Summary
Eukaryotic proteasome consists of a core particle (CP), which degrades unfolded protein, and a regulatory particle (RP), which is responsible for recognition, ATP-dependent unfolding and translocation of polyubiquitinated substrate protein. In the archaea Methanocaldococcus jannaschii, the RP is a homohexameric complex of proteasome-activating nucleotidase (PAN). Here we report the crystal structures of essential elements of the archaeal proteasome: the CP, the ATPase domain of PAN, and a distal subcomplex that is likely the first to encounter substrate. The distal subcomplex contains a coiled-coil segment and an OB-fold domain, both of which appear to be conserved in the eukaryotic proteasome. The OB domains of PAN form a hexameric ring with a 13-Å pore, which likely constitutes the outermost constriction of the substrate translocation channel. These studies reveal structural codes and architecture of the complete proteasome, identify potential substrate-binding sites, and uncover unexpected asymmetry in the RP of archaea and eukaryotes.
Introduction
ATP-dependent proteases are found universally in living cells, and are responsible for rapid and selective turnover of many intracellular proteins (Baker and Sauer, 2006; Baumeister et al., 1998; Goldberg, 1992; Inobe and Matouschek, 2008). These proteases mediate the degradation of misfolded, mistranslated, and mutant proteins, which may be toxic if allowed to accumulate, and regulate numerous key proteins such as cyclins and NF-κB, which are degraded in response to specific conditions or stimuli. Unfolding of the substrate protein, an energy-requiring process, is a required early step in the degradation of many substrates. The requirement for unfolding owes to the basic structure of these proteases, in which a proteolytically active topological compartment is linked to a substrate-recognizing ATPase module via a narrow, gated channel through which protein substrates are translocated. The complex architecture of these proteases, and especially their sequestered proteolytic sites, endow them with high specificity, processivity, and capacity for regulation.
Bacteria contain an array of ATP-dependent proteases, such as ClpAP, ClpXP, and HslUV (Baker and Sauer, 2006; Groll et al., 2005). These are typically derived from two gene products, which assemble into stacked ring complexes, thus forming a homo-oligomeric protease component and a homo-oligomeric ATPase component. In contrast, the eukaryotes contain only one known ATP-dependent protease, known as the proteasome (Baumeister and Lupas, 1997; Bochtler et al., 1999; Goldberg, 2007; Hanna and Finley, 2007; Pickart and Cohen, 2004). The proteasome contains 33 distinct subunits, and is thus architecturally different from the bacterial proteases. This complexity results in part from the use of ubiquitin as a substrate recognition tag for the eukaryotic proteasome. The proteasome is composed of a 20S core particle (CP) and a 19S regulatory particle (RP). The CP is responsible for the degradation of unfolded substrate protein, while the RP recognizes polyubiquitinated protein, unfolds the substrate in an ATP-dependent manner, and translocates the unfolded substrate into the CP for degradation (Elsasser and Finley, 2005). The RP contains at least 19 different subunits, which are organized into a CP-proximal base and a distal lid that is connected to the base on the side (Glickman et al., 1998; Walz et al., 1998). The base includes six essential AAA+ ATPases (hereafter the Rpt proteins) that are responsible for key functions of the RP.
The archaeal proteasome has the architectural simplicity of bacterial ATP-dependent proteases, but shares key features of the eukaryotic particle and may represent its evolutionary precursor. The CP assemblies of both archaea and eukaryotic proteasomes are barrel-like structures of four stacked heptameric rings of subunits (Groll et al., 1997; Lowe et al., 1995; Smith et al., 2005; Zwickl et al., 1999). The outer rings are composed of α–type subunits, the inner rings of β-type subunits. In eukaryotes each ring is hetero-oligomeric, and in archaea they are homo-oligomeric, but the overall architecture is very similar. Degradation of the substrate protein occurs in the central chamber formed by the two β-heptamer rings (Groll et al., 1997; Lowe et al., 1995).
The proteasome RP in archaea is assembled from a single protein named proteasome-activating nucleotidase (PAN) (Zwickl et al., 1999). Although PAN can form dodecamers after overexpression in E. coli, it is thought to function as a hexamer (Smith et al., 2005). Unlike the ATPase components of the bacterial proteases, PAN shares extensive sequence homology with the six Rpt proteins of the eukaryotic proteasome (Zwickl et al., 1999). The PAN complex is sufficient to execute essential functions shared with eukaryotic RP, such as substrate recognition, unfolding, and translocation (Smith et al., 2006).
