Abstract
In shotgun proteomics, the quality of a hypothesized match between an observed spectrum and a peptide sequence is quantified by a score function. Because the score function lies at the heart of any peptide identification pipeline, this function greatly affects the final results of a proteomics assay. Consequently, valid statistical methods for assessing the quality of a given score function are extremely important. Previously, several research groups have used samples of known protein composition to assess the quality of a given score function. We demonstrate that this approach is problematic, because the outcome can depend on factors other than the score function itself. We then propose an alternative use of the same type of data to assess the quality of a given score function. The central idea of our approach is that database matches that are not explained by any protein in the purified sample comprise a robust representation of incorrect matches. We apply our alternative assessment scheme to several commonly used score functions, and we show that our approach generates a reproducible measure of the calibration of a given peptide identification method. Furthermore, we show how our quality test can be useful in the development of novel score functions.
Introduction
A shotgun proteomics experiment involves tryptic digestion of a complex protein mixture and subsequent detection of the resulting peptides via liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). A central step in the analysis of spectra produced by such an experiment is peptide identification. This step is usually achieved by searching each spectrum against a peptide database. During the search, the fragmentation spectrum is matched to theoretical spectra derived from a target database, comprised of peptides from the analyzed organism. The resulting matches between experimental and theoretical spectra are denoted peptide-spectrum matches (PSMs), and a score function assigns a score to each PSM, indicative of the quality of the match.
A good score function will exhibit two complementary properties. First, the function should be discriminative, meaning that it successfully separates correct from incorrect PSMs. In general, the search engine identifies a single target peptide that best explains each observed spectrum, a top-scoring PSM. However, some of these hypotheses are incorrect, often because a given spectrum does not stem from a peptide in the database. Given a large set of spectra, a highly discriminative score function will assign higher scores to correct PSMs than to incorrect PSMs.
Second, the score function should be well calibrated, meaning that the scores have well defined and accurate semantics. For clarity, we distinguish between raw score functions that output uncalibrated scores and statistical score functions that estimate a probabilistic measure of the error associated with a PSM. Examples of raw scores are SEQUEST’s XCorr1 and X!Tandem’s hyperscore.2 To facilitate interpretation, a statistical score function is often derived from a given raw score function by appending a post-processing step. For example, target-decoy analysis3 can be used to derive statistical scores for any given raw score function, or the hypergeometric distributions of hyperscores in X!Tandem can be used to estimate statistical scores such as q values or expectation values (E values). The quality of the calibration of a statistical score function can be very important. For example, if a collection of PSMs has an estimated false discovery rate (FDR) of 5%, and if the FDR estimate is well calibrated, then no more than approximately 5% of the PSMs in the collection should be incorrect. A well calibrated score allows the researcher to design follow-up experiments with an accurate estimate of the probability of false positive identifications. Conversely, a poorly calibrated score may lead to overoptimistic or conservative conclusions, in the worst case, invalidating an entire study.
In this study, we focus on methods for assessing the calibration of a given statistical score function. The common approach for evaluating either the discrimination or the calibration normally requires spectra derived from samples of known, purified proteins. Such spectra can be matched to a bipartite database, used specifically for a sample with known protein content. First, sequences corresponding to the small number of known proteins in the mixture, along with sequences of known or expected contaminants, make up what we here refer to as the sample sequences of the bipartite database. Second, a large number of entrapment sequences, representing proteins highly unlikely to be found in the sample, such as those obtained from an evolutionarily distant organism or shuffled versions of the sample sequences, are appended to the database. To avoid confusion with concatenated target and decoy databases, we want to clarify that the bipartite databases are not used to estimate error rates for regular shotgun proteomics experiments of unknown samples. For those purposes one might use a decoy database. A bipartite database is solely used for benchmarking purposes, using samples of known protein content.
The de facto standard method for performing such an assessment of a score function is what we here refer to as the fully labeled method. The fully labeled method assigns a label to every PSM, assuming that any top-scoring matches to the sample sequences are correct, whereas matches against the entrapment sequences are incorrect. Using these labels, theoretical error rates can be calculated and compared to the reported statistical score whose calibration we want to test. This approach has previously been used to validate and compare a variety of methods, including several inter-lab benchmarking studies.4–14 Here, we demonstrate that the reported performance—both the discrimination and the calibration—of a score function evaluated using the fully labeled method depends strongly on the choice of entrapment database used in the evaluation. This dependency makes accurate conclusions from a fully labeled assessment difficult, if not impossible, to draw.
