is the only known exception) but instead relies on RNA trans-splicing to generate a complete mRNA from cox3 gene fragments (Jackson et al. 2007). Curiously, initial attempts to amplify full-length cob and cox1 generated multiple PCR products but those of correct length were revealed, by cloning and sequencing, as further linked gene fragments similar to those described above (but containing the cob or cox1 termini as single fragments). Only when gel isolation of these PCR products of appropriate size range was used as template for secondary nested PCRs were copies of the full-length cob and cox1 genes recovered. Comparison of the full-length gene sequences to the gene fragments showed that fragments represent near identical (on average 99.9%) copies despite their truncated form.
Intergenic Sequence in mtDNA Amplicons
The noncoding mtDNA sequence between gene fragments was compared between PCR amplicons and found to be highly duplicated and occurring in multiple genomic contexts, similar to that of the coding sequence (fig. 1B). Duplicates are also variable in length from one copy to another, but within copies, the sequence identity is typically close to 100%. The intergenic regions of other characterized dinoflagellate mtDNAs are known to contain conspicuous inverted repeat sequences predicted to be able to form secondary structures such as stem–loops (Norman and Gray 2001; Jackson et al. 2007; Nash et al. 2007). In Hematodinium sp., we find no evidence of this character with only very occasional inverted repeats occurring in both genic and intergenic sequence, likely by chance.
Characterization of mtDNA Transcripts
RNA was harvested from in vitro cultured cells representing multiple aflagellate trophont stages, and a cDNA library was generated using the SMARTer PCR cDNA synthesis kit (clonetech) that selects for polyadenylated, 5′-complete RNA copies. This library was 454 sequenced (Genome Quebec; 65,833 reads). Other dinoflagellate and apicomplexan mtDNA transcripts are known to be oligoadenylated and therefore, these data were used as a springboard for studies of Hematodinium sp. mitochondrial transcripts.
Protein-Coding cDNAs
From the 454 sequencing of the cDNA library, transcripts were found for the three protein-encoding genes: cob, cox1, and cox3 (147, 304, and 67 reads, respectively). Contigs made for each of these genes appeared to cover the full coding sequence when compared with homologues from other organisms. To verify the transcript extremities, circular reverse transcriptase PCR (cRT-PCR) was performed. Hematodinium sp. RNAs were first circularized using RNA ligase to join the 3′ transcript end to its 5′ end. Outward directed primers in the gene termini were then used to RT-PCR across the linked termini, and the 3′ oligo-A tail was used to identify the boundary. In each case, the mRNA termini recovered by cRT-PCR matched the full-length termini in the 454-sequenced cDNAs. Furthermore, the length of the oligo-A tail could be determined ranging from 5 to 17 nt for the three gene transcripts. In the basal dinoflagellate O. marina, each of its two mtDNA-encoded mRNAs (cox1 and a cob–cox3 fusion) receive a 5′ cap of approximately eight to nine U nucleotides, which are not encoded in the genome (Slamovits et al. 2007). In Hematodinium sp., examination of the 5′ ends of each transcript did not reveal any evidence of 5′ capping.
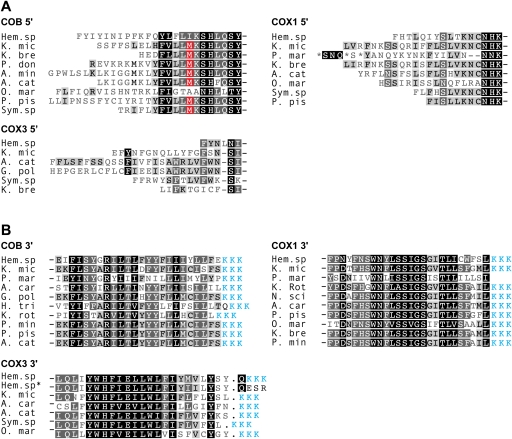
No dinoflagellates studied to date appear to employ standard AUG start codons for their cox1, cob, or cox3 mRNAs (Nash et al. 2008; Waller and Jackson 2009). We examined the 5′ ends of both genomic and cDNA sequences for each gene in Hematodinium sp. and found that AUG codons are absent in both the genome and gene transcript (fig. 2A). A conserved AUG present at the 5′ end of cob cDNAs in several dinoflagellate taxa (fig. 2A red boxes) is absent in Hematodinium sp. cob, and this is also absent in O. marina cob cDNAs. Thus, it is likely that these AUGs are not conserved functional start codons in those species that contain them and that translation begins further upstream in these transcripts (Jackson et al. 2007).
FIG. 2.—
Absence of conventional start and stop codons represented in protein alignments of dinoflagellate COB, COX3, and COX1. Predicted amino acid sequence termini represent (A) 5′ and (B) 3′ sequences from cDNAs. Sequence marked with “*” represents gDNA. Blue sequence represents the conceptual translation of 3′ oligo-A tails of mRNAs. Identical and similar residues are represented by black and gray background, respectively. Methionine residues highlighted in red correspond to potential start codons in cob cDNAs (see text). Dinoflagellate taxa and accession numbers: K. mic, Karlodinium micrum, this study; Hem. sp, Hematodinium sp., this study; K. bre, Karenia brevis, CO065693, EX967108, CO517356, CO059546; P. don, Prorocentrum donghaiense, FJ418633; A. min, Alexandrium minutum, GW798579; A. cat, Alexandrium catenella, AB374233, AB374235, AB374233; O. mar, Oxyrrhis marina, EF680823, EF680822; P. pis, Pfiesteria piscicida, AF463413, AF357518; Sym. sp., Symbiodinium sp., EH037582, EH037973, FE864081; G. pol, Gonyaulax polyedra, AF142470; A. car, Amphidinium carterae, CF064846, CF065669, CF064811; L. pol, Lingulodinium polyedrum, CD810189; K. rot, Katodinium rotundatum, EF036556, EF036582; H. tri, Heterocapsa triquetra, EF036554; P. min, Prorocentrum minimum, AY030285, AF463415; N. sci, Noctiluca scintillans, EF036583.