Mechanistic understanding of proteasome function requires elucidation of its three-dimensional structure, particularly that of the RP, which is considerably more complex than the CP. Structures of the 20S CP have been elucidated for Thermoplasma acidophilum (Lowe et al., 1995), Archaeoglobus fulgidus (Groll et al., 2003), Mycobacteria tuberculosis (Hu et al., 2006), Saccharomyces cerevisiae (Groll et al., 1997), and mammals (Unno et al., 2002a, b). Structures of two RP components, Rpn13 (Schreiner et al., 2008) and the C-terminus of Rpt3 (Nakamura et al., 2007), are also known. However, vigorous effort has failed to unveil any atomic structure of the intact eukaryotic RP or its archaeal homolog. In this manuscript, we report three crystal structures from the archaea Methanocaldococcus jannaschii (M. jannaschii): the CP, the adjacent ATPase domain of PAN, and the distal subcomplex of PAN.
Results
Identification of two subcomplexes in the archaeal RP
The three gene products that compose the M. jannaschii proteasome, PAN and the α and β subunits of the CP, were individually expressed and purified to homogeneity. The α and β subunits were subsequently assembled into a stable 20S core particle (data not shown). Despite years of intense effort, crystals of the PAN regulatory complex failed to diffract X-rays beyond 8 Å resolution. Reasoning that the poor diffraction quality of the crystals might be in part due to flexible surface loops, we subjected the oligomeric PAN complex to limited proteolysis, which led to the generation of two non-overlapping subcomplexes.
Analysis by mass spectroscopy and N-terminal peptide sequencing revealed that subcomplex I spans residues 74-150, which comprise portion of a predicted coiled-coil domain and a predicted globular domain. Subcomplex I was stable and exhibited an apparent molecular mass of approximately 50-kDa by gel filtration (Supplementary Figure 1), suggesting a hexameric assembly of this 8.4-kDa fragment. Subcomplex II extends from residue 155 to the C-terminus of PAN (residue 430), and represents the entire nucleotidase domain. In contrast to subcomplex I, subcomplex II dissociated slowly on gel filtration and was eluted in a broad range of molecular weights from 200-kDa, which corresponds to a hexameric assembly, to 30-kDa, which corresponds to a monomer (Supplementary Figure 1). These observations are consistent with previous studies using electron cryo-microscopy (Smith et al., 2005) and further indicate that residues 74-150 make a major contribution to the formation of the PAN hexamer.
Structure of subcomplex I of the archaeal RP
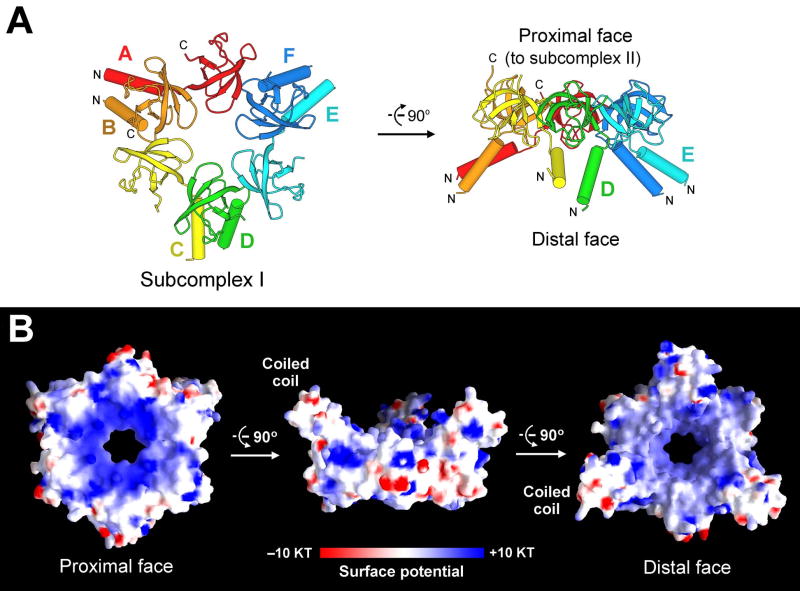
We succeeded in crystallization of subcomplex I and determined its structure at 2.1 Å resolution by multi-wavelength anomalous dispersion (Figures 1 and 2, Table 1, Supplementary Table 1). Each PAN fragment contains an N-terminal α-helix (residues 74-89) followed by a compact β-domain (residues 90-150) (Figures 1A and 2). The 6 β-domains, each comprising a 5-stranded β-sandwich, associate with each other with a pseudo six-fold symmetry to form a donut-shaped structure with an axial pore of 13 Å (Figure 1B), which, as discussed below, is presumably the first constriction through which substrates must pass to be degraded. Three pairs of the asymmetric dimers associate with each other to form the hexameric subcomplex I (Figure 1A), which has a diameter of 70 Å and a height of 40 Å. Within each dimer, the two α-helices pair up to form a coiled-coil, but the coiled coil is skewed towards one member of the dimeric pair. The position of the coiled coils at the distal face of PAN suggests that they may assist in substrate recognition.