We then suggest an alternative method for evaluating the calibration of a score function using a sample of known protein composition. The approach, which we refer to as the semi-labeled method, relies on the observation that, for the purposes of assessing calibration, it is sufficient to have an accurate model of the scores associated with incorrect PSMs. This observation is beneficial because the digestion of a small set of known proteins generates a limited set of peptides and hence a relatively low number of fragmentation spectra that stem from actual peptides in the sample. Furthermore, because many of these spectra correctly match the sample sequences of the bipartite database, correct PSMs are weeded out. The remaining entrapment PSMs correspond either to spectra with incorrectly assigned charge states, spectra derived from unanticipated (contaminant) proteins, or spectra that do not originate from a peptide at all. They also contain some PSMs of true peptide spectra that are simply incorrectly matched to the entrapment sequences.
We argue that these “unexplained” entrapment PSMs provide a relatively unbiased null model. Hence, a set of entrapment PSMs serves as a powerful method to assess the calibration of a score function. We demonstrate that this method can detect statistical biases of scores during the development of novel score functions. Finally, we determine the calibration of some commonly used score functions, including SEQUEST’s XCorr coupled with target-decoy analysis, X!Tandem’s E values and MS-GFDB p values.
Material and Methods
Experimental spectra
Fragmentation spectra were obtained from the The Standard Protein Mix Database of the Seattle Proteome Center.15 In the remainder of this article, we refer to this standard protein mixture as the ISB18 mix. The Orbitrap spectra used here were taken from runs 2–10 of mixture 7 of the ISB18 mix.
Composition of protein sequence databases
Bipartite sequence databases were assembled as described in Klimek et al.15 The first part of the database—the sample partition—consists of the ISB18 mix protein sequences and a list of contaminants. The remainder of the database consists of entrapment sequences, either from Haemophilus influenzae proteins or from shuffled versions of the sequences of the standard protein mixture with contaminants. The standard protein mixture sequences yielded ~4,000 tryptic peptides. As in Klimek et al., the number of tryptic peptides in the entrapment database was set to approximately 45,000 in each case, which required the sample sequences to be shuffled repeatedly in the case of shuffled entrapment sequences.
Some of the examined statistical scoring systems required decoy sequences.3 In these cases, decoy databases were generated by reversing the full bipartite target databases, both the sample and the entrapment sequences.
Peptide matching and scoring
All database searches were conducted using monoisotopic masses with a ±50 ppm mass tolerance window on tryptic peptides in the database. Non-tryptic searches were carried out for searches that were followed by post-processing using Percolator.16 To perform SEQUEST-like searches, RAW format files of the ISB18 mix spectra were converted to the ms2 file format using MakeMS217 and searched with Crux 1.22 in the sequest-search mode.18 Target and decoy searches were done separately. When target-decoy competition was performed, the competition was carried out after the search by comparing the scores of the top-scoring target and decoy PSMs for each spectrum.
MS-GFDB searches were run using version 2010092119 on the mzXML files available directly from The Standard Protein Mix Database.15 X!Tandem2 was obtained from the Trans-Proteomic Pipeline (TPP 4.3.1).20 The mzXML files of The Standard Protein Mix Database were used directly, without conversion.
To re-score PSMs after database searching, we used Percolator 1.14.16 In experiments based on past versions, Percolator 1.03 was used.
Calculations of score function p values
In this article, we consider three statistical score functions, each of which produces a p value, defined as the probability that an incorrect top-scoring PSM would score as well or better than the observed PSM by chance.
The first method for generating p values uses target-decoy analysis to post-process the scores produced by the raw score function XCorr. For separate target-decoy searches, all top-scoring target and decoy PSMs were considered, calculating the p value for a target PSM with score x as (r + 1)/(n + 1),21 where r is the number of decoy PSMs scoring ≥ x, and n is total number of decoy PSMs. For competitive target-decoy searches, p values were calculated similarly, but only considering the top-scoring target or decoy PSM for each spectrum.
