Abstract
Comparative genomics of closely related bacterial strains can clarify mutational processes and selective forces that impact genetic variation. Among primary bacterial endosymbionts of insects, such analyses have revealed ongoing genome reduction, raising questions about the ultimate evolutionary fate of these partnerships. Here, we explored genomic variation within Blochmannia vafer, an obligate mutualist of the ant Camponotus vafer. Polymorphism analysis of the Illumina data set used previously for de novo assembly revealed a second Bl. vafer genotype. To determine why a single ant colony contained two symbiont genotypes, we examined polymorphisms in 12 C. vafer mitochondrial sequences assembled from the Illumina data; the spectrum of variants suggests that the colony contained two maternal lineages, each harboring a distinct Bl. vafer genotype. Comparing the two Bl. vafer genotypes revealed that purifying selection purged most indels and nonsynonymous differences from protein-coding genes. We also discovered that indels occur frequently in multimeric simple sequence repeats, which are relatively abundant in Bl. vafer and may play a more substantial role in generating variation in this ant mutualist than in the aphid endosymbiont Buchnera. Finally, we explored how an apparent relocation of the origin of replication in Bl. vafer and the resulting shift in strand-associated mutational pressures may have caused accelerated gene loss and an elevated rate of indel polymorphisms in the region spanning the origin relocation. Combined, these results point to significant impacts of purifying selection on genomic polymorphisms as well as distinct patterns of indels associated with unusual genomic features of Blochmannia.
Keywords: genome reduction, strand asymmetry, mutational bias, simple sequence repeats, variant detection, next-generation sequencing
Many insect species harbor bacterial symbionts, which enable adaptation of hosts to different environmental niches and affect diversification of insect lineages (Moran et al. 2008; Ferrari and Vavre 2011). Obligate primary endosymbionts are strictly vertically inherited and evolve in parallel with their hosts. Evidence from genomics supports a nutritional role for primary symbionts. For example, Buchnera and Blochmannia synthesize essential amino acids for their aphid and ant hosts, respectively (Shigenobu et al. 2000; Gil et al. 2003), and Wigglesworthia produce vitamins missing from the blood diet of their tsetse fly hosts (Akman et al. 2002).
Pathways to synthesize key nutrients are retained in primary endosymbionts despite extensive gene loss. Other hallmarks of obligate primary endosymbionts include high AT content, accelerated evolutionary rates, lack of horizontal gene transfer or phage-related genes, and conservation of gene order (Moran et al. 2008). In the absence of recombination, functions cannot be reacquired after they are lost, which implies that ongoing gene erosion may lead to an evolutionary dead end (Latorre et al. 2005).
Our understanding of metabolic streamlining in primary endosymbionts of insects has benefited tremendously from comparative genomics. Genome sequences from symbiotic systems of diverse phylogenetic lineages have elucidated ancient and ongoing genome reduction (Tamas et al. 2002; Sabree et al. 2010). Within single species, genome comparisons have revealed mechanisms of gene erosion. Analysis of seven strains of Buchnera aphidicola from pea aphids (Acyrthosiphon pisum) showed that purifying selection purged most indels and nonsynonymous substitutions from protein-coding genes (Moran et al. 2009). Despite this, homopolymers within coding regions provide hot spots for slippage-induced indel mutations, and the resulting frameshifts may lead to gene degradation and loss. Similar processes may influence intraspecific variation in Blochmannia, obligate primary mutualists of ants of the tribe Camponotini. Comparisons of 16 intergenic regions from nine Blochmannia floridanus strains emphasized repetitive sequences as indel hot spots (Gomez-Valero et al. 2008). These studies illustrate that DNA sequence composition, particularly high abundance of slippage-prone regions, may contribute to ongoing gene inactivation and erosion in endosymbionts.
We recently reported the genome of Bl. vafer using Illumina sequencing (Williams and Wernegreen 2010). Here, we analyze variation between the published genome and a second genotype of Bl. vafer detected in the same Illumina read data set. This is the first intraspecific genome-wide comparison for Blochmannia. We also assembled and analyzed 12 protein-coding genes from the mitochondrial genome of the ant host Camponotus vafer. These data demonstrate the power of deep sequencing to explore genomic variation in a system with multiple players.
