Abstract
Objectives
Altered signaling in B-cells is a predominant feature of systemic lupus erythematosus (SLE). The genes BANK1 and BLK were recently described as associated with SLE. BANK1 codes for a B-cell-specific cytoplasmic protein involved in B-cell receptor signaling and BLK codes for an Src tyrosine kinase with important roles in B-cell development. To characterize the role of BANK1 and BLK in SLE, we performed a genetic interaction analysis hypothesizing that genetic interactions could reveal functional pathways relevant to disease pathogenesis.
Methods
We Used the method GPAT16 to analyze the gene-gene interactions of BANK1 and BLK. Confocal microscopy was used to investigate co-localization, and immunoprecipitation was used to verify the physical interaction of BANK1 and BLK.
Results
Epistatic interactions between BANK1 and BLK polymorphisms associated with SLE were observed in a discovery set of 279 patients and 515 controls from Northern Europe. A meta-analysis with 4399 European individuals confirmed the genetic interactions between BANK1 and BLK.
As BANK1 was identified as a binding partner of the Src tyrosine kinase LYN, we tested the possibility that BANK1 and BLK could also show a protein-protein interaction. We demonstrated co-immunoprecipitation and co-localization of BLK and BANK1. In a Daudi cell line and primary naïve B-cells the endogenous binding was enhanced upon B-cell receptor stimulation using anti-IgM antibodies.
Conclusions
Here, we show a genetic interaction between BANK1 and BLK, and demonstrate that these molecules interact physically. Our results have important consequences for the understanding of SLE and other autoimmune diseases and identify a potential new signaling pathway.
Keywords: systemic lupus erythematosus, genetics, polymorphism, B-cells, autoantibodies
INTRODUCTION
Systemic lupus erythematosus (SLE) is a complex autoimmune disease where B-cell activity plays a major role in its development and clinical expression through the production of auto-antibodies and antigen presentation. Therefore, susceptibility genes co-expressed in B-cells are interesting candidates to be tested for genetic and functional interactions.
In humans, polymorphisms of the BANK1 gene have been associated with susceptibility for SLE in European and Asian populations (1–3). BANK1 is located on chromosome 4q24 and codes for an adaptor/scaffold protein of 785aa (full length isoform) primarily expressed in B cells. BANK1 protein has 13 tyrosines susceptible of phosphorylation, two ankyrin repeats, a conserved Dof, BCAP, and BANK (DBB) domain, and a coiled-coil motif. It was identified as a binding partner of LYN, and it is also phosphorylated by SYK (4). BANK1 protein binds the IP3 receptors type 1 (IP3R-1) and 2 (IP3R-2) and promotes their LYN-mediated phosphorylation to induce Ca2+ mobilization from endoplasmic reticulum stores (4). However, Ca2+ mobilization was not impaired in a Bank1 knock-out mouse (5). Further, the Bank1 deficient mouse showed slight increase in germinal center formation and increased T-dependent responses with activation of Akt dependent on CD40 signaling. These features were subtle and no autoimmune phenotype was investigated. BLK was also recently identified as a susceptibility gene for SLE (6–8). The genetic polymorphisms of BLK associated with SLE, rs1327713 and its proxy rs2736340, are located in the promoter of BLK and the risk genotypes are correlated with reduced gene transcript levels. BLK is a Src tyrosine kinase specifically expressed in the B cell lineage (9). A knockout mouse for Blk did not show any phenotype and BLK was deemed to be redundant in B cell development and immune responses (10).
In this study we tested whether BANK1 and BLK polymorphisms associated with SLE showed a genetic epistatic interaction, but we also extended our study to analyze whether BANK1 and BLK, like LYN and BANK1 (4), could show a protein-protein interaction. While we identified an interaction between polymorphisms in both genes, we also found that both proteins immunoprecipitated and their co-expression influenced the sub-cellular location of the kinase. As the genetic interaction involves risk variants correlated with gene expression, the genetic interaction might reflect an imbalance in gene expression. The relative amounts of the gene products could be important to maintain the homeostasis of a common pathway.
MATERIALS AND METHODS
Patients and controls
We extracted data from an Affymetrix® 100k SNPs genome-wide association scan conducted in 279 cases with SLE and 515 controls from Northern Europe (1). Individuals used for the 100k GWAS have been described (1). Two independent sets of cases and controls were used for replication. Set 1 (“USA”) is a European-American multicenter cohort of 621 cases and 774 controls. The second set (“Europe”) comprised 1697 SLE cases and 1550 sex- and ethnically-matched controls from a European multicenter collection (BIOLUPUS) including Germans, Italians, Argentineans and Spanish individuals.
Genetic outliers with <90% European ancestry were removed, as estimated using principal component analysis and the clustering algorithms implemented in EIGENSTRAT and STRUCTURE software, respectively, based on genotype data from 350 Ancestry Informative Markers or genome-wide data (available for the Argentineans and North Europeans). All SLE cases met at least 4 of the 11 classification criteria of the American College of Rheumatology (11). All individuals provided informed consent as approved by the recruiting site Institutional Review Boards at each of the affiliate Institutions. All clinical investigation has been conducted according to the Declaration of Helsinki.