Standard stop codons are also not encoded in the mtDNA of other dinoflagellates for cob, cox1, or cox3, and so we examined the 3′ ends of genomic and cDNA Hematodinium sp. sequences for evidence of termination signals. Comparison of genomic and cDNA sequences revealed that cob and cox1 transcripts are oligoadenylated upstream of any standard stop codon present in the genome and therefore, these transcripts lack a stop (fig. 2B). We also searched for a codon that occurred only at the 3′ end of these genes and that could represent an alternative stop codon, but none were found. For cox3, however, a standard UAA stop codon is encoded in the mtDNA and oligoadenylation of cox3 transcripts occurs after this stop (by one nucleotide, fig. 2B). This contrasts with the absence of an encoded stop codon, but the de novo generation of a UAA stop codon, in cox3 transcripts in other dinoflagellates, and is therefore the first genome-encoded stop codon to be detected in any dinoflagellate mitochondrion.
rRNA cDNAs
Many short cDNAs corresponding to the fragmented dinoflagellate and apicomplexan mitochondrial rRNAs were identified in the Hematodinium sp. 454 data. In total, we identified cDNAs corresponding to seven LSU fragments, five SSU fragments, and two unassigned rRNA fragments (table 1). The fragmented rRNA species identified in Hematodinium sp. include all of the fragments cumulatively identified from all other dinoflagellate taxa (Waller and Jackson 2009), as well as five further fragments known from apicomplexans but not previously found in dinoflagellates: SSU rRNA fragments SSUA, SSUB, SSUD, and SSUF and unassigned rRNA fragment RNA6 (fig. 3). These data provide further strong support for the pattern of rRNA fragmentation being well conserved between dinoflagellates and apicomplexans. Figure 4 indicates the positions of these rRNA fragments on standardized rRNA structures.
Table 1.
Plasmodium falciparum rRNA Fragments, and Corresponding Homologues Represented by cDNA and gDNA Sequence in Hematodinium sp.
| P. falciparum rRNA Gene | Hematodinium sp. Homologue |
| LSUA | cDNA (159 bp) gDNA |
| LSUD | cDNA (77 bp) |
| LSUE | cDNA (180 bp) gDNA |
| LSUF | cDNA (112 bp) gDNA |
| LSUG | cDNA (103 bp) gDNA |
| RNA2 | cDNA (63 bp) |
| RNA10 | cDNA (103 bp) |
| SSUA | cDNA (96/157 bp) gDNA |
| SSUB | cDNA (135/110 bp) gDNA |
| SSUD | cDNA (51 bp) |
| SSUF | cDNA (65 bp) |
| RNA8 | cDNA (91 bp) gDNA |
| RNA6 | cDNA (63 bp) |
| RNA7 | cDNA (72 bp) gDNA |
Note.—Numbers in brackets correspond to Hematodinium rRNA fragments lengths, derived from 454-sequenced cDNAs (excluding poly-A tails).
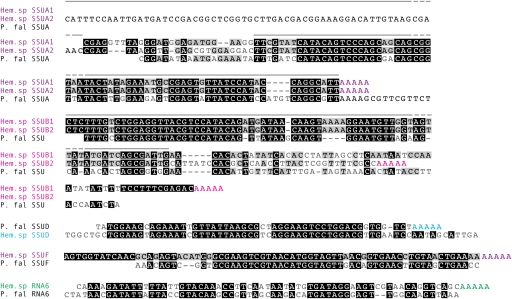
FIG. 3.—
Hematodinium sp. rRNA sequences aligned to those of their fragmented apicomplexan counterparts. Color groups indicate distinct rRNA fragments, with oligoadenylation also shown in color. Gray lines indicate positions of Northern blot probes. Hem. sp, Hematodinium sp.; P. fal, Plasmodium falciparum.
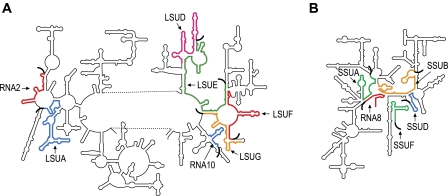
FIG. 4.—
Predicted locations of dinoflagellate LSU (A) and SSU (B) rRNA fragments, relative to Escherichia coli LSU and SSU secondary structures. The approximate locations of dinoflagellate rRNA fragment sequences are shown in color. Oligo(A) tails of dinoflagellate rRNA transcripts are represented as thick curved black lines.
Two of the new SSU rRNA gene fragments, SSUA and SSUB, were unusual in that each were represented by two different sets of transcript sequences with divergent sequence ends. SSUA1 and SSUA2 (represented by 46 and 19 reads, respectively) share 70 identical nucleotides and a conserved poly-A site that correspond well to apicomplexan SSUA but contain divergent sequence at the 5′ ends. Here, they share a nonidentical although semiconserved ∼25 nucleotide sequence, and SSUA2 contains a further 62 nt upstream of this region (fig. 3). SSUB1 and SSUB2 (each represented by four reads) present a similar situation, although the sequence divergence between these two is at the 3′ end. Again an identical sequence region (first 75 nt) is followed by a semiconserved region before complete sequence divergence, and the lengths of these extensions differ by ∼30 nt (fig. 3).