Figure 1.
Structure of subcomplex I of the PAN regulatory particle. (A) Structure of the hexameric subcomplex I. The 6 Pan fragments are labeled A through F. Each Pan fragment (residues 74-150) contains an N-terminal α-helix followed by a β-domain that is structurally homologous to the oligonucleotide/oligosaccharide binding fold (OB-fold). Two α-helices from adjacent domains form a coiled coil. The distal and proximal faces refer to their relative orientation with respect to the 20S core particle. (B) Surface potential of subcomplex I. The positive and negative charges are colored blue and red, respectively.
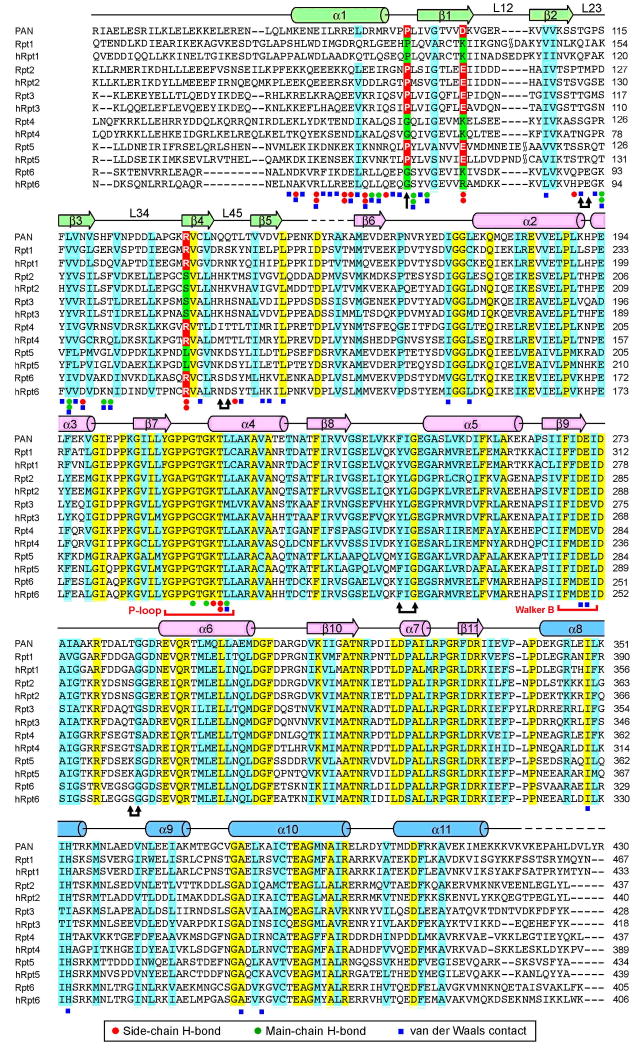
Figure 2.
Structure-based sequence alignment between PAN and the AAA+ ATPases in yeast and human proteasomes. Rpt1 through Rpt6 are derived from S. cerevisiae. Secondary structural elements of PAN are indicated above the sequences. Invariant and conserved amino acids are colored yellow and cyan, respectively. Residues that correspond to Pro91, Asp99, and Arg134 of PAN, are highlighted in red and green. Doubled arrows indicate residues that are predicted to line the substrate translocation channel. The loop that falls between β8 and α5 is known as pore loop 1 (pore-1 loop, or Ar-Φ loop), and that between α8 and α9 is pore loop 2 (pore-2 loop). The OB domain pore loops are termed L23 and L45.