Genotyping
The Swedish individuals were genotyped using the 100k Affymetrix® SNP array as described (1). The previously associated SNPs for BLK (rs2736340), which is not included in the 100k Affymetrix® SNP array, was genotyped by TaqMan® (ABI, Foster City, CA) pre-designed genotyping assays. SNPs showing genetic interaction with BANK1 in the 100k were selected for replication. The replication set 1 (“USA”) SNPs were genotyped on the BeadExpress Illumina system. SNP rs10516483 (BANK1) was not available for this data set. Genotyping of set 2 (“Europe”) was performed for SNPs rs10516487 and rs10516483 (BANK1), rs1478895 and rs2736340 (BLK) also using TaqMan®. Only individuals with a genotyping rate >90% were used for analysis.
Statistical Analysis
From the 100k Affymetrix® SNP array data, nine tag SNPs in BANK1 (rs7675129, rs10516487, rs10516483, rs2850390, rs1872701, rs10516490, rs1395306, rs871153 and rs238486) were individually tested for 16 types of interaction against 7 tag SNPs in BLK (rs1478895, rs1478890, rs2252534, rs1382566, rs9329246, rs7014565 and rs2061830). SNPs were filtered as following Hardy-Weinberg equilibrium (HWE) in controls (p>0.01) and having missing data rate per SNP <5%. Only markers with minor allele frequencies >30% in controls and >10% in cases, and minor genotype frequencies >10% in controls and >5% in cases were used. The rationale was that we wanted to screen only common variants of the general population (controls) in order to have enough 2-SNPs combinations and we did not want to miss some SNP that would be less common in the SLE population. Linkage disequilibrium (LD) blocks were determined using the method of Gabriel et al (12) and tag SNPs were selected not to be in strong LD (r2 <.80). BLK SNPs covered 22% while BANK1 SNPs covered 44% of the alleles in each genomic region at a r2 >95%.
For the replication stage, SNPs following Hardy-Weinberg Equilibrium (HWE) in controls (p>0.001) and with missing data rates per SNP <10% were included in the analysis. None of the SNPs had significant differences in missing data between cases and controls (p>0.05).
Genetic Interaction Analysis
We used the GPAT16 method of Wirapati et al. (13). In brief, this method tests the genetic interaction between every pair of non-correlated SNPs (r2<0.8) by recording the 16 possible contingency tables formed by the combinations or co-occurrences of alleles or genotypes of both SNPs under dominant and recessive models. For each contingency table, a Pearson score S is computed with its corresponding P value. A P<1×10−5 was considered significant. A significant interaction reflects the sum of additive (or main effects) and epistatic effects for a specific genotype combination (dominant or recessive). In this particular experiment our total number of tests performed was 504 (9 BANK1 bait SNPs x 7 BLK SNPs x 16 tests / 2). GPAT16 makes 16 tests, but the total number is divided by 2 because each interaction is tested only in one direction. To determine the epistatic effect, that is, the increase in risk and an association odds ratio higher than expected under the null hypothesis of independence, each interaction is computed as the difference between the observed Pearson score S of each contingency table and the expected Pearson score S0 under the null hypothesis of no epistasis (14). By doing so, it derives an epistasis-like score (Se=S−S0). An epistasis P value (Pe) is obtained through permutation. A Pe<1×10−3 was considered significant. This score is the difference of two dependent scores, each one following asymptotically a 1-df c2. Therefore it does not follow any known statistical law and p-values pet have to be empirically determined by permutation. If two genotypes when combined have a significant association (S score significant, P<1×10-5) but there is no significant epistatic effect (Pe>1×10-3) we conclude that such association is mainly due to the sum of the individual or marginal effects of the associated genotypes. If the epistatic effect is significant (Pe<1×10-3) we then refer to it as a genetic epistatic interaction.
Protein Interaction Experiments
Antibodies
The synthesized peptide ETKHSPLEVGSESSC was used to immunize rabbits to generate polyclonal anti-human BANK1 anti-sera (ET-BANK antibody) and affinity purified using the SulfoLink Kit (Pierce). Additional antibodies include anti-mouse and anti-rabbit Alexa Fluor488, anti-mouse and anti-rabbit Alexa Fluor647, anti-V5 (Invitrogen, Carlsbad, CA); anti-Flag M2 monoclonal and rabbit anti-Flag (Sigma); anti-rabbit and anti-mouse IgG HRP (Zymed, San Francisco, CA). Mouse anti-human BLK antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA) and anti-β-tubulin from Sigma-Aldrich (St. Louis, MO).
DNA Cloning
BANK1 and BLK sequences were amplified by PCR using cDNAs from human blood and the BJAB cell line, respectively and ORFs were cloned in pcDNA3.1D/V5-His (Invitrogen) and confirmed by sequencing. Proteins tagged by V5 and His epitopes at the C-terminal were produced by stop codon deletion. The N-terminal FLAG-tagged BANK plasmids were constructed by sequential PCR using overlapping primers. The amplified product coding FLAG fused to BANK1 variants was cloned into pCR4-TOPO (Invitrogen) excised by EcoRI and BamHI and directionally sub-cloned into pIRESS2-EGFP (Clontech, Mountain View, CA). Sequences of the constructs are available upon request.
Co-immunoprecipitation and immunoblot analysis
Embryonic kidney HEK293T cells were seeded on 6-well plates and transfected with 4 μg of expression plasmids containing FLAG-tagged BANK1 and V5-tagged BLK using Lipofectamine 2000. At 40 hrs cells were solubilized in Triton X-100 buffer (1% Triton X-100, 50mM HEPES pH 7.1, 150 mM Nacl, 1 mM EDTA, 2 mM Na3VO4, 10 % Glycerol, 0.1% SDS) containing protease inhibitors (Roche, Indianapolis, IN) and 1mM PMSF. Aliquots of pre-cleared lysates were saved for input analysis and the remaining lysate was incubated with rabbit anti-FLAG or mouse anti-V5 and immobilized with A or G-Sepharose beads (GE Heathcare, Uppsala, Sweden), respectively. Beads were washed with 1:1 Triton X100 buffer:PBS and immunoprecipitates eluted with SDS sample buffer boiling 5min. SDS-PAGE and immmunoblotting were carried out using standard protocols.