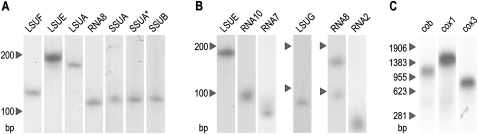
Total RNA was probed on Northern blots to determine if the rRNA fragments occur as the short molecules predicted from cDNA data or whether fragments are joined to form more conventional longer rRNA molecules. RNA was prepared from cultured trophonts stages. All probes (LSUA, LSUE, LSUF, RNA8, SSUA, SSUB) labeled a single band on Northern blots (fig. 5A). The observed sizes correspond with the predicted rRNA fragment lengths, although there is some uncertainty of poly-A length (the dinophycean dinoflagellate K. micrum indicates poly-A tails ranging from 1 to 18 bases on rRNAs, [Jackson CJ, Waller RF, unpublished data]). For SSUA and SSUB, only a single band was observed from trophont RNA, and for SSUB, size estimation suggests this is the smaller cDNA transcript type (SSUB2). A probe to conserved SSUA sequence and a specific probe to the 5′ extension of SSUA2 (fig. 3) labeled bands of the same size, indicating that the longer SSUA2 transcript is dominant in trophonts, but that this is running slightly faster than its true size (potentially due to persistent secondary structure despite denaturing conditions). The Hematodinium sp. Northern blots provided no evidence of splicing or ligation of rRNA fragments to form larger rRNAs. These data were compared with equivalent Northern blots of K. micrum, which confirmed the presence of small rRNA fragments (fig. 5B) corresponding in length to cDNA data and their counterparts in apicomplexan (Feagin et al. 1997; Jackson et al. 2007). Only one rRNA probe, for the SSU fragment RNA8, bound to a second larger band in addition to the predicted RNA8 band (of 88 nt).
FIG. 5.—
Northern blot analysis of Hematodinium sp. and Karlodinium micrum total RNA, probed with 32P labeled probes specific to Hematodinium mtDNA rRNA genes LSUF, LSUE, LSUA, RNA8, SSUA, SSUA2, SSUB (A), K. micrum rRNA genes LSUE, RNA10, RNA7, LSUG, RNA8, RNA2 (B), and Hematodinium cob, cox1, and cox3 (C). Total RNA was separated by electrophoresis. Blots were hybridized with gene-specific probes as labeled. The lane labeled “SSUA*” was hybridized with a probe specific to SSUA2 sequence. Size markers (indicated in nucleotides) are shown to the left of each individual blot.
RNA Editing
Substitutional RNA editing of mtDNA transcripts is known to occur in diverse dinoflagellates although it is thought to be absent from basal taxa (Lin et al. 2008). We searched for evidence of RNA editing in Hematodinium sp. mitochondrial transcripts by comparing genomic and cDNA sequences. Multiple, independently generated sequences for both genomic and cDNA sequences were made to verify sequence accuracy, before any edited changes were inferred. By this method, substitutional RNA editing was detected in cob, cox1, and cox3 transcripts, with six substitution types detected including transitions and transversions. The density and types of edits are shown for the protein-coding genes in table 2. Consistent with RNA editing in other dinoflagellate mtDNA transcripts (Lin et al. 2008), the majority of editing types in Hematodinium sp. are A to G, most editing affects the first and second codon positions, and most result in an encoded amino acid change.
Table 2.
Number, Density, and Type of RNA Editing Changes in cox1, cob, and cox3 from Hematodinium sp.
| Gene | Number/Density of Editing Sites | % at First or Second Codon Positions | % Causing an Amino Acid Change | Editing Type and % of Total for Individual Genes |
| Cox1 | 20 (1 per 72 bp) | 91% | 91% | A→G (60%), A→U (5%), G→A (5%), C→U (15%), C→G (5%), U→C (10%) |
| Cox3 | 10 (1 per 76 bp) | 90% | 100% | A→G (100%) |
| Cob | 11 (1 per 93 bp) | 75% | 70% | A→G (73%), C→U (9%), U→C (18%) |
Transcription of Gene Fragments and Polycistronic Transcripts
The majority of reads from the 454-sequenced cox1, cox3, and cob cDNAs corresponded to full-length transcripts, however, for each gene, several reads indicated the presence of truncated transcripts. For example, for cox1 (1,430 nt in length), we found reads polyadenylated at position 1220, 910, 590, and 450 (10, 3, 4, and 1 reads, respectively) (fig. 6). Similarly for cob (1,023 nt), premature poly-A sites occurred at positions 1000, 475, 415, and 205 (1, 9, 3, and 1 reads, respectively), and for cox3 (760 nt), truncation occurred at positions 655 and 445 (3 and 1 reads, respectively). These premature poly-A tails did not appear to be artifactual due to mispriming of the poly-T primer during cDNA synthesis because: 1) they did not occur in A-rich regions and 2) where A-rich gene regions did occur no premature polyadenylation was found.
FIG. 6.—
Schematic of Hematodinium sp. cox1 (A) and cob (B) truncated transcripts generated by 454 sequencing and cRT-PCR. For 454 reads, unique 5′ adaptors are shown by red squares. Broken lines at the 5′ ends of 454 reads indicate sequences that did not have a unique 5′ adaptor sequence (see text), and so the 5′ termini of these sequences is uncertain. Broken lines at the 3′ ends of 454 reads indicate reads that lacked an oligo-A tail. In cRT-PCR products, primer sites are shown by black arrowheads. Underlined sequence at the 3′ end of cob cRT-PCR products represents sequence that does not encode cob and is presumably intergenic.
Evidence of 5′-truncated gene transcripts was also observed. cDNA synthesis entailed adding a short unique adaptor sequence to the 5′ ends of completed reverse transcripts (incomplete transcripts were excluded from this process). Although this adaptor was evident at the 5′ end of full-length transcripts, 5′-truncated transcripts were evident by the presence of the adaptor sequence at intermediate positions in each gene sequence (fig. 6). To further test that these 5′-truncated and prematurely oligoadenylated cDNAs are present in the Hematodinium sp. mitochondrion, cRT-PCR was carried out on Hematodinium RNA to independently generate transcript end data. Outward directed primers were used from within several regions of cob and cox1 coding sequence. This method recovered products that corresponded to either 5′-truncations, premature polyadenylation or both (fig. 6). Interestingly, all of these truncated transcripts exhibit postedited nucleotides at RNA editing sites.