The MS-GFDB method directly reports spectral probabilities, i.e., the probability that a spurious peptide would score as well or better against the same spectrum as a given peptide. These probabilities were converted to p values as previously described by Gupta et al.22 Accordingly, we use the Šidák correction23 to calculate the p value, p = 1 − (1 − Ps)N, associated with the spectral probability Ps, where N is the number of tested peptides (which corresponds to the number of amino acids in the searched database for the method MS-GFDB).
X!Tandem’s E values were converted to p values first by division by the number of candidate peptides, N, considered for each spectrum, followed by the Šidák correction described above using N as the number of candidate peptides.
Calibration test
To assess the calibration of a given score function f (·), we perform a search on a bipartite database, with a sample and an entrapment partition, to obtain a single PSM for each spectrum, along with a corresponding p value. The p values of the PSMs associated with entrapment peptides represent an unbiased collection of null p values. This procedure is outlined in Algorithm 1.
Algorithm 1. The algorithm for searching the bipartite database using spectra of known protein samples.
The procedure collects a set of p values, R, that follows the null model, reported from a score function f (·). As an input we use a set
 of MS/MS spectra from an experiment involving a known protein mixture as well as a bipartite database. The bipartite database is made up of sequences from the known proteins and likely contaminants, D, and entrapment sequences, E.
of MS/MS spectra from an experiment involving a known protein mixture as well as a bipartite database. The bipartite database is made up of sequences from the known proteins and likely contaminants, D, and entrapment sequences, E.
| 1: |
procedure GatherNullPValues(f (·),
 , D, E) , D, E) |
| 2: | R ← { } |
| 3: | for S ∈
 do do
|
| 4: | (P, p) ← BestScore(f (·), S, D∪E) ▹ Store top-scoring PSM as a tuple (peptide, score) |
| 5: | if P ∈ E then |
| 6: | R ← R∪{p} |
| 7: | end if |
| 8: | end for |
| 9: | return (R) |
| 10: | end procedure |
A collection of null p values are by definition uniformly distributed. Therefore, to qualitatively measure the uniformity of a given set of null p values, we plot quantile-quantile (Q-Q plots), with a uniform distribution over the interval [0, 1] on the x-axis and the reported p value distribution on the y-axis. If the p values are well calibrated, then the resulting points should lie close to the line y = x. We use logarithmic axes on the Q-Q plot because we are primarily interested in the calibration of the left tail of the p value distribution.
As a quantitative measurement of the uniformity of a set of p values, we employ a two-sample Kolmogorov-Smirnov (K-S) test between the p values and a uniform distribution over [0, 1]. Both these samples contain the same number of values. The test calculates the maximum difference, a D value, between the two samples’ cumulative frequencies. A large value of D implies that the two sample distributions are dissimilar.
Results
The results of a fully labeled analysis depend on the composition of the entrapment database
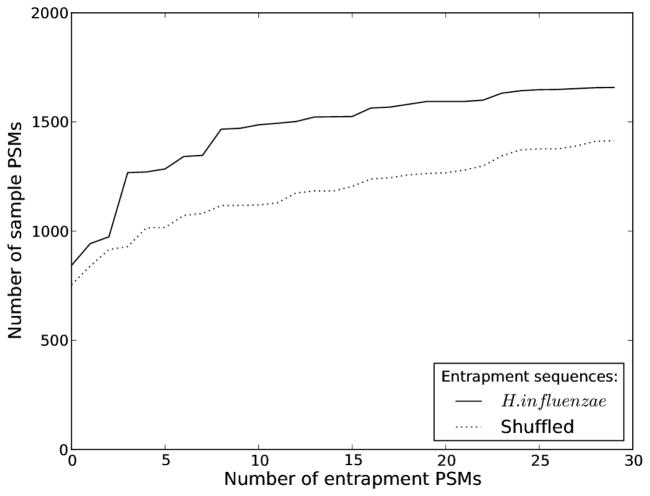
To demonstrate that the traditional, fully labeled method for assessing the performance of a score function is problematic, we searched a set of spectra against two different protein databases. A SEQUEST search was carried out using Crux on Orbitrap spectra from the ISB18 mix. First, we used a bipartite database containing the sample sequences of the ISB18 mix and entrapment sequences of H. influenzae. Second, we replaced the entrapment sequences with shuffled versions of the sample sequences. Figure 1 shows that replacing the entrapment database dramatically affects the number of matches to the (fixed) sample database. For example, allowing for 10 H. influenzae entrapment PSMs means that we accept 1487 sample PSMs. On the other hand, allowing for 10 shuffled entrapment PSMs, we only accept 1120 sample PSMs. We have observed the same effect for all other score functions used in this article (data not shown).