Primary B-cell separation and purification
Peripheral blood mononuclear cells (PBMCs) from buffy coats were isolated by Ficoll-Paque Plus (GE Healthcare, Uppsala, Sweden) density gradient centrifugation. For preparation of purified, unmanipulated naïve B cells, PBMCs were subjected to negative selection using naïve B cells isolation kit II (Miltenyi Biotec, Auburn, CA). For depletion of CD10+ transitional B cells from negatively selected CD19+CD27- naïve B cells, selected cells were incubated with anti-human CD10 microbeads (Miltenyi, Biotec, Auburn, CA). Cells were magnetically separated with MACS Columns and MACS Separator (Miltenyi, Biotec, Auburn, CA). The negatively selected naïve B cells consisted of >95% CD19+CD27- cells.
Endogenous Co-immunoprecipitation
Primary naïve B-cells (3×106 per condition) were treated without (−) or with (+) aIgM (10μm/ml) for 10 minutes in serum-free RPMI medium. Cell extracts were made from the treated cells and subjected to immunoprecipitation and Western analysis. Antibodies against human BANK1 and BLK were purchased from Santa Cruz Biotech, Inc., and Abnova Corporation (Heidelberg, Germany), respectively. Recombinant Protein–G sepharose 4B beads were obtained from Invitrogen. Cell extracts were prepared using the lysis buffer containing 1% TritonX100, 50mM Tris pH7.4, 50 mM NaCl, 1 mM EDTA 2 mM Na3V04 and protease inhibitor cocktail from Roche. Immunoprecipitation was carried out using anti-human BANK1 antibody overnight. The immunocomplexes were precipitated using Protein-G beads and washed three times with lysis buffer.
The precipitated complexes were mixed with SD-PAGE sample buffer from Invitrogen and the proteins were resolved in 4–12% gradient NuPAGE gel (Invitrogen). Western blot was carried out using standard protocols.
Confocal Microscopy
Transfected cells were fixed for 20 min at room temperature (RT) with 3,7% paraformaldehyde in PBS/0.18% Triton-X and permeabilized in ice-cold 50:50 methanol-acetone at −20°C for 10 min. After blocking in 3% BSA, 3% goat serum in PBT antibodies were diluted in blocking buffer and incubated overnight at 4°C. Fluorochrome-conjugated secondary antibodies were incubated for 2 hrs at RT and counterstained with SlowFade antifade with DAPI (Invitrogen). Fluorescence fusion proteins were visualized directly after fixation, FX enhancer treated (Invitrogen) and mounted in Vectashield (Vector Laboratories, Burlingame, CA).
Confocal microscopy was performed using a Zeiss 510 Meta confocal scanning microscope with Zeiss plan-Apochromat 63x oil-immersion objective. Dual- or triple- color images were acquired by consecutive scanning with only 1 laser line active per scan to avoid cross-excitation. Image analysis was prepared using ImageJ and Adobe Photoshop.
RESULTS
Genetic interactions with BANK1
In the initial gene interaction analysis performed in the North European set, we observed a genetic interaction between BLK and BANK1, with the strongest epistatic effect between the BANK1 SNP rs10516483 and the BLK SNP rs1478895 (Pe = 0.0001) (Table 1). Two SNPs in BANK1 (rs10516483 and rs10516487, D′=0.86, r2=0.36) and two in BLK (rs1478895 and rs2736340, D′=0.93, r2=0.06) were involved in significant interactions although they did not reach the Pe<10−3 threshold. Given the moderate sample size of the North European data set, we chose these four SNPs for replication in two larger and independent sets of cases and controls of European ancestry. We observed significant interactions between BANK1 and BLK across all data sets (Table 1). The strongest association was displayed by the combination of recessive genotypes of BANK1 rs10516487 (GG) and dominant genotypes of BLK rs2736340 (TT+TC) (Pmeta-analysis =1.75 x 10−15) by using a total of 4399 samples. A significant Pe was demonstrated for this association in the replication set 2 from Europe (Pe=0.0013) (Table 1). In this set, a significant epistatic interaction was also observed between BANK1 rs10516483 (CC) and BLK rs2736340 (TT+TC) genotypes (Pe = 0.0024).
Table 1.