Table 1. Summary of data collection and structure refinement.
| Data Collection | |||
| Data set | Subcomplex I | PAN nucleotidase | Core Particle |
| Wavelength (Å) | 0.9791 | 1.10 | 1.08 |
| Resolution (Å) | 50-2.1 | 50-3.10 | 50-4.10 |
| Unique reflections | 58,588 | 22,762 | 54,076 |
| Redundancy | 6.4 | 6.6 | 4.8 |
| I/σ (outer shell) | 11.0 (2.8) | 19.7 (3.1) | 9.5 (3.5) |
| Completeness (%) (outer shell) | 99.3 (95.4) | 99.8 (100.0) | 99.8 (100.0) |
| Rsym (outer shell) | 0.100 (0.376) | 0.084 (0.523) | 0.126 (0.488) |
| Refinement | |||
| Resolution (Å) | 50-2.1 | 50-3.1 | 50-4.1 |
| Reflections (work/free) | 55,430/2,931 | 19,605/1,092 | 51,010/2,737 |
| Completeness (%) | 98.8 | 95.8 | 99.8 |
| R-work/R-free | 0.212/0.259 | 0.221/0.277 | 0.265/0.321 |
| RMSD bond lengths (Å) | 0.0084 | 0.011 | 0.009 |
| RMSD bond angles (°) | 1.52 | 1.43 | 1.26 |
Rsym = ΣhΣi | Ih,i - Ih | / ΣhΣi Ih,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h. R = Σ |Fobs - Fcalc | / ΣFobs, where Fobs = FP, and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections). R.m.s.d. in bond lengths and angles are the deviations from ideal values, and the r.m.s.d. deviation in B factors is calculated between bonded atoms.
The extensive interactions among the 6 PAN fragments within the hexameric assembly result in the burial of approximately 9100 Å2 of otherwise exposed surface area. The inter-subunit interactions are rich in hydrogen bonds (H-bonds), which are susceptible to distance changes. These structural features, together with the observation that subcomplex I consists exclusively of rigid β-strands at the center (Figure 1A), suggest that, if the central ring formed by the β-domains were to undergo conformational changes upon substrate binding or during ATP hydrolytic cycles, such changes must be coordinated and likely affect the overall conformation.
The N-terminal face of subcomplex I is on the same side as the coiled coils and is hereafter referred to as the distal face, as it is farther away from the CP in the assembled proteasome. Similarly, the C-terminal face is spatially connected to subcomplex II and referred to as the proximal face (Figure 1A). The proximal face contains a high density of positively charged amino acids surrounding the axial pore whereas the distal face has a mixture of positively charged and hydrophobic residues (Figure 1B).
The observed structural features of subcomplex I in the PAN regulatory complex appear to be generally conserved in eukaryotes. Sequence alignment of PAN with Rpt1-6, the six AAA+ ATPases in the base of the RP from S. cerevisiae, indicates that each of the Rpt subunits contains a coiled-coil domain followed by a highly conserved β-domain (Figure 2). Thus, the 6 putative β-domains in Rpt1-6, as well as their counterparts from humans, are predicted to form the central ring of a hexameric complex, with the adjacent 6 α-helices forming three pairs of coiled-coils.
