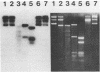
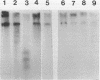
Abstract
Hemimethylated duplex DNA of the bacteriophage phi X 174 was synthesized using primed repair synthesis is in vitro with E. coli DNA polymerase I followed by ligation to produce the covalently closed circular duplex (RFI). Single-stranded phi X DNA was used as a template, a synthetic oligonucleotide as primer and 5-methyldeoxycytidine-5'-triphosphate (5mdCTP) was used in place of dCTP. The hemimethylated product was used as substrate for cleavage by various restriction enzymes. Out of the 17 enzymes tested, only 5 (BstN I, Taq I, Hinc II, Hinf I and Hpa I) cleaved the hemimethylated DNA. Two enzymes (Msp I and Hae III) were able to produce nicks on the unmethylated strand of the cleavage site. Msp I, which is known to cleave at CCGG when the internal cytosine residue is methylated, does not cleave when both cytosines are methylated. Another enzyme, Apy I, cleaves at the sequence CCTAGG when the internal cytosine is methylated, but is inactive on hemimethylated DNA in which both cytosines are methylated. Hemimethylated molecules should be useful for studying DNA methylation both in vivo and in vitro.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Cedar H., Solage A., Glaser G., Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6(6):2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godson G. N., Roberts R. J. dna, single stranded/*metab. Virology. 1976 Sep;73(2):561–567. doi: 10.1016/0042-6822(76)90421-9. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Razin A., Sedat J. W., Sinsheimer R. L. Structure of the DNA of bacteriophage phiX174. VII. Methylation. J Mol Biol. 1970 Oct 28;53(2):251–259. doi: 10.1016/0022-2836(70)90298-6. [DOI] [PubMed] [Google Scholar]
- Razin A., Urieli S., Pollack Y., Gruenbaum Y., Glaser G. Studies on the biological role of dna methylation; IV. Mode of methylation of DNA in E. coli cells. Nucleic Acids Res. 1980 Apr 25;8(8):1783–1792. doi: 10.1093/nar/8.8.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakazawa K., Shinomiya T. A DNA methylase from Thermus thermophilus HB8. J Biochem. 1980 Sep;88(3):737–747. doi: 10.1093/oxfordjournals.jbchem.a133026. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]