Variant Analysis Reveals Second Bl. vafer Genotype and Mitochondrial Genotypes
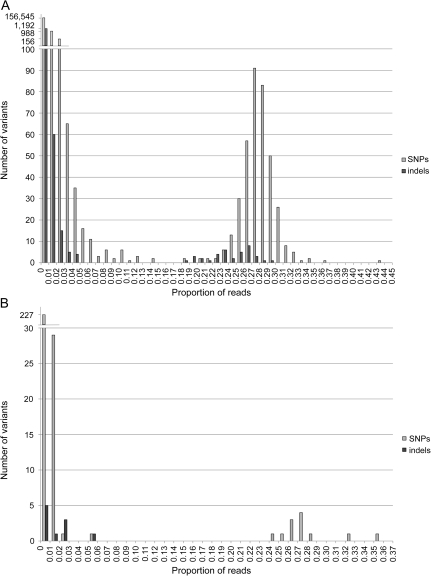
To evaluate polymorphisms in Bl. vafer, we aligned the Illumina reads (used previously for de novo assembly) to the published genome (NC_014909), which generated average coverage of 930× after duplicate removal. Distributions of single nucleotide polymorphisms (SNPs) and indels are bimodal (fig. 1A). We detected a set of SNPs occurring in 18–44% of reads and an overlapping set of indels in 18–30% of reads. The transition/transversion ratio differs markedly for SNPs with frequencies below versus above 18%. Among the 382 SNPs occurring in >18% of reads, transitions exceed transversions (Ts/Tv = 3.20), as expected for biologically real polymorphisms. By contrast, SNPs occurring in <18% of reads consist of more transversions (Ts/Tv = 0.52), even when we considered only SNPs in 10–15% of reads (Ts/Tv = 0.09). For further analyses, we considered variants in 18–44% of reads as comprising the second genotype. Variants in <18% of reads are likely mostly sequencing errors, although we cannot rule out the possibility that some fraction of these variants belong to the second genotype but were omitted due to the cutoff used.
FIG. 1.—
Distribution of variants in Blochmannia vafer and Camponotus vafer mitochondrial genes. (A) Distribution of variants detected in an alignment of the Illumina read data set to the Bl. vafer genome (NC_014909). (B) Distribution of variants detected in an alignment of the Illumina read data set to 12 C. vafer mitochondrial protein-coding genes. In both panels, the x axis shows the proportion of reads with a given variant out of the total reads covering the position in the alignment. SNPs and indels are represented by light gray and dark gray bars, respectively.
The second Bl. vafer genotype differs from the published genotype by 419 variants (382 SNPs and 37 indels) (supplementary table 1, Supplementary Material online). To test for linkage among variants, we examined the 65 variants located within 100 bp (the Illumina read length) of at least one other variant and determined how frequently nearby variants are found on the same read. Most of the 65 variants occur within 100 bp of one other variant (24 pairs); in six cases, three or four variants occur within 100 bp. For all but 1 of the 30 groups of variants, 97–100% of reads show linkage of variants within the group, supporting the hypothesis that these variants are part of a single genotype. The single exception is a group of three variants within a long (>100 nt) palindrome, which is one of only four such palindromes in Bl. vafer. Average coverage within this palindrome (276×) is lower than the overall average (930×), and few reads span all three variants, which limits our ability to accurately test linkage among this group of variants.
We collected ants from a single C. vafer colony and thus we expected one Blochmannia genotype because Camponotus is generally considered monogynous (e.g., Gadau et al. 1996) and a single maternal lineage is thought to possess one symbiont genotype. Explanations for the presence of two Bl. vafer genotypes in one ant colony may include 1) dual endosymbiont infection in one host lineage or 2) two C. vafer lineages, each harboring a distinct Bl. vafer strain, residing in the same colony. These hypotheses predict different patterns of mitochondrial polymorphisms. A dual Blochmannia infection in one host lineage predicts a single mitochondrial genotype, reflected in a unimodal distribution of mitochondrial variants with most occurring at low frequency due to sequencing errors. By contrast, the presence of two C. vafer lineages predicts two mitochondrial genotypes, leading to a bimodal distribution of mitochondrial variants similar to that observed for Blochmannia. Because mitochondria are also isolated during preparation of symbiont cells for genome sequencing, the Illumina reads include mitochondrial sequence information, and we used this to test the above predictions.