OB fold in the RP
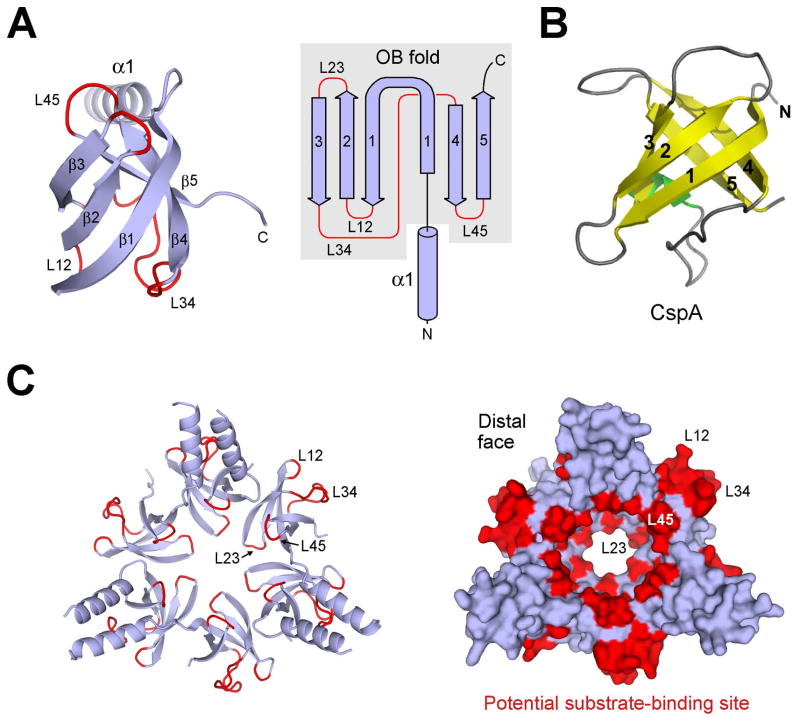
An unanticipated finding is that structure of the β-domain conforms to the oligonucleotide/oligosaccharide-binding fold, known as the OB-fold (Agrawal and Kishan, 2003; Arcus, 2002; Theobald et al., 2003) (Figure 3A). Similar to other OB-fold proteins such as CspA (Schindelin et al., 1994) (Figure 3B), strand β1 in the OB-fold domain of PAN participates in two β-sheets, with its N-terminal portion pairing up with strand β4 and C-terminal half interacting with β2 (Figure 3A). Residues 74-150 of PAN are hereafter referred to as the coiled-coil OB-fold domain, or CC-OB domain. Previous studies of OB-fold proteins revealed that the surface loops connecting the 5 β-strands generally play an important role in ligand binding (Agrawal and Kishan, 2003; Arcus, 2002; Theobald et al., 2003). For example, loop L23 in Cdc13, which connects β-strands β2 and β3, makes crucial contacts with telomeric DNA elements (Mitton-Fry et al., 2002). In subcomplex I, these surface loops are concentrated in two areas: L23 and L45 line the inner surface of the central passage whereas L12 and L34 are located on the outer surface between coiled-coils (Figure 3C). Thus, two of the surface loops are properly positioned to contact or transiently bind the protein substrate as it enters the translocation channel. In agreement with this scenario, residues in L23 appear to impose the narrowest passage in the central pore of subcomplex I (Figure 3C).
Figure 3.
The β-domain of PAN exhibits an oligonucleotide/oligosaccharide binding fold (OB-fold). (A) Ribbon (left panel) and topology (right panel) diagrams of the β-domain of PAN reveal an OB-fold. The surface loops connecting adjacent β-strands are highlighted in red. (B) Structure of a representative OB-fold protein CspA (Schindelin et al., 1994). (C) Candidate substrate-binding site in subcomplex I. Subcomplex I is represented in ribbon diagram (left panel) and surface potential (right panel). The surface loops are concentrated in two areas: inner surface of the central passage (L23 and L45) and outer surface between coiled-coils (L12 and L34).
t- and c-type subunits in the RP
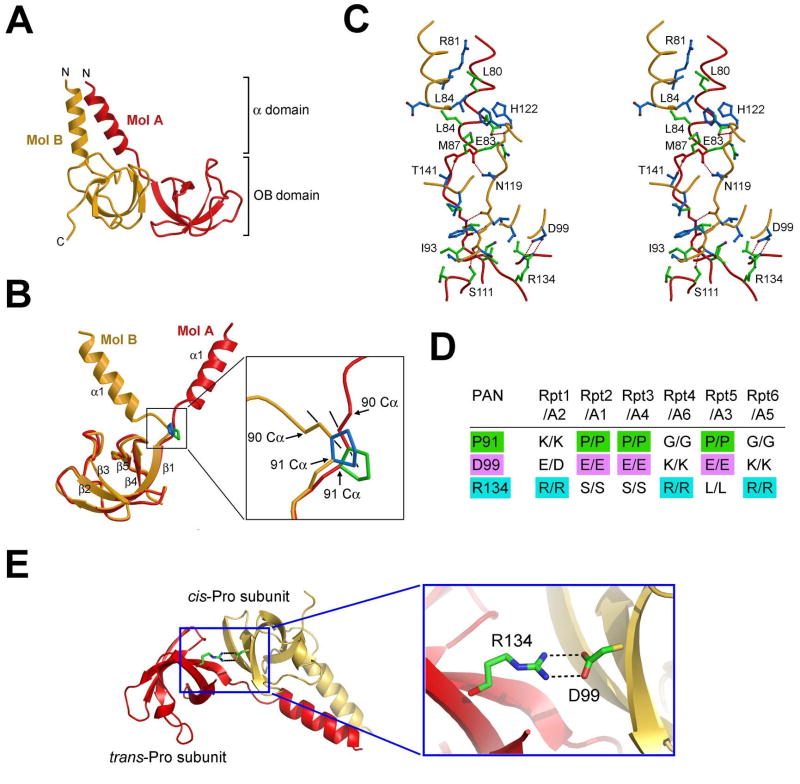
Subcomplex I is a trimer of asymmetric dimer (Figure 1A). To facilitate description of subcomplex I, the 6 CC-OB domains are designated molecules A through F (Figure 1A). An asymmetric dimer (such as that involving molecules A and B) contains a coiled-coil formed by two α-helices (Figure 4A). The underlying key to this asymmetry is the main chain configuration of Pro91, which alternates across subunits within a hexameric ring. Configuration of the peptide bond preceding Pro91 is trans in three subunits (molecules A, C, and E), and cis in the other three (molecules B, D, and F) (Figure 4B). Hence, we designate the first set as trans-Pro