Figure 1. The dependency on entrapment sequences.
Spectra derived from a known protein mixture were scored against bipartite databases consisting of the known sample sequences and two different compositions of entrapment sequences. From the resulting PSMs we plotted the number of sample PSMs accepted for increasing numbers of entrapment PSMs, considering their XCorr score. The two different entrapment versions of the database had the same number of peptides, and sequences were either H. influenzae or shuffled versions of the sample proteins.
The above experiment shows that using PSMs from a standard mixture as a fully labeled set is problematic because the outcome can be very sensitive to random variations in the entrapment sequences. Here, we have kept the size of the two entrapment databases equal, whereas in practice, different database sizes would introduce additional variation to the results. We fear that, with this protocol to evaluate PSMs of known protein samples, entrapment databases could be tailored to generate results for almost any purpose. An alternative approach for testing the performance of a given score function is thus desirable.
An alternative use of known protein mixtures
Although we cannot use the conventional, fully labeled approach to assess the discriminative capabilities of a score function, we propose an alternative assessment protocol that also makes use of shotgun proteomics data derived from a known sample and searched against a bipartite target databases. Our proposed semi-labeled method relies on the observation that the partition of the bipartite database into sample and entrapment sequences is effective at identifying many correct PSMs. The remaining spectra, representing molecules not found in the sample, are likely to match the much larger entrapment portion of the database. Thus, when applied to samples of known mixtures, entrapment sequences serve as a robust trap for spurious matches in the database.
Importantly, and perhaps counterintuitively, the distribution of scores of the entrapment PSMs are not very sensitive to the composition of the entrapment database. Using the same sets of PSMs as for Figure 1, we compared the XCorr scores obtained from the two different groups of entrapment PSMs. Figure 2 shows a Q-Q plot of these XCorr scores, obtained from searches through the two different databases, a H. influenzae entrapment database, and a shuffled entrapment database. A straight line represents a situation where both sets of scores are distributed identically. Interestingly, although some deviation is seen, the graph shows that the two entrapment databases yield highly similar score distribution. Given that the entrapment PSMs comprise a robust sample of incorrect matches, we may use them to evaluate any given score function that outputs a statistical score defined by the behavior of random incorrect PSMs.
Figure 2. The similarity between PSMs obtained from two different entrapment databases.
The XCorr scores of entrapment PSMs obtained using either a H. influenzae entrapment database or a shuffled entrapment database were compared in a Q-Q plot. The filled circles represent the quantiles of the two groups of entrapment XCorr scores, D value = 0.023. The black line represents the x = y diagonal corresponding to perfect identity between the quantiles of the two groups. Both entrapment databases matched nearly 30,000 spectra.
Many statistical scores are defined with respect to the score distribution produced by random incorrect PSMs. Our proposed method can evaluate the calibration of any score function that reports such statistical scores, including E values and p values. For consistency throughout this article we consistently use the p value, which estimates the probability that an incorrect top-scoring PSM would obtain the observed score or higher. This definition implies that the calibration of p values produced by a score function can be evaluated using only the matches to the entrapment sequences, because these matches represent incorrect PSMs. By definition, accurate p values of incorrect PSMs must follow a uniform distribution between 0 and 1; therefore, the calibration of a score function can be tested directly by investigating the uniformity of the p values reported for entrapment PSMs, using either a quantile-quantile plot or a K-S test (see Material and Methods). In contrast, the p values of correct PSMs do not necessarily follow a predictable distribution and therefore are excluded from the calibration test.