We assembled and analyzed 12 protein-coding genes from the C. vafer mitochondrial genome. Average coverage of each gene ranges from 160× to 400× with read duplicates removed. Polymorphism analysis of the mitochondrial genes revealed a bimodal distribution of SNPs (fig. 1B), with 12 SNPs occurring in 24–36% of reads. No indels occur in >6% of reads, suggesting that indels in protein-coding genes were generally eliminated by selection. The 12 SNPs comprise ten transitions and two transversions, and all but one are synonymous differences. These patterns are consistent with biologically real polymorphisms. This supports the hypothesis that the C. vafer colony included two maternal haplotypes, perhaps due to the presence of two queens. Many instances of polygyny have been documented in Camponotus, including in species closely related to C. vafer (Goodisman and Hahn 2005).
Purifying Selection Shapes Polymorphisms within Bl. vafer
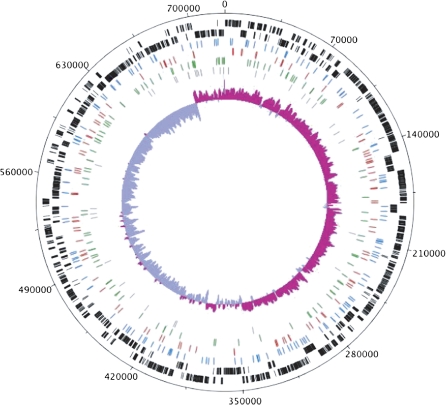
We analyzed the 419 variants that differentiate the published Bl. vafer genotype from the second genotype. The 382 SNPs are generally distributed evenly across the chromosome (fig. 2); however, SNP density is higher in intergenic regions (0.922 SNPs/kb) compared with protein-coding genes (0.452 SNPs/kb) (table 1). Of the 267 SNPs in protein-coding genes, 108 are nonsynonymous and 159 are synonymous. We calculated genome-wide values of 0.00022 nonsynonymous SNPs per nonsynonymous site (dN) and 0.00166 synonymous SNPs per synonymous site (dS), which indicate that approximately 90% of nonsynonymous SNPs were removed by purifying selection. Almost all indels (34/37) occur in intergenic regions (table 1), suggesting that most indels in protein-coding genes were removed by selection. The two indels within protein-coding genes are three nucleotides each; therefore, they do not disrupt the reading frame.
FIG. 2.—
Genome map of Blochmannia vafer. Tickmarks on the outermost circle show nucleotide positions in 35 kb increments. The two outermost tracks in black show coding sequences on the plus strand (track 1) and the minus strand (track 2). The next three tracks display positions for synonymous SNPs (blue), nonsynonymous SNPs (red), and intergenic SNPs (green). Indel positions are shown in gray. The innermost track shows GC skew calculated with window size of 1,000 bp and step size of 10 bp. Pink shading above the center line indicates GC skew values greater than the genome average, whereas purple shading below the center line indicates GC skew values less than the genome average. Position 0 on the genome map corresponds to the location of the origin of replication in Blochmannia pennsylvanicus and Escherichia coli. The figure was generated using DNAPlotter (Carver et al. 2009).
Table 1.
Polymorphisms in Blochmannia vafer
| SNPs | SNPs/kb | Indels | Indels/kb | Indel bpa | Indel bpa/kb | SNP/Indel | |
| Protein coding | 267 | 0.452 | 2 | 0.003 | 6 | 0.010 | 133.5 |
| RNA coding | 1 | 0.118 | 1 | 0.118 | 1 | 0.118 | 1 |
| Intergenic | 114 | 0.922 | 34 | 0.275 | 64 | 0.517 | 3.35 |
| Total | 382 | 0.529 | 37 | 0.051 | 71 | 0.098 | 10.32 |
Total bp contained in indels.
Patterns of variation between the two Bl. vafer genotypes are broadly similar to those observed among seven Bu. aphidicola strains (Moran et al. 2009), which also showed signatures of purifying selection. There were 166 times as many SNPs as indels in Buchnera protein-coding genes, which is close to the 133.5:1 ratio in Bl. vafer (table 1). The SNP/indel ratio in intergenic regions is also similar between Bl. vafer (3.35 SNPs/indel) and Bu. aphidicola (3.1 SNPs/indel). Overall SNP/indel ratios of Bl. vafer and Bu. aphidicola are 10.32 and 16.8, respectively. By comparison, SNP/indel ratios calculated by Chen et al. (2009) from pairs of bacterial strains with similar nucleotide divergence (≤0.05%) ranged from 1.8 to 12.4, with an average of 4.8.