Summary of the two-gene interactions between BANK1 and BLK in three independent sets of cases and controls
| Gene SNP1 | Genotype SNP1 | Gene SNP2 | Genotype SNP2 | Population set | OR | OR L95 | OR U95 | P | Se | Pe | Freq. Cases | Freq. Controls | a | b | c | d | N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BANK1 | BLK | ||||||||||||||||
| rs10516483 | CC | rs1478895 | CC | North Europe 100k | 2.38 | 1.69 | 3.36 | 4.83E-07 | 8.9 | 0.0001 | 35% | 18% | 88 | 167 | 92 | 416 | 763 |
| Set 1 (USA) | - | - | - | - | - | - | - | - | - | - | - | ||||||
| Set 2 (Europe) | 1.42 | 1.17 | 1.72 | 0.0003 | −0.3 | 0.8256 | 25% | 19% | 261 | 773 | 285 | 1200 | 2519 | ||||
| Meta-analysis b | 1.60 | 1.36 | 1.89 | 7.61E-12 | nc | 349 | 940 | 377 | 1616 | 3282 | |||||||
| rs10516487 | GG | rs1478895 | CC | North Europe 100k | 1.82 | 1.35 | 2.45 | 8.27E-05 | 3.7 | 0.0063 | 48% | 33% | 131 | 144 | 171 | 342 | 788 |
| Set 1 (USA) | 1.33 | 1.06 | 1.66 | 0.0124 | 2.7 | 0.0170 | 38% | 31% | 233 | 388 | 241 | 533 | 1395 | ||||
| Set 2 (Europe) | 1.33 | 1.13 | 1.57 | 0.0005 | −1.0 | 0.9578 | 42% | 35% | 434 | 605 | 527 | 980 | 2546 | ||||
| Meta-analysis a | 1.40 | 1.24 | 1.58 | 1.97E-10 | nc | 798 | 1137 | 939 | 1855 | 4729 | |||||||
| rs10516483 | CC | rs2736340 | TT+TC | North Europe 100k | 2.32 | 1.44 | 3.76 | 4.68E-04 | 4.6 | 0.0111 | 24% | 12% | 56 | 178 | 31 | 229 | 494 |
| Set 1 (USA) | - | - | - | - | - | - | - | - | - | ||||||||
| Set 2 (Europe) | 1.70 | 1.36 | 2.13 | 3.65E-06 | 6.3 | 0.0024 | 18% | 12% | 184 | 824 | 169 | 1287 | 2464 | ||||
| Meta-analysis b | 1.80 | 1.47 | 2.21 | 1.18E-11 | nc | 19% | 12% | 240 | 1002 | 200 | 1516 | 2958 | |||||
| rs10516487 | GG | rs2736340 | TT+TC | North Europe 100k | 1.82 | 1.22 | 2.72 | 0.0031 | 2.6 | 0.0373 | 32% | 21% | 80 | 170 | 54 | 209 | 513 |
| Set 1 (USA) | 1.57 | 1.22 | 2.02 | 4.36E-04 | −1.6 | 0.9850 | 27% | 19% | 168 | 453 | 148 | 626 | 1395 | ||||
| Set 2 (Europe) | 1.62 | 1.34 | 1.95 | 3.55E-07 | 6.7 | 0.0013 | 29% | 20% | 293 | 720 | 297 | 1181 | 2491 | ||||
| Meta-analysis a | 1.63 | 1.41 | 1.87 | 1.75E-15 | nc | 29% | 20% | 541 | 1343 | 499 | 2016 | 4399 | |||||
|
| |||||||||||||||||
| BANK1 | GG | rs10516487 | Meta-analysis a | 1.37 | 1.22 | 1.54 | 4.33E-08 | 56% | 48% | 4791 | |||||||
| CC | rs10516483 | Meta-analysis b | 1.52 | 1.31 | 1.76 | 1.66E-12 | 36% | 27% | 3334 | ||||||||
| BLK | TT+TC | rs2736340 | Meta-analysis a | 1.33 | 1.21 | 1.46 | 4.31E-14 | 30% | 24% | 8862 | |||||||
The epistasis P value (Pe) measures only the statistical significance of the epistatic effect of the interaction. For every 2 SNP-genotype combination tested, a contingency table under the null hypothesis of independence between both SNPs (no epistasis) is derived and an expected Pearson S score is calculated (S0). The epistatic score is then defined as Se = S − S0. This score is the difference of two dependent scores, each one following asymptotically a one-degree of freedom chi-square distribution. Pe are empirically determined by permutations (100,000 case/control label shuffling). nc: Due to the nature of the Pe statistic, no permutation p-value can be calculated for meta-analysis.
North Europe 100k: discovery set, genotype data extracted from a 100.000 SNPs genome-wide scan in Swedish. The SNP rs2736340 was genotyped a posteriori. Replication Set 1 (USA): European-American set. Replication Set 2 (Europe): A combined set of German, Italian, European-Argentine and Spanish cases and controls.
a = cases with genotype SNP1 AND genotype SNP2, b = cases with other genotype, c = controls with genotype SNP1 AND genotype SNP2, d = controls with other genotype. N: number of cases and controls with non-missing genotypes for both SNP 1 and SNP 2. Frequencies of the 2 SNP-genotype combinations in cases and controls are shown.
Biochemical Interaction between BANK1 and BLK Proteins
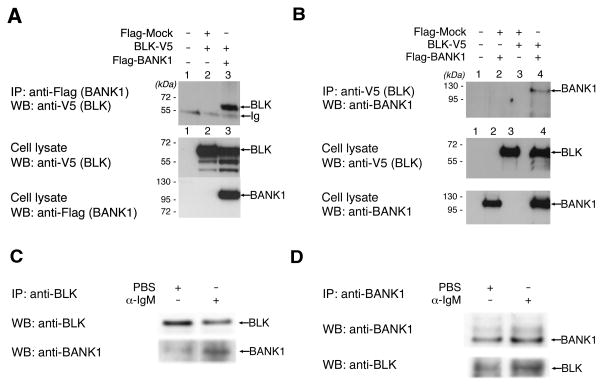
The fact that BANK1 was identified as a partner of LYN (4), a Src tyrosine kinase, led us to test whether BANK1 would show a similar interaction with Blk, also a Src tyrosine kinase. We found that BANK1 and BLK co-immunoprecipitated each other in co-transfected HEK293T cells (Figure 1a,b). As the products of co-transfection could result in an enhanced artifactual binding, we then tested whether the endogenous proteins co-immunoprecipitate in the B-cell line Daudi and in isolated naïve B cells. We demonstrated co-immunoprecipitation between the endogenous BANK1 and BLK in the B-cell line (Figure 1c) and in primary, naïve B-cells (Figure 1d). We further showed that the binding was enhanced by stimulation through the B-cell receptor using anti-IgM antibodies (Figure 1c,d) suggesting that activation of BANK1 or BLK may be required to enhance protein-protein interaction.