A K-S test derives the maximum distance, D, between the cumulative frequencies of two samples, relating to their similarity (or dissimilarity). More precisely, for each value, x, found among any of the two samples, we evaluate the proportion of each sample with values less than or equal to x. Naturally, two identical samples will have equal such proportions for every x. If the sample distributions are different, however, the D value is defined as the largest difference between the two proportions, using all possible thresholds of x. Thus, a D value of 0.1 means that there exists a value of x below which, for example, the first sample has 50% of its values, and the other sample has only 40% of its values. Applying this test to our proposed semi-labeled method of reported entrapment PSM p values, D = 0.1 means that for some score threshold of x, 10% of the entrapment PSMs score too well, or too poorly, compared to the ideal p value distribution. This calibration value allows us to easily estimate the implications of a worst case scenario, in which the x generating the maximum difference, D, is also used as our threshold value to separate correct from incorrect PSMs. Given that a score function, calibrated to a D value of, say, 0.01, is used to score 30,000 incorrect PSMs, we risk that 0.01 ×30, 000 = 300 incorrect PSMs score too well (or too poorly) in comparison with the ideal p value distribution. Distinguishing between these two cases—p values that are too high or too low—can been done by examining the Q-Q plot.
Demonstration of biased features
A case in which our semi-labeled method for evaluating score function calibration would have been very useful occurred during the development of early versions of Percolator.16 Percolator is a machine learning algorithm that collects a variety of properties (called features) of target and decoy PSMs and uses a support vector machine classifier to discriminate between correct and incorrect PSMs. In addition to the features employed in recent versions, early releases of Percolator used three so-called intraset features that used information about other PSMs in the given data set to describe the PSM at hand. At first glance, these features seemed to improve Percolator’s ability to discriminate between correct and incorrect PSMs, leading to an increase in the number of target PSMs accepted with respect to a fixed statistical score threshold. However, the intraset features turned out to be biased in the sense that they led Percolator to systematically assign higher scores to target PSMs than to decoy PSMs. The apparent improvement in discriminative performance was thus a result of biased scoring, rather than a result of better separation between incorrect and correct matches. To avoid this bias, the intraset features were removed from the algorithm. In general, deceptive statistical scores may lead to incorrect conclusions, and make comparisons between studies impossible.
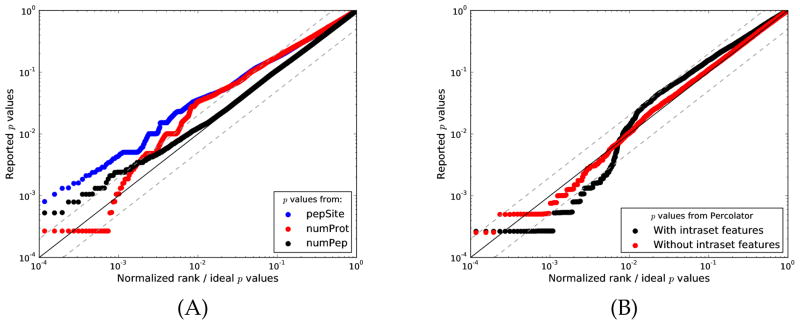
Using entrapment PSMs of spectra from known protein samples, biases like this one can be easily identified. Figure 3A shows how a Q-Q plot quickly exposes the bias of each individual intraset feature (denoted pepSite, numProt and numPep), and Figure 3B shows how the distribution of reported p values of Percolator with intraset features differs from a uniform distribution. Plotted in the same figure is Percolator without intraset features, reporting p values that are considerably more uniformly distributed. Clearly, this type of evaluation would have been beneficial in the early development of Percolator, because biased features would have been identified immediately.
Figure 3. Biased features of early versions of Percolator.
We scored Orbitrap spectra of 9 runs of the ISB18 mix against a bipartite target database containing shuffled entrapment proteins. A decoy search was made against a reversed decoy database. (A) Empirical p values for the entrapment PSMs were calculated using only the individual intraset feature values normally given as an input to the Percolator 1.03 algorithm. A Q-Q plot was drawn with these p values against hypothetical p values of an ideal, unbiased scenario. (B) The scores reported from Percolator 1.03 were used to calculate empirical p values for the entrapment PSMs with the intraset features activated. Percolator 1.14 calculates empirical p values without using the intraset features. We plot the reported p values against hypothetical, perfectly uniform p values. (K-S test goodness-of-fit p values: 2.0 ×10−16 with intraset features, 0.001 without intraset features.) The solid line corresponds to y = x; dashed lines y = 2x and y = x/2.