Despite the effects of purifying selection, we identified three nonsynonymous differences in each of dnaE, murG, nuoL, and yjgP and four nonsynonymous differences in ilvE. Using the binomial test, we found that the number of nonsynonymous differences in each of these genes is unlikely to occur by chance (P < 0.03 for dnaE and P < 0.01 for the remaining genes), given the number of nonsynonymous sites in each gene and the overall level of nonsynonymous differences across the genome. These genes may be under relaxed purifying selection or possibly positive selection. IlvE catalyzes the final step of leucine, isoleucine, and valine biosynthesis, which are essential amino acids required by insects. Changes in ilvE could impact the nutritional contributions of Bl. vafer to its ant host.
Multimeric Simple Sequence Repeats Are Indel Hot Spots
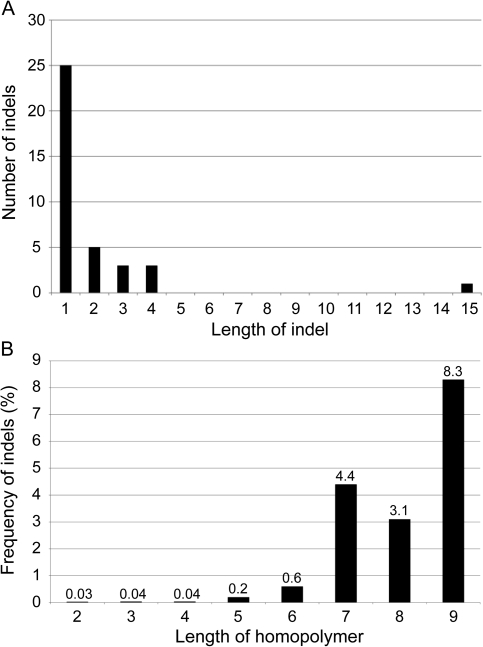
Most of the 37 indel polymorphisms are 1 nt long, but one indel is exceptionally long at 15 nt (fig. 3A). Thirty of 37 indels (81%) are located in repetitive sequences, indicating the important role of repeats as indel hot spots. Twenty-three indels (62%) occur in homopolymers ≥2 nt in length, all of which are in intergenic regions. Indel frequency increases with homopolymer length, occurring more frequently in long (≥6 nt) intergenic homopolymers (fig. 3B). Whereas the role of homopolymers as indel hot spots is well documented (Tamas et al. 2008; Moran et al. 2009), we discovered that other types of simple sequence repeats (SSRs) play a role in generating variation within Bl. vafer. Specifically, many indel polymorphisms occur in multimeric SSRs, which have repeat units of 2–5 nt. Of the 14 indels not located in a homopolymer, seven (50%) are found in multimeric SSRs.
FIG. 3.—
Indels in Blochmannia vafer. (A) Size of indels and (B) frequency of indels calculated by dividing the number of indels occurring in homopolymers of a particular length by the total number of homopolymers of that length in Bl. vafer intergenic regions.
Multimeric SSRs are more abundant in Bl. vafer and Bl. floridanus compared with the more distantly related Blochmannia pennsylvanicus and Bu. aphidicola of Acyrthosiphon pisum (NC_002528), even when we account for differences in genome size (table 2). By contrast, long (≥6 nt) homopolymers are considerably more abundant in Bu. aphidicola than in the Blochmannia lineages (table 2). Long homopolymers outnumber multimeric SSRs in Blochmannia and Buchnera and likely account for the majority of indel polymorphisms in both species; however, the observed discrepancies in relative abundance of the two types of SSRs suggest that contribution of multimeric SSRs to genomic variation may be more pronounced in Blochmannia than in Buchnera.
Table 2.
SSRs in Blochmannia and Buchnera
| Number of Multimeric SSRs | Multimeric SSR kb/kb Genome | Number of Longa Homopolymers | Longa Homopolymer kb/kb Genome | |
| Blochmannia vafer | 2,038 | 0.030 | 3,718 | 0.034 |
| Blochmannia floridanus | 2,030 | 0.031 | 3,083 | 0.029 |
| Blochmannia pennsylvanicus | 1,834 | 0.024 | 3,713 | 0.031 |
| Buchneraaphidicola of A. pisum | 1,385 | 0.023 | 6,395 | 0.068 |
Long homopolymers are defined as ≥6 nt in length.