Figure 1. BANK1 and BLK Display a Protein-Protein Interaction.
a) Immunoprecipitation and western blot showing protein-protein binding of BANK1 and BLK. FLAG-BANK1 and BLK-V5 were co-transfected into HEK293T cells and immunoprecipitation was done using anti-FLAG antibodies. Western blot was performed using anti-V5 antibodies and confirmed with anti-FLAG antibodies. Lanes show: 1. Untransfected cells; 2. Co-transfection of FLAG-mock vector and BLK-V5; 3. Co-transfection of FLAG-BANK1 and BLK-V5.
b) Immunoprecipitation of cell extracts from co-tranfections showing recovery of BANK1 with the anti-V5 antibody directed to BLK-V5. Lanes show: 1. Untransfected cells; 2. Co-transfection of FLAG-Mock vector and FLAG-BANK1; 3. Co-transfection with FLAG-Mock and BLK-V5; and 4. Co-transfection of BLK-V5 and FLAG-BANK1.
c) Immunoprecipitation of endogenous BANK1 and BLK in the human cell line Daudi. Cell extracts were immunoprecipitated using anti-human BLK and the immunoprecipitates analyzed by Western blot.
d) Immunoprecipitation of endogenous BANK1 and BLK in naïve primary B cells. Cells were treated with anti-human IgM (SouthernBiotech) in a final concentration of 10 ug/ml for 10 minutes in serum-free RPMI medium or left unstimulated. Cell extracts were immunoprecipitated with anti-human BANK1 antibody (sc-133357, Santa Cruz Biotech) and analyzed by Western blot.
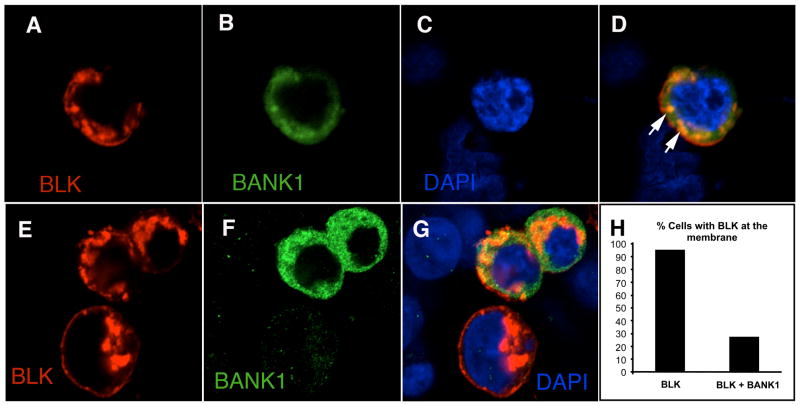
BANK1 is classified as an adaptor/scaffold protein and as such, could function to direct other molecules towards specific sub-cellular compartments. Confocal microscopy showed that both BANK1 and BLK co-localized in the cytoplasm when co-expressed (Figure 2a–d). Interestingly, BLK localized preferentially to the plasma membrane in the absence of BANK1 (Figure 2e–g) while it was mostly retained in the cytoplasm when BANK1 was co-expressed in the same cell (Figure 2g). In fact, BLK was located at the plasma membrane in 95% of cells when the protein was expressed alone contrary to 27% of cells co-expressing both BLK and BANK1 (Figure 2h). Our results suggest that BANK1 could modulate the subcellular localization of BLK, which would be in agreement with the function of BANK1 as an adaptor/scaffold protein.
Figure 2. BANK1 co-localization with BLK and modulation of the subcellular localization of BLK by BANK1.
Confocal images of HEK293 cells co-transfected with plasmids expressing BLK-V5 and BANK1 detected with immuno-fluorescence staining using antibodies against V5 and BANK1. Figures 2a–2d and 2e–2g represent two separate selected fields in two separate experiments. a) BLK (in red), b) BANK1 (in green); c) The nucleus stained with DAPI; d) merging showing co-localization of BANK1 and BLK in cytoplasmic compartments (arrows) and some BLK in the plasma membrane; e) Three cells expressing BLK (in red); f) Of the three cells, two co-express BANK1 (in green); g) Cell not expressing BANK1 shows BLK at the plasma membrane, while co-expression leads to its retention in cytoplasmic compartments, reduced at the plasma membrane. h) Diagram showing the proportion of cells harboring BLK at the plasma membrane when co-expressing or not BANK1. Approximately 200 cells were counted blindly in two independent experiments. BLK was detected with mouse anti-V5 followed by anti-mouse Alexa-647; BANK1 was detected using the rabbit anti-human BANK1 polyclonal antibody ET-BANK and anti-rabbit Alexa-488.
DISCUSSION
Here, we demonstrate that two SLE susceptibility genes showing a genetic interaction, namely BANK1 and BLK, also interact physically.