This example provides two important lessons. First, judging from only the theory behind a feature (or any other method), biases can be difficult to predict or detect. From only the description of the intraset features of early Percolator, the developers did not recognize the bias. Consequently, we recommend using the semi-labeled method to evaluate the calibration of any novel score function. Second, the Percolator example shows us one of the main problems with poor calibration: biased methods often appear to give very significant results. In reality, however, the apparently strong performance is an artifact of a biased score function. This is a very important point, because such statistical scores can result in costly, misleading conclusions.
Investigation of the calibration of some commonly used statistical scores
To emphasize the range of peptide identification methods to which our proposed calibration test is applicable, we analyzed the calibration of a few well-known score functions. Here, we used the entrapment PSMs of nine runs of ISB18 mix Orbitrap spectra, and we summarized the calibration results in Table 1. The table shows results using two types of entrapment databases also used to generate Figure 1. Our suggested use of data from known protein samples evaluates the calibration without considerable database composition sensitivity, as seen for the traditional approach. Additionally, Figure 4 shows Q-Q plots of p values reported from target-decoy analyses, X!Tandem and MS-GFDB.
Table 1. K-S test evaluations of the calibration of different score functions.
Here, we report the D values from K-S tests performed on nearly 30,000 entrapment PSMs from the concatenated runs of ISB18 mix Orbitrap spectra searched through two different bipartite databases. In the first experiment, we used a shuffled entrapment database; in the second, we used an entrapment database of H. influenzae sequences.
| Method | Shuffled entrapment K-S D value |
H. influenzae entrapment K-S D value |
|---|---|---|
| Separate target-decoy analysis, XCorr | 0.028 | 0.028 |
| Target-decoy competition, XCorr | 0.011 | 0.013 |
| Percolator 1.14, without intraset features | 0.026 | 0.035 |
| Percolator 1.03, with intraset features | 0.087 | 0.116 |
| X!Tandem | 0.747 | 0.738 |
| MS-GFDB | 0.210 | 0.225 |
Figure 4. Examples of score function calibrations.
Orbitrap spectra from 9 runs of the ISB18 mix were scored by a few commonly used score functions. A bipartite database of known sample sequences and entrapment sequences made up of the shuffled sample sequences was used. For each score function, we plotted reported p values vs. a ranked list from 0 to 1 representing ideally distributed p values. The solid line indicates the y = x diagonal, dashed lines y = 2x and y = x/2. (A) Separate and competitive target-decoy analyses (TDA) were performed based on SEQUEST’s XCorr and a reversed decoy database. All p values were calculated as described earlier.21 (B) X!Tandem’s E values and MS-GFDB’s spectral probabilities were converted to p values using a Šidák correction as described under Material and Methods.
The K-S test indicates that both X!Tandem and MS-GFDB with Šidák corrections produce poorly calibrated scores. For both methods, the K-S maximum distance D values are considerably higher than, for instance, any of the two approaches to target-decoy analysis using raw XCorr scores. High D values indicate that the observed p value distribution is not close to uniform. D values of around 0.7, as for X!Tandem, implies that up to 70% of all incorrect identifications score above or below the threshold erroneously, as compared to the ideal p value distribution. The Q-Q plot in Figure 4B provides further evidence of this poor calibration. The plot also shows that the p values produced by X!Tandem are too conservative (too high), whereas the p values from MS-GFDB are anti-conservative (too low). Conservative p values underestimate the significance of PSMs, increasing the risk that truly correct peptide identifications are missed. Anti-conservative p values, on the other hand, overestimate the significance of a match. This is highly undesirable because accepted PSMs will be incorrect to a larger extent than specified.
Discussion
In this paper, we propose an alternative use of known protein mixtures to evaluate the calibration of PSM score functions for shotgun proteomics. We have used a procedure where negative findings—entrapment PSMs—are treated as a robust representation of incorrect matches. In our scheme, the statistical scores of entrapment PSMs are compared to theoretical, ideally distributed null scores. If these two samples are unlikely to come from the same distribution, then the score function is assumed to be poorly calibrated.
We have demonstrated that it can be hard to obtain a representative measure of the discriminative capabilities of a score function using samples of known protein content, because correct and incorrect PSMs are more easily singled out under some conditions than under others. Instead, we propose an alternative method to assess the discrimination. Once we have assured that the calibration of our method is accurate, we can safely measure the discriminative performance of our score function on any dataset. A well-calibrated score can be trusted to examine how many identifications we obtain at any threshold defined in terms of our statistical score. This procedure enables us to measure our score function’s discriminative performance directly for the conditions we are interested in.