Relocation of the Origin of Replication May Contribute to Genome Reduction
In bacterial genomes, the two replicating strands are typically differentiated by an excess of G over C on the leading strand compared with the lagging strand, resulting in a shift in GC skew ((G − C)/(G + C)) at the origin of replication (Rocha 2004). In Bl. pennsylvanicus and Escherichia coli, the shift in GC skew occurs near mnmG, but in Bl. vafer and Bl. floridanus, it occurs ∼31.5 kb upstream within the yibN-hldD intergenic sequence (fig. 2) (Williams and Wernegreen 2010). We propose that this reflects relocation of the origin of replication in the lineage leading to Bl. vafer and Bl. floridanus prior to their divergence. Although Blochmannia lacks dnaA, we searched both genomes for the E. coli dnaA box conserved motif TTATCCACA, but we found no matches near the positions of the relocated or ancestral origins, even when we allowed one mismatch. Allowing two mismatches generated hits in both genomes at both locations; therefore, we could not confirm the position of the origin using dnaA boxes. Despite this, GC skew provides strong evidence for relocation of the origin.
Compared with Bl. pennsylvanicus, Bl. vafer is missing seven genes in the 31.5-kb region spanning the origin relocation, and Bl. floridanus is missing four genes. Specific gene losses and their functional implications are detailed in our previous publication (Williams and Wernegreen 2010). Here, we did logistic regression to test if gene loss in the lineage leading to Bl. vafer (or, in a separate test, Bl. floridanus) is predicted by whether the gene is located in the 31.5-kb region or not. Based on the results, we can reject the null hypothesis for both Bl. vafer (P < 0.001) and Bl. floridanus (P = 0.01) and conclude that the probability of deletion is higher for genes in the 31.5-kb region in both genomes.
Importantly, relocation of the origin of replication implies that genes once positioned on the leading strand now occur on the lagging strand and vice versa. Klasson and Andersson (2006) showed that up to 90% of G versus C bias in Bl. floridanus third codon positions is explained by asymmetric mutational bias on leading and lagging strands. Therefore, genes whose strand switched due to relocation of the origin likely experienced strong shifts in mutational pressure. In other bacterial genomes, genes that switched strands due to inversions rapidly acquired the compositional bias of the new strand (Szczepanik et al. 2001) and showed higher amino acid divergence than genes retained on the same strand (Tillier and Collins 2000). Based on this evidence, we hypothesize that changes in strand-associated mutational pressure arising from relocation of the origin resulted in higher mutation rates, accumulation of amino acid changes, and potentially accelerated gene losses in the 31.5-kb region spanning the origin relocation in Bl. vafer and Bl. floridanus.
Comparing the two Bl. vafer genotypes may reveal whether an ancient relocation of the origin continues to influence recent variation. Although SNP density is slightly elevated in the 31.5-kb region compared with the rest of the genome (table 3), this difference is not statistically significant (Fisher's exact test of independence, P > 0.05). We also compared proportions of nonsynonymous, synonymous, and intergenic SNPs separately between the two regions (supplementary table 2, Supplementary Material online), but we did not find any statistically significant differences (Fisher's exact test, P > 0.05). Therefore, relocation of the origin does not appear to affect the rate of SNPs in the 31.5-kb region when considering recent variation. This is likely because most substitutions arising from asymmetric mutational bias occurred in the ancestor of Bl. vafer and Bl. floridanus shortly after origin relocation.
Table 3.
Variant and Repeat Densities in the Region Spanning the Origin Relocation and in the Rest of the Blochmannia vafer Genome
| Variant Density |
Repeat Density |
||||||||
| SNPs/kb | Indels/kb | Indels/kb igsa | Number of SSRsb/kb igsa | Number of Multimeric SSRsc/kb igsa | Number of Homopolymers/kb igsa |
||||
| ≥6 nt |
<6 nt |
||||||||
| A/T | C/G | A/T | C/G | ||||||
| 31.5-kb region | 0.636 | 0.286 | 0.907 | 206.6 | 5.2 | 6.9 | 0.2 | 176.7 | 17.6 |
| Rest of genome | 0.524 | 0.041 | 0.220 | 204.4 | 6.2 | 6.0 | 0.1 | 175.7 | 16.4 |
igs indicates intergenic sequence.
SSRs with repeat units of 1–5 nt (includes homopolymers and multimeric SSRs).
Multimeric SSRs with repeat units of 2–5 nt.