We used the GPAT16 method to test for associated genotypic interactions, a method in principle similar to the Multifactor Dimensionality Reduction (MDR) (16) and slightly more powerful than standard algorithms (17). According to simulations, GPAT16 is at least as powerful as the method of Marchini, et al. (18). The GPAT16 method enumerates exhaustively genetically relevant genotype combinations under dominant and recessive inheritance models, resembling the Batesonian definition of epistasis. This method is different from methods that consider the Fisherian definition of epistasis such as that implemented in PLINK (19), which test the interaction term in a logistic regression model.
The genetic interactions between BANK1 and BLK observed in the North European and European data sets follow a recessive model for the BANK1 genotypes (rs10516483 CC or rs10516487 GG) and a dominant model for BLK (rs2736340 TT+TC) genotypes. The interactions described here were not observable using logistic regression as implemented in PLINK (19) (supplementary Table 2), except for a weak significant interaction using the discovery set.
True epistatic interactions have been very difficult to detect and replicate (20, 21). We observed in the North European set a strong epistatic effect. As there is no established P-value for genetic interaction analysis, we used replication with independent sets of cases and controls. We replicated some of the epistatic effects (represented by the Pe value) that however did not reach our stringent Pe limit of <10−3 (22). Due to the computational characteristics of the method, a meta-analysis cannot be done.
We chose to study the interaction between BANK1 and BLK because of their functional interest in relation with SLE and their role in B-cell signaling. We believe that this way of analyzing genetic interactions fits our purpose of prioritizing candidate interacting genes for biological validation (23–25). In fact, a recent paper by Sun, et al. (25) analyzed human genome protein-protein interactions and found that physical connections were preferentially involved in gene-gene interactions. Thus, we believe that statistical genetics may guide the identification of true functional pathways in complex diseases.
Our findings point to a B-cell specific pathway that might be relevant in lupus pathogenesis. We showed that B-cell receptor stimulation enhances BANK1 and BLK binding. Because the engagement of the B-cell receptor with anti-IgM leads to tyrosine phosphorylation of numerous proteins including BANK1, it is likely that the interaction between BANK1 and BLK is regulated by cellular kinases. In chicken cell lines, SYK is a major player in phosphorylation of BANK1 upon BcR stimulation (4). BANK1 is a proline- and tyrosine-rich protein containing several predicted motifs for binding the SH2 and SH3 domains of Src-kinases. The binding of BANK1 to the Src-kinase LYN has been demonstrated but the precise protein domains involved in the interaction have not been defined (4). Detailed mutational analyses of BANK1 and BLK would be required to understand how BANK1 interacts with this family of kinases.
The change in sub-cellular distribution when BLK and BANK1 are expressed simultaneously suggests two possible functional scenarios. First, BANK1 as an adaptor protein could curb the positioning of BLK at the BcR by arresting it in intracellular compartments or, alternatively BANK1 could remove BLK from the BcR to restrict a sustained signaling. In both cases BANK1 could play an inhibitory role in B-cell activation. Supporting this idea, the bank1 deficient mouse shows an increase in B-cell activation illustrated by an increased IgM production in response to T-dependent antigens (5).
It is important though to remember that the interacting SNPs in BLK are located in non-coding regions. The risk genotypes of rs2736340 in BLK correlate with gene expression (6). The interacting SNP of BANK1 rs10516487 is located in exon 2 and leads to a R61H substitution but it is also a proxy of an intron 1 variant (rs17266594, r2 = 0.90) associated with higher level of expression of BANK1 (1). In summary, the risk allele of BLK is associated with lower level of gene expression while the risk alleles of BANK1 are coupled with higher level of their own gene expression (Supplementary Figure 3). At this point we are unable to draw the precise mechanistic pathway to explain how the risk allele interactions lead to B-cell abnormalities. A hypothesis is that alleles affecting gene expression could impair the homeostasis of the B-cell by a combinatorial inhibition model as proposed by Ferrell (26). This model claims that the signaling is impaired due to alteration of the relative concentration of the interacting proteins.
The interacting variants of BANK1 and BLK presented in this study might not be the functional variants responsible for the biological interaction effect as more extensive fine mapping and re-sequencing are required. Also, the SNP coverage would need to be increased although in detriment of multiple testing issues, particularly for whole-genome interaction analyses, which will be possible with new high-density arrays, so replication of the interactions will become even more important.
In summary, we describe here the use of a genetic interaction approach to reveal biologically relevant interactions and demonstrate that such approach can serve to define new pathways of disease, in this particular case a B cell-specific signaling pathway, which might be impaired in lupus patients.
Supplementary Material
Acknowledgments
The authors are indebted to the patients that have consented to have their samples used for this and other lupus genetics studies.