subunits, or t-type subunits, and the second set as cis-Pro subunits, or c-type subunits. Although the OB domains of t-type and c-type subunits are nearly identical, with a root-mean-squared deviation (rmsd) of ∼0.7 Å, their α-helices are nearly perpendicular to each other relative to the aligned OB domains (Figure 4B). The divergent trajectories of the α-helices are linked to the configuration of the peptide bond preceding Pro91, and the cis configuration in the c-type subunit likely plays an important role in allowing its α-helix to form a coiled-coil with the corresponding α-helix in the t-type subunit. Apart from Pro91, all amino acids in the two molecules have virtually identical conformations. To confirm the importance of Pro91, we generated three mutations: P91A, P91G, and P90A/P91A. Unfortunately, none of these mutant proteins was stable enough to allow biochemical characterization, as all mutant proteins were readily degraded during expression and purification (data not shown). This observation highlights the structural role of Pro91 and further demonstrates its critical importance in maintenance of the correct conformation for PAN.
Figure 4.
Conserved asymmetry in subcomplex I of the PAN regulatory particle. (A) Interface between molecules A and B involves both the coiled-coil and the OB-fold domains. (B) Structural difference between molecules A and B is rooted in the peptide bond configuration preceding Pro91. This bond exists in trans configuration in molecule A and cis in molecule B. (C) A stereo view of the A-B interface. Side chains from molecules A and B are colored green and blue, respectively. H-bonds are represented by red dashed lines. (D) A conserved pattern of asymmetric amino acids that play key roles in stabilizing the structure of subcomplex I. The asymmetry defines two types of subunits in subcomplex I: cis-Pro subunits (c-type) and trans-Pro subunits (t-type). (E) A conserved pair of H-bonds at the interface between molecules B and C. Arg134 and Asp99 come from the t- and c-type subunits, respectively.
There are two types of interface within the same hexameric assembly: the interface within the asymmetric dimer (Figure 4C) and the interdimer interface (Supplementary Figure 2). The intradimer interface involves both the OB domains and the coiled-coils, while the less intimate interdimer interface involves only the OB domains. The intradimer interface contains 9 inter-molecular H-bonds and extensive van der Waals interactions, involving 2214 Å2 surface area (Figure 4C). Within the OB domain, Asp99 of the c-type subunit accepts a pair of charge-stabilized H-bonds from Arg134 of the t-type subunit (Figure 4C). Within the CC domain, hydrophobic amino acids Leu80, Leu84, and Met87 of the t-type subunit stack closely against Leu80, Leu84, and the aliphatic side chain of Arg81 of the c-type subunit.