Throughout the study, we have used statistical score functions that produce p values. Under the null model, p values follow a uniform distribution, which is easily compared to the empirical distribution of entrapment PSM scores. However, our proposed semi-labeled method works for any statistical score that is defined in terms of the behavior of incorrect PSMs (a null hypothesis). For example, the calibration of X!Tandem could be evaluated using its reported E values directly, without prior conversion to p values. In those cases, the ideal null distribution of E values would replace the uniform null distribution of p values. Our scheme thus requires the score function to report statistical scores with a known null model distribution. Regardless of the statistical score used to test the calibration, in order to avoid misinterpretations, we want to emphasize that the entrapment null model concerns only top-scoring PSMs. Consequently, the reported statistical score must be defined in terms of top-scoring PSMs as well. This is the reason behind the Šidák correction explained in “Materials and Methods”. Ideally, p values should be reported from all statistical score functions. Assuming that other statistical scores reported from the same score function contain the same bias, this would enable a straightforward evaluation of the calibration.
To examine the extent of similarity between entrapment and ideal null p values, we have used the K-S statistic D. As explained earlier, D represents the maximum difference between two cumulative frequencies. This value can be interpreted as the proportion of incorrect PSMs that we risk erroneously placing either above or below a specified threshold score. The K-S test, as it is most commonly used, calculates a goodness-of-fit p value which we have not used here. Such p values represent the probability that two samples are drawn from the same distribution. However, this measure depends on the number of spectra we score. Due to the large number of entrapment PSMs, even small differences between the samples will result in a seemingly significant goodness-of-fit p value. On the other hand, the large number of PSMs grants a D value highly invariant to varying sample sizes. Thus, it represents a robust measure of the sample similarity. Additionally, as described above, the D value helps interpreting the implication of the calibration directly.
As mentioned earlier, the purpose of the bipartite database is to efficiently separate between correct and incorrect PSMs. Thus, the size of the entrapment partition is preferably many times larger than the size of the sample partition of a bipartite database. However, an entrapment database infinitely larger than the sample partition is likely to capture all top-scoring PSMs, making it equivalent to a normal decoy database. The ideal proportion of sample and entrapment sequences, for the purpose of creating the optimal null model, thus remains to be elucidated. In this study, we have, somewhat arbitrarily, used the size of the H. influenzae database, as in ref.,15 for the entrapment databases. Furthermore, how to preferably set a K-S statistic D value threshold for acceptable level of score function calibration, have not yet been examined.
Compared to previous methods, our proposed semi-labeled method to assess score function calibration helps overcome some computational problems relating to database searching. On the other hand, our method does not make up for low sample complexity and other experimental drawbacks of using samples of known protein mixtures. Hence, a low K-S statistic D value is a necessary but not sufficient requirement for a score function. On the other hand, it is a necessary requirement that few score functions live up to.
Synopsis.
The quality of a match between an observed spectrum and a peptide sequence is quantified by a score function. Previously, samples of known protein composition have been used to assess the quality of a given score function. We demonstrate that this method is problematic and propose an alternative approach. We show that our approach generates a reproducible measure of the calibration of a given peptide identification method.
Acknowledgments
This work was supported by grants from the Swedish Research Council, The Swedish Foundation for Strategic Research and the Lawski foundation.