FUNDING
This work has been supported in part by grants from the European CVDIMMUNE project from the European Commission LSHM-CT-2006-037227, the Swedish Research Council for Medicine, the Swedish Association against Rheumatism, the Magnus Bergwalls Foundation, the Gustaf V:e 80th-year Jubilee, the Torsten and Ragnar Söderbergs Foundation and the Marcus Borsgtröms Foundation, the NIH-NCRR/COBRE grant P20 RR020143 to MEAR (PI JBH), the OCAST grant HR09-106 and the Instituto de Salud Carlos III partly financed through FEDER funds of the European Union to MEAR. This work was also partially supported by FISM, Regione Piemonte (CIPE and grant 2008) to SDA, the BMBF Kompetenznetz Rheuma C2.12, Germany to TW, grants SAF2006-00398, CTS-1180 and RETICS Program, RD08/0075 (RIER) from Instituto de Salud Carlos III (ISCIII) to JM. Dr. Pons-Estel is the coordinator of the Argentine Collaborative group and his work was in part supported by the Federico Wihelm Agricola Foundation Research grant. National Institutes of Health RR020143 (JMG and JBH), RR015577 (JMG, JBH, JAJ), N01 AI050026-001 (JMG and JAJ), AR053483 (JMG and JAJ), AI063274 (PMG), AI031584 (JBH, JMG, JAJ), AR052125 (PMG), AR043247 (KLM), Kirkland Scholar awards (JBH, LAC and JAJ), AR049084 (JBH), AR42460 (JBH), AR62277 (JBH), AI24717 (JBH), AR048940 (JBH, JAJ), AI083194 (JBH), R01 DE018209 (JBH), AI082714 (JBH), Alliance for Lupus Research (JBH), the US Department of Veterans Affairs (JBH), and OHRS award for project number HR08-037 from the Oklahoma Center for the Advancement of Science & Technology (JMG).
APPENDIX: THE LIST OF PARTICIPANTS
The Argentine Collaborative Group Participants are:
Hugo R. Scherbarth MD, Pilar C. Marino MD, Estela L. Motta MD Servicio de Reumatología, Hospital Interzonal General de Agudos “Dr. Oscar Alende”, Mar del Plata, Argentina; Susana Gamron MD, Cristina Drenkard MD, Emilia Menso MD Servicio de Reumatología de la UHMI 1, Hospital Nacional de Clínicas, Universidad Nacional de Córdoba, Córdoba, Argentina; Alberto Allievi MD, Guillermo A. Tate MD Organización Médica de Investigación, Buenos Aires, Argentina; Jose L. Presas MD Hospital General de Agudos Dr. Juán A. Fernandez, Buenos Aires, Argentina; Simon A. Palatnik MD, Marcelo Abdala MD, Mariela Bearzotti PhD Facultad de Ciencias Medicas, Universidad Nacional de Rosario y Hospital Provincial del Centenario, Rosario, Argentina; Alejandro Alvarellos MD, Francisco Caeiro MD, Ana Bertoli MD Servicio de Reumatología, Hospital Privado, Centro Medico de Córdoba, Córdoba, Argentina; Sergio Paira MD, Susana Roverano MD, Hospital José M. Cullen, Santa Fe, Argentina; Cesar E. Graf MD, Estela Bertero PhD Hospital San Martín, Paraná; Cesar Caprarulo MD, Griselda Buchanan PhD Hospital Felipe Heras, Concordia, Entre Ríos, Argentina; Carolina Guillerón MD, Sebastian Grimaudo PhD, Jorge Manni MD Departamento de Inmunología, Instituto de Investigaciones Médicas “Alfredo Lanari”, Buenos Aires, Argentina; Luis J. Catoggio MD, Enrique R. Soriano MD, Carlos D. Santos MD Sección Reumatología, Servicio de Clínica Medica, Hospital Italiano de Buenos Aires y Fundación Dr. Pedro M. Catoggio para el Progreso de la Reumatología, Buenos Aires, Argentina; Cristina Prigione MD, Fernando A. Ramos MD, Sandra M. Navarro MD Servicio de Reumatología, Hospital Provincial de Rosario, Rosario, Argentina; Guillermo A. Berbotto MD, Marisa Jorfen MD, Elisa J. Romero PhD Servicio de Reumatología Hospital Escuela Eva Perón. Granadero Baigorria, Rosario, Argentina; Mercedes A. Garcia MD, Juan C Marcos MD, Ana I. Marcos MD Servicio de Reumatología, Hospital Interzonal General de Agudos General San Martín, La Plata; Carlos E. Perandones MD, Alicia Eimon MD Centro de Educación Médica e Investigaciones Clínicas (CEMIC), Buenos Aires, Argentina; Cristina G. Battagliotti MD Hospital de Niños Dr. Orlando Alassia, Santa Fe, Argentina.
The German Collaborative Group Participants:
K. Armadi-Simab, MD, Wolfgang L. Gross, MD, Abteilung Rheumatologie, University Hospital of Schleswig-Holstein, Campus Luebeck, Rheumaklinik Bad Bramstedt, Luebeck, Germany, Erika Gromnica-Ihle, MD, Rheumaklinik Berlin-Buch, Berlin, Germany, Hans-Hartmut Peter, MD, Medizinische Universitaetsklinik, Abteilung Rheumatologie und Klinische Immunologie, Freiburg, Germany, Karin Manger, MD, Medizinische Klinik III derFAU Erlangen-Nuernberg, Erlangen, Germany, Sebastian Schnarr, MD, Henning Zeidler, MD, Abteilung Rheumatologie, Medizinische Hochschule Hannover, Hannover, Germany, Reinhold E. Schmidt, MD, Klinik fûr Immunologie und Rheumatologie, Medizinische Hochschule Hannover, Hannover, Germany.