Conserved asymmetry in the RP
Among the 20 naturally occurring amino acids, only Pro allows the preceding peptide bond to populate the cis configuration. Thus, because the six yeast Rpt subunits are thought to form a structure similar to that of subcomplex I from M. jannaschii (Figure 2), we predicted that three of the six Rpt subunits should belong to the c-type, namely, three should contain a Pro residue that corresponds to Pro91 of PAN. Sequence alignment confirmed this prediction (Figure 2). Rpt2, Rpt3, and Rpt5 each have a conserved Pro at this position; consequently, these Rpt subunits may correspond to c-type subunits. Similar analysis confirmed the same prediction in the human proteasomal RP (Figures 2 and 4D).
We have described two asymmetries in the CC-OB structure: the positioning of the coiled-coils and the alternating cis-trans configuration of the peptide bond preceding Pro91. A third asymmetry in the CC-OB structure further strengthens the physiological relevance of these asymmetries and their generality to the eukaryotic proteasome. This more subtle asymmetry is a direct result of asymmetric dimer formation – which leads to a slight intradimer rotation of the two OB domains relative to the central axis of the hexamer. Consequently, stabilizing contacts are established between the OB domains of intradimeric but not interdimeric neighbors: Asp99 of a c-type subunit accepts two H-bonds from Arg134 of the t-type subunit (Figure 4E).
If, as suggested above, Rpt2, Rpt3, and Rpt5 are c-type subunits, they should each contain a conserved acidic residue in the position corresponding to Asp99. Indeed, this prediction was verified by the identification of an invariant Glu residue (Figure 4D). This analysis further suggests that Rpt1, Rpt4, and Rpt6 should be t-type subunits. Consistent with this view, Rpt1, Rpt4, and Rpt6 each contain a conserved Arg residue that aligns with Arg134 of PAN. Moreover, the position of Pro91 is not strictly conserved in these predicted t-type subunits (Figure 2), as they have no need to form a cis peptide bond. In summary, we propose that PAN and Rpts of the eukaryotic proteasome share key structural features in the CC-OB element, especially a linked ensemble of asymmetries that may be relevant to the functioning of these complexes. Remarkably, the asymmetries that are embedded in the sequence features of the hetero-oligomeric Rpt ring are exactly reflected in the structure of the homo-oligomeric PAN complex.
Subcomplex II of the archaeal RP
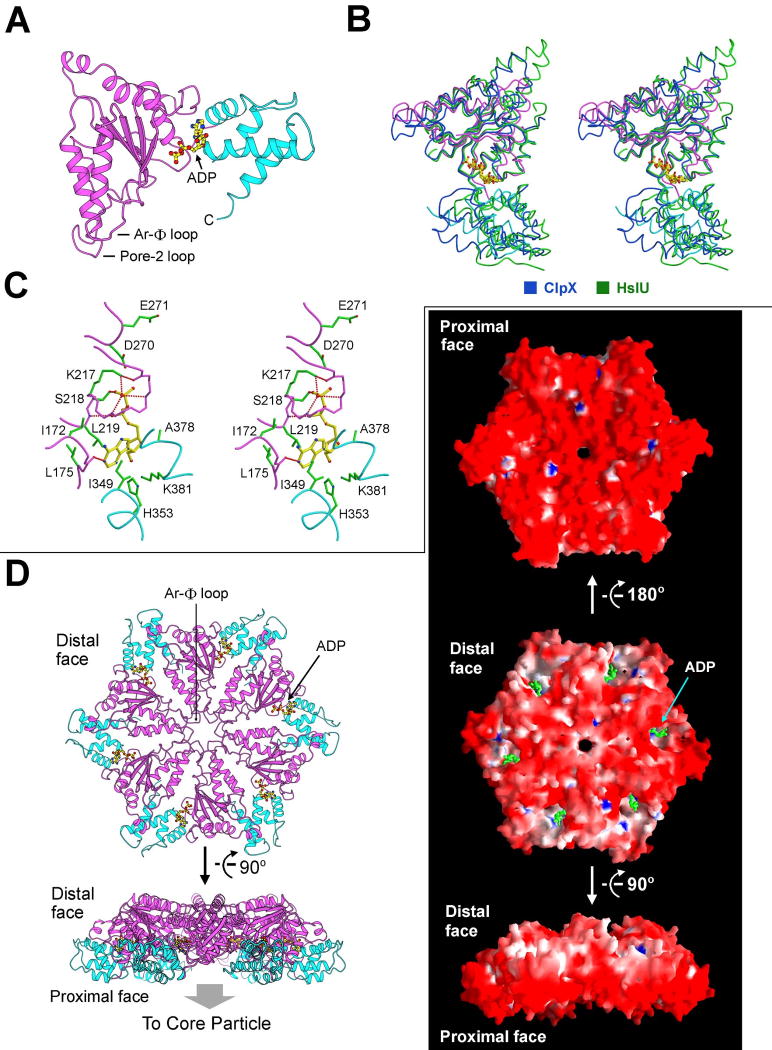
Compared to subcomplex I, subcomplex II was unstable and dissociated slowly (Supplementary Figure 1). We combined the fractions containing subcomplex II, generated crystals, and determined the structure at 3.1 Å resolution (Table 1, Supplementary Table 2). There are three molecules of the PAN nucleotidase domain (residues 155-430) in one asymmetric unit (Supplementary Figure 3). The lack of 6-fold symmetry confirms that subcomplex II had dissociated during purification and crystallization. The structure of the PAN nucleotidase domain conforms to the canonical fold of AAA+ ATPases, with a nucleotide-binding α/β fold and a characteristic small helical domain toward the C-terminus (Lupas and Martin, 2002) (Figure 5A). Interestingly, the bound nucleotide is ADP, not ATP. To obtain the ATP-bound structure, we attempted to crystallize the walker B mutant nucleotidase domain in the presence of ATP but were only able to obtain the ADP-bound crystals. These observations suggest that ADP-binding may be favored over ATP, which may have interesting implications for PAN function.