In addition to cDNAs corresponding to single gene products, a number of Hematodinium sp. cDNAs were identified which contained fragments of more than one mitochondrial gene. In the 454 data, we identified >10 different polycistronic transcripts (fig. 7). Typically, these transcripts contained fragments rather than complete genes, and although oligo(A) tails occurred in some cases at the correct 3′ terminus of a gene fragment (if that was present), other times it occurred at seemingly random points in gene and intergenic sequence. The presence of polycistronic transcripts was independently verified by using RT-PCR to amplify between gene sequences (not shown). Using this approach identical linkages to those seen in the 454 data were observed. Within gene sequence of the polycistronic transcripts, some RNA editing sites exhibited postedited nucleotides, whereas some sites contain the preedited nucleotide, indicating incomplete RNA editing of some of these molecules. In most cases, gene fragments were encoded on the same DNA strand, although on two transcripts antisense sequence for one gene fragment was found.
FIG. 7.—
Schematic of Hematodinium sp. polycistronic cDNAs from Hematodinium 454 data. Mitochondrial sequences represent individual 454 reads. Mitochondrial sequences are drawn to scale, with coding sequence in on either the forward or reverse strand indicated above or below the line, respectively. Colored blocks indicate protein-coding genes, textured black boxes represent rRNA genes. Gene sequence lengths are indicated in nucleotides and are relative to cDNAs in figure 1A.
To assess the relative abundances of the truncated and/or polycistronic transcripts compared with full-length transcripts, Northern blots of total Hematodinium sp. RNA were probed for cox1, cob, and cox3 sequence. For each gene, the probes produced a single dominant band of a size consistent with full-length gene transcripts, as predicted from cDNAs (fig. 5C).
Discussion
In this study, we have examined the organization and expression of the mitochondrial genome of Hematodinium sp. and find that although several character states of this taxon are shared with later-branching dinoflagellates, others are missing and may indicate either ancestral or further divergent characters.
The phylogeny of the dinoflagellate lineage has been notoriously difficult to resolve by either morphologic or molecular characters and points to an explosive radiation of diversity within this group (Hoppenrath and Leander 2010). Nevertheless, the most basal taxon of the dinoflagellate lineage is reliably identified as the Perkinsidae, both by molecular phylogenies (Saldarriaga et al. 2003, 2004; Bachvaroff et al. 2011) and by the presence of some dinoflagellate derived characters (e.g., trans-splicing leaders onto nuclear mRNAs) but lack of others (e.g., dinokaryon including permanently condensed chromosomes and deviant histones) (Zhang, Campbell, et al. 2011; Zhang, Dungan, et al. 2011). Relatively little is known of the mitochondrial gene copy number or genome organization in Perkinsus, however, RNA editing is apparently absent suggesting that this is a latter derived character in dinoflagellates. Perkinsus mitochondria, however, do possess their own eccentricities with a novel trait of frameshifts occurring in protein-encoding genes (cox1 and cob), and transcripts apparently require a unique translation decoding mechanism to generate meaningful proteins (Masuda et al. 2010; Zhang, Campbell, et al. 2011).
Among other basal dinoflagellate members, although later-branching than Perkinsus, is the parasitic order Syndiniales within which Hematodinium sp. is classified and, separately, the free-living heterotrophic genus Oxyrrhis. The inclusion of Hematodinium sp. in the Syndiniales is supported by a common multiphase lifecycle including a multinucleate plasmodial stage, feeding trophont and sporont stages, and a flagellated dispersal stage (dinospore) (Appleton and Vickerman 1998; Stentiford and Shields 2005). Although molecular phylogenies also give support to Hematodinium sp. being a member of this group (Saldarriaga et al. 2004), syndinians are generally very poorly represented in current molecular data sets, and the monophyly of this diverse clade requires further testing. Furthermore, the relative positions of Oxyrrhis and the Syndiniales are poorly resolved. The dinokaryotic state of each group's nucleus is equivocal, with lack of the inflated DNA content seen in most dinophycean dinoflagellates, but evidence of deviant histones and/or nonhistone basic proteins as dominant nuclear proteins (Cachon and Cachon 1987; Fensome et al. 1993; Kato et al. 1997). Oxyrrhis uses an intranuclear mitotic spindle, an apparently ancestral state, whereas syndinians share the extranuclear spindle of dinophyceans that traverses channels in the nucleus (Ris and Kubai 1974; Saldarriaga et al. 2004). This latter trait is also known from Perkinsus, however, so Oxyrrhis mitosis might represent a reversion. Recent concatenated phylogenies (17 proteins) continue to be unable to resolve the relative positions of Oxyrrhis and syndinians (Bachvaroff et al. 2011).
Despite the branching order of Hematodinium sp. relative to Oxyrrhis remaining unresolved, our study of the mitochondrial genome of Hematodinium sp. offers significant insight into the development of this highly divergent genome. The use of deep 454 sequencing of the transcriptome in combination with extensive amplification of the genome has provided the most thorough coverage of dinoflagellate mitochondrial genes to date. We have recovered more of the mitochondrial rRNA fragments but confirmed that no further coding sequences beyond the LSU and SSU rRNAs and protein genes cob, cox1, and cox3 are found in dinoflagellate mitochondria.
Early Rearrangements of the Dinoflagellate Mitochondrial Genome
The genome organization of Hematodinium sp. shows abundant evidence of a high rate of recombination with copious copies of gene fragments occurring in seemingly limitless combinations. High sequence fidelity of both the gene fragments with their complete copies, and the multiply represented intergenic spaces, suggests that recombination events are frequent enough that they occur ahead of the accumulation of mutations. This duplicated and highly recombined genome form is equivalent to that seen in dinophyceans Amphidinium carterae, Crypthecodinium cohnii, and K. micrum (Norman and Gray 2001; Jackson et al. 2007; Nash et al. 2007, 2008; Kamikawa et al. 2009; Waller and Jackson 2009). Numerous points of recombination were observed (e.g., 10 within both cox1 and cob sequences), but only in one case were both flanking sequences of a single recombination point observed (3′ and 5′ fragments separated at cob nucleotide 633; fig. 1 clone v, and supplementary fig . 1 clone x, Supplementary Material online). This suggests that many further sites of recombination within this genome would likely be observed with further sampling. We also examined the sequences surrounding the recombination points in the protein-encoding genes (50 nt on either side) for any common motifs, potential secondary structures, or nucleotide composition bias that could facilitate the recombination events, but none were found.