The Spanish Collaborative Group participants are:
Norberto Ortego-Centeno (Servicio Medicina Interna, Hospital Clínico San Cecilio, Granada); Juan Jiménez-Alonso and Mario Sabio (Servicio de Medicina Interna, Hospital Virgen de las Nieves, Granada); Julio Sánchez-Román and Francisco J Garcia-Hernandez (Servicio de Medicina Interna, Hospital Virgen del Rocio, Sevilla); Enrique de Ramón y Mayte Camps (Servicio Medicina Interna, Hospital Carlos Haya, Malaga); Miguel Angel López-Nevot (Servicio de Inmunología, Hospital Virgen de las Nieves, Granada); Maria F. González-Escribano (Servicio de Inmunología, Hospital Virgen de las Nieves, Sevilla); Carmen Gutierrez and Ana Suarez (Hospital Universitario Central de Asturias, Oviedo); Miguel A Gonzalez-Gay (Hospital Xeral-Calde, Lugo); Carles Tolosa (Servicio Medicina Interna, Hospital Parc Taulí, Sabadell); Luisa Micó (Servicio Medicina Interna, Hospital La Fe, Valencia).
The Italian collaborative participants are:
Maria Giovanna Danieli (Dipartamento di Scienze Mediche e Chirurgiche, Universitá Politecnica delle Marche, Ancona, Italy), Gian Domenico Sebastiani (U.O.C. di Reumatologia Ospedale San Camillo, Roma – Italy), Enrica Bozzolo (IRCCS San Raffaele Hospital, Milan, Italy), Mauro Galeazzi, (Siena University, Siena, Italy), Sergio Migliaresi (Rheumatology Unit Second University of Naples, Naples, Italy). Also we would like to thank Prof. Armando Gabrielli, Clinica Medica di Scienze Mediche e Chirurgiche, Universitá Politecnica delle Marche.
Footnotes
Competing Interest
Please list Competing Interests if they exist if not please include the following statement; Competing Interest: Dr Jerome Wojcik is an employee of Merck Serono International, SA.
CONFLICT OF INTEREST STATEMENT
JW is an employee of MerckSerono Inc, and as such, the data or software he has developed belongs to MerckSerono Inc.
Licence for Publication
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd to permit this article (if accepted) to be published in ARD and any other BMJPGL products and sublicences such use and exploit all subsidiary rights, as set out in our licence (http://group.bmj.com/products/journals/instructions-for-authors/licence-forms).
References
- 1.Kozyrev SV, Abelson AK, Wojcik J, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 2.Guo L, Deshmukh H, Lu R, et al. Replication of the BANK1 genetic association with systemic lupus erythematosus in a European-derived population. Genes Immun. 2009;10:531–538. doi: 10.1038/gene.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang YK, Yang W, Zhao M, et al. Association of BANK1 and TNFSF4 with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2009;10:414–420. doi: 10.1038/gene.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama K, Su Ih IH, Tezuka T, et al. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. Embo J. 2002;21:83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aiba Y, Yamazaki T, Okada T, et al. BANK negatively regulates Akt activation and subsequent B cell responses. Immunity. 2006;24:259–268. doi: 10.1016/j.immuni.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Hom G, Graham RR, Modrek B, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Ng P, Zhao M, et al. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong. Chinese Genes Immun. 2009;10:219–226. doi: 10.1038/gene.2009.1. [DOI] [PubMed] [Google Scholar]
- 8.Ito I, Kawasaki A, Ito S, et al. Replication of the association between the C8orf13-BLK region and systemic lupus erythematosus in a Japanese population. Arthritis Rheum. 2009;60:553–558. doi: 10.1002/art.24246. [DOI] [PubMed] [Google Scholar]
- 9.Dymecki SM, Niederhuber JE, Desiderio SV. Specific expression of a tyrosine kinase gene, blk, in B lymphoid cells. Science. 1990;247:332–336. doi: 10.1126/science.2404338. [DOI] [PubMed] [Google Scholar]
- 10.Texido G, Su IH, Mecklenbrauker I, et al. The B-cell-specific Src-family kinase Blk is dispensable for B-cell development and activation. Mol Cell Biol. 2000;20:1227–1233. doi: 10.1128/mcb.20.4.1227-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 12.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 13.Wirapati P, Forner K, Delgado-Vega A, et al. Detecting epistasis with restricted response patterns in pairs of biallelic loci. Ann Hum Genet. 2011;75:133–145. doi: 10.1111/j.1469-1809.2010.00625.x. [DOI] [PubMed] [Google Scholar]
- 14.VanderWeele TJ. Epistatic interactions. Stat Appl Genet Mol Biol. 2010;9:Article 1. doi: 10.2202/1544-6115.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattin KA, White BC, Barney N, et al. A computationally efficient hypothesis testing method for epistasis analysis using multifactor dimensionality reduction. Genet Epidemiol. 2008;33:87–94. doi: 10.1002/gepi.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller DJ, Zhang Y, Yu G, et al. An algorithm for learning maximum entropy probability models of disease risk that efficiently searches and sparingly encodes multilocus genomic interactions. Bioinformatics. 2009;25:2478–2485. doi: 10.1093/bioinformatics/btp435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimherr M, Nicolae DL. You’ve gotta be lucky: Coverage and the elusive gene-gene interaction. Ann Hum Genet. 2011;75:105–111. doi: 10.1111/j.1469-1809.2010.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2011;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossin EJ, Lage K, Raychaudhuri S, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandyopadhyay S, Mehta M, Kuo D, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun YV, Kardia SL. Identification of epistatic effects using a protein-protein interaction database. Hum Mol Genet. 2010;19:4345–4352. doi: 10.1093/hmg/ddq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrell JE. What do scaffold proteins really do? Sci STKE. 2000:pe1. doi: 10.1126/stke.2000.52.pe1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.