References
- 1.Eng J, McCormack A, Yates J, Eng JK, McCormack AL, Yates JR, et al. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Craig R, Beavis R. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004:921. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 3.Moore R, Young M, Lee T. Qscore: an algorithm for evaluating SEQUEST database search results. Journal of the American Society for Mass Spectrometry. 2002;13:378–386. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 4.Keller A, Purvine S, Nesvizhskii A, Stolyar S, Goodlett D, Kolker E. Experimental protein mixture for validating tandem mass spectral analysis. OMICS: A Journal of Integrative Biology. 2002;6:207–212. doi: 10.1089/153623102760092805. [DOI] [PubMed] [Google Scholar]
- 5.Keller A, Nesvizhskii A, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 6.Nesvizhskii A, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 7.Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Prediction of error associated with false-positive rate determination for peptide identification in large-scale proteomics experiments using a combined reverse and forward peptide sequence database strategy. J Proteome Res. 2007;6:392–8. doi: 10.1021/pr0603194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi H, Ghosh D, Nesvizhskii A. Statistical validation of peptide identifications in large-scale proteomics using the target-decoy database search strategy and flexible mixture modeling. Journal of proteome research. 2007;7:286–292. doi: 10.1021/pr7006818. [DOI] [PubMed] [Google Scholar]
- 9.Choi H, Nesvizhskii A. Semisupervised model-based validation of peptide identifications in mass spectrometry-based proteomics. Journal of proteome research. 2008;7:254. doi: 10.1021/pr070542g. [DOI] [PubMed] [Google Scholar]
- 10.Bell A, Deutsch E, Au C, Kearney R, Beavis R, Sechi S, Nilsson T, Bergeron J, Beardslee T, Chappell T, Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJM, Beardslee TA, Chappell T, et al. A HUPO test sample study reveals common problems in mass spectrometry–based proteomics. Nature methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulovich A, Billheimer D, Ham A, Vega-Montoto L, Rudnick P, Tabb D, Wang P, Blackman R, Bunk D, Cardasis H, Paulovich AG, Billheimer D, Ham AJL, Vega-Montoto L, Rudnick PA, Tabb DL, Wang P, Blackman RK, Bunk DM, Cardasis HL, et al. Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS platform performance. Molecular & Cellular Proteomics. 2010;9:242. doi: 10.1074/mcp.M900222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabb D, Vega-Montoto L, Rudnick P, Variyath A, Ham A, Bunk D, Kilpatrick L, Billheimer D, Blackman R, Cardasis H, Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJL, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, et al. Repeatability and Reproducibility in Proteomic Identifications by Liquid Chromatography- Tandem Mass Spectrometry. Journal of proteome research. 2009;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett L, Bierl C, Master S. Unbiased Statistical Analysis for Multi-Stage Proteomic Search Strategies. Journal of Proteome Research. 2010;9:700–707. doi: 10.1021/pr900256v. [DOI] [PubMed] [Google Scholar]
- 14.Yang C, Yang C, Yu W. A Regularized Method for Peptide Quantification. Journal of Proteome Research. 2010;9:2705–2712. doi: 10.1021/pr100181g. [DOI] [PubMed] [Google Scholar]
- 15.Klimek J, Eddes J, Hohmann L, Jackson J, Peterson A, Letarte S, Gafken P, Katz J, Mallick P, Lee H, Klimek J, Eddes JS, Hohmann L, Jackson J, Peterson A, Letarte S, Gafken PR, Katz JE, Mallick P, Lee H, et al. The standard protein mix database: a diverse data set to assist in the production of improved peptide and protein identification software tools. J Proteome Res. 2008;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Käll L, Canterbury J, Weston J, Noble W, MacCoss M. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 17.McDonald W, Tabb D, Sadygov R, MacCoss M, Venable J, Graumann J, Johnson J, Cociorva D, Yates J., III MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Communications in Mass Spectrometry. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 18.Park C, Käll L, Klammer A, MacCoss M, Noble W. Rapid and accurate peptide identification from tandem mass spectra. Journal of proteome research. 2008;7:3022. doi: 10.1021/pr800127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Mischerikow N, Bandeira N, Navarro J, Wich L, Mohammed S, Heck A, Pevzner P. The generating function of CID, ETD and CID/ETD pairs of tandem mass spectra: Applications to database search. Molecular & Cellular Proteomics. 2010 doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller A, Eng J, Zhang N, Li X, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular systems biology. 2005;1 doi: 10.1038/msb4100024. year. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davison AC, Hinkley DV. Bootstrap Methods and Their Application. Cambridge University Press; Cambridge: 1997. [Google Scholar]
- 22.Gupta N, Pevzner P. False discovery rates of protein identifications: a strike against the two-peptide rule. Journal of Proteome Research. 2009;8:4173–4181. doi: 10.1021/pr9004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Šidák Z. Rectangular confidence regions for the means of multivariate normal distributions. Journal of the American Statistical Association. 1967;62:626–633. [Google Scholar]