The frequent recovery of multiple different linked genes on genomic elements indicates a high density of coding sequences in this genome. O. marina has been shown to share this character of duplicated and fragmented genes, although it is unclear if recombination might take a slightly different, less random form. In O. marina, no genes were found on genome elements linked to different genes, but rather only cases of a gene being linked to another copy of itself, in either orientation and including gene fragments (Slamovits et al. 2007). It remains possible that a more dispersed gene density could have limited the sampling method of PCR amplification, and larger contiguous mitochondrial sequences will be required to discern if this truly represents a different form of recombination in O. marina. Regardless, it is clear from Hematodinium sp. that the scrambling and inflation of the mitochondrial genome occurred relatively early in the dinoflagellate lineage, and examination of Perkinsus for this trait is warranted.
Another conspicuous feature of dinophycean mitochondrial genomes (seen in A. carterae, C. cohnii, K. micrum) is the presence of inverted repeats at gene boundaries and within intergenic sequence (Norman and Gray 2001; Jackson et al. 2007; Nash et al. 2007). These repeats have the potential to form stem/loop secondary structures, and in other systems are implicated in genome regulatory functions and as potential points of recombination (see Waller and Jackson 2009). We found no evidence of inverted repeats such as these in Hematodinium sp. (and none were evident from O. marina mtDNA Slamovits et al. 2007). Therefore, these repeats likely evolved independently from the genome duplication and recombinogenic scrambling and are seemingly a later-derived character in dinophyceans.
Gene Modifications and Rearrangements
The use of alternative initiator codons to the standard AUG apparently developed before dinoflagellates and apicomplexans diverged, and even some (although not all) ciliate mitochondrial genes use alternative initiators (Edqvist et al. 2000). In Plasmodium and O. marina, isoleucine codons AUA and/or AUU have been suggested as alternative initiators (Feagin 1992; Slamovits et al. 2007), however, these codons are not present in all Hematodinium sp. 5′ coding sequences or in all other dinoflagellates (fig. 2). Either further or alternative initiators therefore must be used in this lineage. Without a known start codon, it is not possible to identify the presence of a 5′ UTR (untranslated region) in dinoflagellates. Protein conservation seen in alignment of protein extremities within alveolates and other organisms suggest, however, that such UTRs are extremely short or even nonexistent (see fig. 2).
Loss of standard stop codons is a dinoflagellate specific trait, and one where Hematodinium sp. provides an interesting twist. Hematodinium sp. cob and cox1 genes lack an encoded stop codon, as is also the case for all other dinoflagellates including O. marina. Hematodinium sp. cox3, however, does encode a TAA stop, and the transcript of this gene is polyadenylated downstream of this codon (albeit only by only one nucleotide). This makes Hematodinium sp. unique amongst dinoflagellates with respect to this character, although curiously this is functionally equivalent to all other dinoflagellates (including O. marina) generating an UAA stop codon in cox3 transcripts by polyadenylation immediately after an inframe U nucleotide. The position of the Hematodinium sp. stop codon is perfectly conserved with the de novo stops in the other taxa. Thus, the importance of cox3 transcripts retaining a stop codon, while cob and cox1 do not, is reinforced in this basal taxon. Studies of Perkinsus mitochondrial genes (cob and cox1 only) have not addressed the use of terminator codons (Masuda et al. 2010; Zhang, Campbell, et al. 2011). We note, however, that transcript data strongly implies that this basal group has also eliminated standard stop codons for these two genes (see fig. 2). Data for Perkinsus cox3 are currently unavailable, so it remains to be determined if the stop codon in Hematodinium sp. cox3 represents retention of the ancestral state in Hematodinium sp., or if it is a chance reversion.
The frequent recombining of dinoflagellate mitochondrial genomic elements presumably allows opportunity for novel combinations of coding sequence to be explored. Generally, however, we see remarkable stability of coding elements across dinoflagellate diversity. Despite prolific gene fragments, cox1 and cob genes copies remain intact in the genome. cox3, on the other hand, is known to have been permanently split in all dinophycean dinoflagellates examined (Jackson et al. 2007 and Jackson CJ, Waller RF, unpublished data) and relies on trans-splicing of two distinct cox3 transcripts to provide a complete mRNA. The position of transcript splicing is always conserved and suggests that this change occurred only once in dinoflagellates. Hematodinium sp. contains an intact cox3 gene so presumably this innovation occurred subsequent to its divergence. O. marina also lacks trans-splicing of cox3, but in this taxon, recombination has resulted in a gene fusion between cob and cox3. There is no evidence of such a fusion in Hematodinium sp., so this fusion event occurred in the Oxyrrhis lineage only.
rRNA genes have also been heavily fragmented in dinoflagellates, and this is an older trait shared with apicomplexans. Previously, the SSU rRNA has been very poorly represented in dinoflagellate data with only one fragment previously identified (Jackson et al. 2007), however, from Hematodinium sp., we find five SSU rRNAs. Overall, the pattern of fragmentation, represented as RNA molecules, shows strong conservation with the ancestral state shared with apicomplexans. Interestingly, all rRNA coding sequences recovered in the Hematodinium sp. mtDNA amplicons represent further fragmentations, but, as for the protein genes, these appear to be pseudogenes. Thus, although ongoing opportunity for fragmentation exists, a maximal state of fragmentation has been reached and further disassembly would presumably impact the ability to selfassociate to form function ribosomes. This study also provides the first direct evidence that in dinoflagellates, the fragmented rRNA gene transcripts are not ligated to reconstruct any larger more conventional rRNA molecules, despite the development of RNA trans-splicing in this group for mRNAs. In other organisms where rRNAs have become fragmented, it is thought that intermolecular base-pairing between rRNA fragment transcripts reconstitutes functional rRNA molecules (Adams and Palmer 2003), and this is also presumably the case in dinoflagellates.