By contrast, indel density is significantly higher in the 31.5-kb region compared with the rest of the genome, even when we adjust for the amount of intergenic sequence in each region (table 3) (Fisher's exact test, P < 0.01). This suggests that relocation of the origin continues to affect the rate of indels in the 31.5-kb region. Because indels are more likely to occur in intergenic repetitive sequences, an alternative explanation is that the 31.5-kb region has more such repeats. Overall, when considering SSRs (repeat unit 1–5 nt) in intergenic regions, there is very little difference between the 31.5-kb region and the rest of the genome (table 3). Because we observed indels more frequently in long (≥6 nt) homopolymers in intergenic regions, we compared densities of long and short intergenic homopolymers separately, but we did not detect any differences that might explain the increased indel mutation rate in the 31.5-kb region (table 3). We also compared multimeric SSRs in the 31.5-kb region and the rest of the genome, but we did not detect any differences in the number of repeat units per multimeric SSR that might lead to more indels in the 31.5-kb region (supplementary fig. 1, Supplementary Material online). The elevated rate of indels in this region may be the result of a higher density of recent pseudogenes because pseudogenes do not serve coding or regulatory functions and are likely more tolerant to indels.
Conclusions
Comparative genomics provides important perspectives on the evolutionary fate of obligate bacterial endosymbionts and their hosts. We mined a single Illumina data set to identify and reconstruct a second Bl. vafer genotype and to assemble C. vafer mitochondrial sequences. Polymorphism analysis of mitochondrial genes suggests the C. vafer colony contained two ant host lineages, demonstrating the power of deep sequencing to clarify host colony structure. Genome evolution in Bl. vafer shares many features with that of the pea aphid endosymbiont Bu. aphidicola; for both, purifying selection plays a significant role in shaping intraspecific variation, and homopolymers serve as hot spots for indels. However, we identified two mechanisms that impact Blochmannia differently than Buchnera. In Blochmannia, multimeric SSRs occur at higher frequencies and may play a more significant role in generating variation. In addition, the apparent relocation of the origin of replication in the ancestor of Bl. vafer and Bl. floridanus led to changes in strand-associated mutational bias for a small region of these genomes. This shift in mutational spectra may explain earlier observations of accelerated gene loss near the origin and contribute to ongoing elevation in indel rates in that region.
Materials and Methods
Symbiont isolation, genomic DNA preparation, and Illumina sequencing were described previously (Williams and Wernegreen 2010).
Alignment of Illumina reads to the Bl. vafer genome (NC_014909) using Mosaik (http://bioinformatics.bc.edu/marthlab/Mosaik, last accessed 2011 Dec 5) yielded 7,922,530 unaligned reads. To assemble mitochondrial sequences from these reads, we removed reads with any base Q < 10 and used the remaining reads in a de novo assembly with Velvet (Zerbino and Birney 2008). We used BlastN to identify >100 bp contigs aligning to mitochondrial sequences in GenBank. The resulting draft assembly of the C. vafer mitochondrial genome had 10 contigs of 800–3,286 bp and totaled 16,415 bp. We annotated protein-coding genes using BlastX.
For polymorphism analysis, we used BWA (Li and Durbin 2009) to align Illumina reads to the Bl. vafer genome and the C. vafer mitochondrial genes. To correct misalignments arising from indels, we performed local realignments with the Genome Analysis Toolkit Indel Realigner (McKenna et al. 2010). We removed duplicate reads with Picard MarkDuplicates (http://picard.sourceforge.net) and calculated coverage using BEDTools (Quinlan and Hall 2010). We detected SNPs and indels using VarScan v.2.2.3 (Koboldt et al. 2009), removing variants that occurred on only one strand. We used BEDTools and VarClassifier (Li and Stockwell 2010) to determine location and effect of each variant. To test linkage, we used the Bambino viewer (Edmonson et al. 2011) to extract reads from the alignment based on two criteria: 1) the read spans all variants in a 100 bp region and 2) the read shows at least one of the variants. We then determined how many extracted reads had all variants and therefore showed linkage. For reads that did not show linkage, we examined sequence quality and excluded reads with base quality Q < 20 at the variant positions.
To calculate genome-wide dN and dS, we used TransAlign (Bininda-Emonds 2005) to generate amino acid–based alignments of protein-coding genes from the published Bl. vafer genotype and the second genotype. Using PAML (Yang 1997), we estimated the number of nonsynonymous and synonymous sites for each protein-coding gene. We then calculated genome-wide dN (108 nonsynonymous differences/492,720.3 nonsynonymous sites) and dS (159 synonymous differences/95,921.7 synonymous sites).