Two Hematodinium sp. rRNA fragments, however, suggest that novelty has been generated. SSUA and SSUB each occur as two classes of molecules in the transcriptome data. For both fragments, the class members have divergent ends from their partners, although semiconservation of portions of these extensions indicates selection acting upon them and that both might provide function. In K. micrum RNA8, Northern data indicate a second size class of this rRNA, and in Plasmodium rRNAs, LSUF and SSUF both exist as two distinct sized bands (Feagin et al. 1997). These longer species are unlikely to represent ligated forms to other rRNA fragments as probes to neighboring rRNAs (or any other rRNAs) do not label a common band (Feagin et al. 1997). Coexistence of different versions of rRNAs suggests that different ribosomes might be made, with up to four different types depending on expression timing due to the SSUA/B variants in Hematodinium sp. alone. The 454 transcriptome data indicate that both forms of SSUA and SSUB occur in trophonts, although the trophont Northern blot detects only one form each. In culture, Hematodinium sp. cycles between several different trophont stages in a largely synchronous manner. RNA used to generate the Northern and 454 data was extracted at different time points, and so might originate from different trophont stages. Therefore, there might be stage-specific expression of some alternative rRNAs. In Plasmodium, alternative nuclear rRNAs are expressed at different parasite lifecycle stages (Gunderson et al. 1987), so adaptation of the translation machinery to different cell requirements is known to occur.
RNA Editing
RNA editing of mitochondrial transcripts has been detected in many dinoflagellates species, where it can affect up to 6% of sequence for a single gene (Jackson et al. 2007; Lin et al. 2008). Up to nine different editing types have been observed, including both transitions and transversions, and for protein-coding genes, edits predominantly occur at the first or second position within codons and usually alter the amino acid that is coded for (Lin et al. 2008). Both the editing mechanism and the functional role of editing in the dinoflagellate mitochondrion remains unclear, however, the conservation of many editing sites throughout dinoflagellates indicates that it is a stable character of this group (Lin et al. 2008). RNA editing is absent from the deeply branching dinoflagellate O. marina (Slamovits et al. 2007) as well as the dinophycean Heterocapsa triquetra, whereas in Amphidinium, editing is sparse and mostly consists of A to G changes (Lin et al. 2008). Some phylogenies position H. triquetra and Amphidinium at relatively basal positions in dinophycean diversity (Zhang et al. 2007), and therefore, these data suggest that editing arose later during dinoflagellate radiation and that A to G transitions were the first to appear followed by additional transitions and transversions. This model is challenged, however, by the presence of both transitions and transversions in the Hematodinium sp. mitochondrion. Either editing has been simplified and even lost in some of the dinophycean dinoflagellates (Heterocapsa and Amphidinium) or editing has arisen independently in Hematodinium sp. mitochondria. To test this we compared the conserved editing sites known from numerous dinophycean taxa with the editing sites in Hematodinium sp. and found that no edits appear to be shared between them. This gives weight to editing having arisen independently in Hematodinium sp. at least, and presence of editing in some plastids of dinoflagellates (Wang and Morse 2006; Dang and Green 2009) argues for a propensity for this trait to develop in the organelles of dinoflagellates.
Managing Transcriptome Complexity
Polycistronic transcripts and truncated versions of monocistronic transcripts have been reported from dinoflagellate mitochondria before (Chaput et al. 2002; Imanian and Keeling 2007; Jackson et al. 2007), however, deep sequencing of the Hematodinium sp. transcriptome has offered an unprecedented view of this transcriptome's complexity. We have found numerous cDNAs containing single and multiple gene fragments. This is consistent with the complexity of the highly recombined mtDNA elements and points to lack of tight transcriptional control. The aberrant transcripts include premature polyadenylation, and curiously most of the coding sequences included in such transcripts have been RNA edited according to the editing sites we have defined. Despite the detectible presence of these aberrant transcripts, Northern blots indicate that the full-length transcripts are the dominant forms, and so some process must exist to identify and eliminate this transcriptional “noise.” It is difficult to imagine how this identification occurs, particularly because in full-length mRNAs untranslated regions (UTRs) appear to be extremely short or perhaps even nonexistent. O. marina has been shown to add a distinctive 5′ cap to mRNAs (Slamovits et al. 2007), which could serve to maintain full-length transcripts. We, however, find no evidence of a modified 5′ cap in Hematodinium sp., nor in dinophyceans (Jackson et al. 2007), so this appears to be a character specific to O. marina. It is unclear what process might be involved in controlling appropriate transcript levels or ensuring that translation is not initiated on the aberrant transcripts.
Conclusions
The mitochondrial genome of Hematodinium sp. shows that much of the radical reorganization of this organelle genome occurred early in dinoflagellate radiation. In particular, the onset of frequent recombination was seemingly a key driver of innovation, resulting in massive genome expansion and both duplication and fragmentation of coding sequences. This recombination has generated lineage-specific novelty, with a split cox3 gene (and a mechanism for trans-splicing) only present in dinophycean dinoflagellates, and fusion of cox3 to cob unique to Oxyrrhis. In Hematodinium sp., we find evidence of novel alternative versions of rRNA fragments SSUA and SSUB. Despite genome scrambling through recombination commencing early in dinoflagellates, Hematodinium lacks the inverted repeats in gene flanking regions of the genome and encodes the only known case of a stop codon, occurring in cox3. The simplest explanation is that these both represent ancestral mitochondrial characters. RNA editing, however, suggests a simple linear model of character evolution for this organelle is not sufficient. Hematodinium sp. shows complex editing of mitochondria transcripts despite later-branching dinophyceans (Heterocapsa, Amphidinium) either lacking or possessing simpler editing capabilities. So for this character, at least, it appears that either reversions to the ancestral state (loss or simplification of editing) or independent gain of complex editing must have occurred during dinoflagellate radiation. In Oxyrrhis, the absence of RNA editing and an apparently different form of recombination of the mitochondrial genome might indicate that its true phylogenetic position is basal to Hematodinium sp. A survey of other syndinian mitochondrial genomic characters will now be useful to more clearly resolve the relationship of Hematodinium sp. within these basal lineages and the patterns of genome character evolution in this most deviant lineage of mitochondria.
Supplementary Material
Supplementary figure 1 is available at Genome Biology and Evolution online (http://gbe.oxfordjournals.org/).
Acknowledgments
We wish to thank Professor Douglas M. Neil (University of Glasgow, Scotland, UK) for kindly providing Hematodinium sp. cells for culture in our laboratory. This work was supported by an Australian Research Council Discovery grant (DP0663590). C.J.J. was supported by the University of Melbourne Science Faculty Scholarship.
References
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Appleton PL, Vickerman K. In vitro cultivation and development cycle in culture of a parasitic dinoflagellate (Hematodinium sp.) associated with mortality of the Norway lobster(\ norvegicus) in British waters. Parasitology. 1998;116:115–130. doi: 10.1017/s0031182097002096. [DOI] [PubMed] [Google Scholar]
- Bachvaroff TR, Handy SM, Place AR, Delwiche CF. Alveolate phylogeny inferred using concatenated ribosomal proteins. J Eukaryot Microbiol. 2011;58:223–233. doi: 10.1111/j.1550-7408.2011.00555.x. [DOI] [PubMed] [Google Scholar]
- Bergholtz T, Daugbjerg N, Moestrup Ø, Fernández-Tejedor M. On the identity of Karlodinium veneficum and description of Karlodinium armiger sp. nov. (Dinophyceae), based on light and electron micropscopy, nuclear encoded LSU rDNA and pigment composition. J Phycol. 2006;42:170–193. [Google Scholar]
- Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Burki F, et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachon J, Cachon M. Parasitic dinoflagellates. In: Taylor FJR, editor. The biology of dinoflagellates. Oxford: Blackwell Scientific Publications; 1987. pp. 571–610. [Google Scholar]
- Cavalier-Smith T. Cell diversification in heterotrophic flagellates. In: Patterson D, Larsen J, editors. The biology of free-living heterotrophic flagellates. Oxford: Clarendon Press; 1991. pp. 113–131. [Google Scholar]
- Chaput H, Wang Y, Morse D. Polyadenylated transcripts containing random gene fragments are expressed in dinoflagellate mitochondria. Protist. 2002;153:111–122. doi: 10.1078/1434-4610-00090. [DOI] [PubMed] [Google Scholar]
- Dang Y, Green BR. Substitutional editing of Heterocapsa triquetra chloroplast transcripts and a folding model for its divergent chloroplast 16S rRNA. Gene. 2009;442:73–80. doi: 10.1016/j.gene.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Edqvist J, Burger G, Gray MW. Expression of mitochondrial protein-coding genes in Tetrahymena pyriformis. J Mol Biol. 2000;297:381–393. doi: 10.1006/jmbi.2000.3530. [DOI] [PubMed] [Google Scholar]
- Fast NM, Xue L, Bingham S, Keeling PJ. Re-examining alveolate evolution using multiple protein molecular phylogenies. J Eukaryot Microbiol. 2002;49:30–37. doi: 10.1111/j.1550-7408.2002.tb00336.x. [DOI] [PubMed] [Google Scholar]
- Feagin JE. The 6-kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol Biochem Parasitol. 1992;52:145–148. doi: 10.1016/0166-6851(92)90046-m. [DOI] [PubMed] [Google Scholar]
- Feagin JE. Mitochondrial genome diversity in parasites. Int J Parasitol. 2000;30:371–390. doi: 10.1016/s0020-7519(99)00190-3. [DOI] [PubMed] [Google Scholar]
- Feagin JE, Mericle BL, Werner E, Morris M. Identification of additional rRNA fragments encoded by the Plasmodium falciparum 6 kb element. Nucleic Acids Res. 1997;25:438–446. doi: 10.1093/nar/25.2.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensome RA, et al. A classification of living and fossil dinoflagellates. Micropaleontology Special Publication 7, Hanover: Sheridan Press; 1993. [Google Scholar]
- Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF. Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Mol Biol Evol. 2008;25:1219–1230. doi: 10.1093/molbev/msn070. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- Gunderson JH, et al. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987;238:933–937. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- Hackett J, Anderson D, Erdner D, Bhattacharya D. Dinflagellates: a remarkable evolutionary experiment. Am J Bot. 2004;91:1523–1534. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- Hoppenrath M, Leander BS. Dinoflagellate phylogeny as inferred from heat shock protein 90 and ribosomal gene sequences. PLoS One. 2010;5:e13220. doi: 10.1371/journal.pone.0013220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Keeling PJ. The dinoflagellates Durinskia baltica and Kryptoperidinium foliaceum retain functionally overlapping mitochondria from two evolutionarily distinct lineages. BMC Evol Biol. 2007;7:172. doi: 10.1186/1471-2148-7-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, et al. Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biol. 2007;5:41. doi: 10.1186/1741-7007-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, Inagaki Y, Sako Y. Fragmentation of mitochondrial large subunit rRNA in the dinoflagellate Alexandrium catenella and the evolution of rRNA structure in alveolate mitochondria. Protist. 2007;158:239–245. doi: 10.1016/j.protis.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Kamikawa R, Nishimura H, Sako Y. Analysis of the mitochondrial genome, transcripts, and electron transport activity in the dinoflagellate Alexandrium catenella (Gonyaulacales, Dinophyceae) Phycol Res. 2009;57:1–11. [Google Scholar]
- Kato KH, et al. Isolation of the major basic nuclear protein and its localization on chromosomes of the dinoflagellate, Oxyrrhis marina. Biol Cell. 1997;89:43–52. doi: 10.1016/s0248-4900(99)80080-x. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhang H, Gray MW. RNA editing in Dinoflagellates and its implications for the evolutionary history of the editing machinery. In: Smith HC, editor. RNA and DNA editing: molecular mechanisms and their integration into biological systems. Hoboken (NJ): John Wiley and Sons, Inc; 2008. pp. 280–309. [Google Scholar]
- Lin S, Zhang H, Spencer DF. Norman JE, Gray MW. Widespread and extensive editing of mitochondrial mRNAS in dinoflagellates. J Mol Biol. 320:727–739. doi: 10.1016/s0022-2836(02)00468-0. [DOI] [PubMed] [Google Scholar]
- Masuda I, Matsuzaki M, Kita K. Extensive frameshift at all AGG and CCC codons in the mitochondrial cytochrome c oxidase subunit 1 gene of Perkinsus marinus (Alveolata; Dinoflagellata) Nucleic Acids Res. 2010;38:6186–6194. doi: 10.1093/nar/gkq449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash EA, et al. Organization of the mitochondrial genome in the dinoflagellate Amphidinium carterae. Mol Biol Evol. 2007;24:1528–1536. doi: 10.1093/molbev/msm074. [DOI] [PubMed] [Google Scholar]
- Nash EA, Nisbet RE, Barbrook AC, Howe CJ. Dinoflagellates: a mitochondrial genome all at sea. Trends Genet. 2008;24:328–335. doi: 10.1016/j.tig.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Norman JE, Gray MW. A complex organization of the gene encoding cytochrome oxidase subunit 1 in the mitochondrial genome of the dinoflagellate, Crypthecodinium cohnii: homologous recombination generates two different cox1 open reading frames. J Mol Evol. 2001;53:351–363. doi: 10.1007/s002390010225. [DOI] [PubMed] [Google Scholar]
- Ris H, Kubai DF. An unusual mitotic mechanism in the parasitic protozoan Syndinium sp. J Cell Biol. 1974;60:702–720. doi: 10.1083/jcb.60.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, et al. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Saldarriaga JF, McEwan ML, Fast NM, Taylor FJ, Keeling PJ. Multiple protein phylogenies show that Oxyrrhis marina and Perkinsus marinus are early branches of the dinoflagellate lineage. Int J Syst Evol Microbiol. 2003;53:355–365. doi: 10.1099/ijs.0.02328-0. [DOI] [PubMed] [Google Scholar]
- Saldarriaga JF, Taylor FJR, Cavalier-Smith T, Menden-Deuer S, Keeling PJ. Molecular data and the evolutionary history of dinoflagellates. Eur J Protistol. 2004;40:85–111. [Google Scholar]
- Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol. 2007;372:356–368. doi: 10.1016/j.jmb.2007.06.085. [DOI] [PubMed] [Google Scholar]
- Spencer DF, Gray MW. Ribosomal RNA genes in Euglena gracilis mitochondrial DNA: fragmented genes in a seemingly fragmented genome. Mol Genet Genomics. 2010;285:19–31. doi: 10.1007/s00438-010-0585-9. [DOI] [PubMed] [Google Scholar]
- Stentiford GD, Shields JD. A review of the parasitic dinoflagellates Hematodinium species and Hematodinium-like infections in marine crustaceans. Dis Aquat Organ. 2005;66:47–70. doi: 10.3354/dao066047. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18S rRNA. J Mol Evol. 1997;45:619–630. doi: 10.1007/pl00006266. [DOI] [PubMed] [Google Scholar]
- Vlcek C, Marande W, Teijeiro S, Lukes J, Burger G. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 2010;39:979–988. doi: 10.1093/nar/gkq883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Jackson CJ. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. Bioessays. 2009;31:237–245. doi: 10.1002/bies.200800164. [DOI] [PubMed] [Google Scholar]
- Wang Y, Morse D. Rampant polyuridylylation of plastid gene transcripts in the dinoflagellate Lingulodinium. Nucleic Acids Res. 2006;34:613–619. doi: 10.1093/nar/gkj438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Williamson DH. Extrachromosomal DNA in the Apicomplexa. Microbiol Mol Biol Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters J. The troublesome parasites—molecular and morphological evidence that Apicomplexa belong to the dinoflagellate-ciliate clade. Biosystems. 1991;25:75–83. doi: 10.1016/0303-2647(91)90014-c. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bhattacharya D, Lin S. A three-gene dinoflagellate phylogeny suggests monophyly of prorocentrales and a basal position for Amphidinium and Heterocapsa. J Mol Evol. 2007;65:463–474. doi: 10.1007/s00239-007-9038-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Campbell DA, Sturm NR, Dungan CF, Lin S. Spliced leader RNAs, mitochondrial gene frameshifts and multi-protein phylogeny expand support for the genus Perkinsus as a unique group of alveolates. PLoS One. 2011;6:e19933. doi: 10.1371/journal.pone.0019933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dungan CF, Lin S. Introns, alternative splicing, spliced leader trans-splicing and differential expression of pcna and cyclin in Perkinsus marinus. Protist. 2011;162:154–167. doi: 10.1016/j.protis.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lin S. Mitochondrial cytochrome b mRNA editing in dinoflagellates: possible ecological and evolutionary associations. J Eukaryot Microbiol. 2005;52:538–545. doi: 10.1111/j.1550-7408.2005.00060.x. [DOI] [PubMed] [Google Scholar]