We used PERL scripts to count homopolymers and Phobos (http://www.ruhr-uni-bochum.de/spezzoo/cm/cm_phobos.htm) to detect multimeric SSRs.
We did the logistic regression using R and used JMP version 9 for the binomial test and Fisher's exact test of independence (supplementary methods, Supplementary Material online).
Supplementary Material
Supplementary tables 1 and 2, figure 1, and methods are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Marine Biological Laboratory colleagues: Adam Lazarus collected ants, James Style prepared genomic DNA, and Sheri Simmons provided a script to extract intergenic sequences. We thank Duke colleagues Alan Cowles, Tom Milledge, and Peyton Vaughn for bioinformatics assistance. We gratefully acknowledge Francesca Petralia and the Duke Statistical Consulting Center for valuable assistance with statistical analysis. We also thank Kenneth Kreuzer for helpful discussions. We acknowledge the valuable comments of three anonymous reviewers. This work was supported by the National Institutes of Health (R01GM062626 to J.J.W.) and the National Science Foundation (MCB-1103113 to J.J.W.).
References
- Akman L, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics. 2005;6:156. doi: 10.1186/1471-2105-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, et al. Variation in the ratio of nucleotide substitution and indel rates across genomes in mammals and bacteria. Mol Biol Evol. 2009;26:1523–1531. doi: 10.1093/molbev/msp063. [DOI] [PubMed] [Google Scholar]
- Edmonson MN, et al. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics. 2011;27:865–866. doi: 10.1093/bioinformatics/btr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari J, Vavre F. Bacterial symbionts in insects or the story of communities affecting communities. Philos Trans R Soc Lond B Biol Sci. 2011;366:1389–1400. doi: 10.1098/rstb.2010.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadau J, Heinze J, Holldobler B, Schmid M. Population and colony structure of the carpenter ant Camponotus floridanus. Mol Ecol. 1996;5:785–792. doi: 10.1111/j.1365-294x.1996.tb00374.x. [DOI] [PubMed] [Google Scholar]
- Gil R, et al. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci U S A. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, et al. Patterns and rates of nucleotide substitution, insertion and deletion in the endosymbiont of ants Blochmannia floridanus. Mol Ecol. 2008;17:4382–4392. doi: 10.1111/j.1365-294x.2008.03912.x. [DOI] [PubMed] [Google Scholar]
- Goodisman MA, Hahn DA. Breeding system, colony structure, and genetic differentiation in the Camponotus festinatus species complex of carpenter ants. Evolution. 2005;59:2185–2199. [PubMed] [Google Scholar]
- Klasson L, Andersson SG. Strong asymmetric mutation bias in endosymbiont genomes coincide with loss of genes for replication restart pathways. Mol Biol Evol. 2006;23:1031–1039. doi: 10.1093/molbev/msj107. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre A, Gil R, Silva FJ, Moya A. Chromosomal stasis versus plasmid plasticity in aphid endosymbiont Buchnera aphidicola. Heredity. 2005;95:339–347. doi: 10.1038/sj.hdy.6800716. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Stockwell TB. VariantClassifier: a hierarchical variant classifier for annotated genomes. BMC Res Notes. 2010;3:191. doi: 10.1186/1756-0500-3-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- Sabree ZL, Degnan PH, Moran NA. Chromosome stability and gene loss in cockroach endosymbionts. Appl Environ Microbiol. 2010;76:4076–4079. doi: 10.1128/AEM.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Szczepanik D, et al. Evolution rates of genes on leading and lagging DNA strands. J Mol Evol. 2001;52:426–433. doi: 10.1007/s002390010172. [DOI] [PubMed] [Google Scholar]
- Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Tamas I, et al. Endosymbiont gene functions impaired and rescued by polymerase infidelity at poly(A) tracts. Proc Natl Acad Sci U S A. 2008;105:14934–14939. doi: 10.1073/pnas.0806554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier ER, Collins RA. Replication orientation affects the rate and direction of bacterial gene evolution. J Mol Evol. 2000;51:459–463. doi: 10.1007/s002390010108. [DOI] [PubMed] [Google Scholar]
- Williams LE, Wernegreen JJ. Unprecedented loss of ammonia assimilation capability in a urease-encoding bacterial mutualist. BMC Genomics. 2010;11:687. doi: 10.1186/1471-2164-11-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]