Figure 5.
Structure of subcomplex II of the PAN regulatory particle. (A) Structure of the nucleotidase domain of PAN. The α/β fold and the helical domain are shown in magenta and cyan, respectively. ADP is bound to the hinge region between these two domains. (B) The nucleotidase domain of PAN is structurally similar to the AAA+ ATPases of ClpX and HslU. Shown here is a stereo comparison of the relevant structures. (C) ADP is coordinated by residues from both the α/β fold and the helical domain. (D) Model of the nucleotidase ring. The atomic coordinates of the PAN nucleotidase were superimposed onto those of the HslU hexamer (Bochtler et al., 2000; Sousa et al., 2000) to generate the nucleotidase ring. Two perpendicular views are shown on the left. The surface representations are shown on the right. Note the highly acidic proximal face.
The core elements of the PAN ATPase can be superimposed with those of the representative AAA+ ATPases ClpX (Kim and Kim, 2003) and HslU (Bochtler et al., 2000; Sousa et al., 2000) with rmsd of 1.65 Å for 115 Cα atoms and 1.92 Å for 140 Cα atoms, respectively (Figure 5B). ADP is bound between the α/β domain and the small helical domain (Figure 5C), suggesting that binding and hydrolysis of ATP may impact on the relative position of these two domains. In addition to the P-loop (Walker A motif) and the Walker B motif in the α/β fold, the small helical domain also contributes four amino acids to bind nucleotide. Ile349 and His353 interact with the adenine moiety whereas Ala378 and Lys381 stabilize the ribose (Figure 5C).
AAA+ ATPases are ATP-dependent molecular machines with highly conserved oligomeric structures (Baker and Sauer, 2006; Davey et al., 2002; Erzberger and Berger, 2006; Lupas and Martin, 2002; Sauer et al., 2004). The conserved structural features between PAN and the bacterial ATPase HslU (Bochtler et al., 2000; Sousa et al., 2000) allowed us to superimpose the atomic coordinates of the PAN nucleotidase domain onto the six molecules of HslU to generate a model for subcomplex II (Figure 5D). The quality of the model is manifested by the paucity of steric clashes in the main chains of adjacent molecules. Interestingly, the modeled subcomplex II contains a very narrow central passage, with a surface diameter of less than 5 Å. As observed in other oligomeric AAA+ ATPases (Hinnerwisch et al., 2005; Lupas and Martin, 2002; Martin et al., 2008a; Wang et al., 2001), the Ar-Φ loop (also known as pore-1 loop) and the pore-2 loop (Figure 5A, D) line the central passage and define the constriction points. The bound nucleotide is partially exposed on the distal face, which may closely approach the proximal face of subcomplex I. Electrostatic interactions may promote such contacts, because the negative charges at the distal face of the modeled subcomplex II (Figure 5D) are complementary to the positive charges at the proximal face of subcomplex I (Figure 1B).
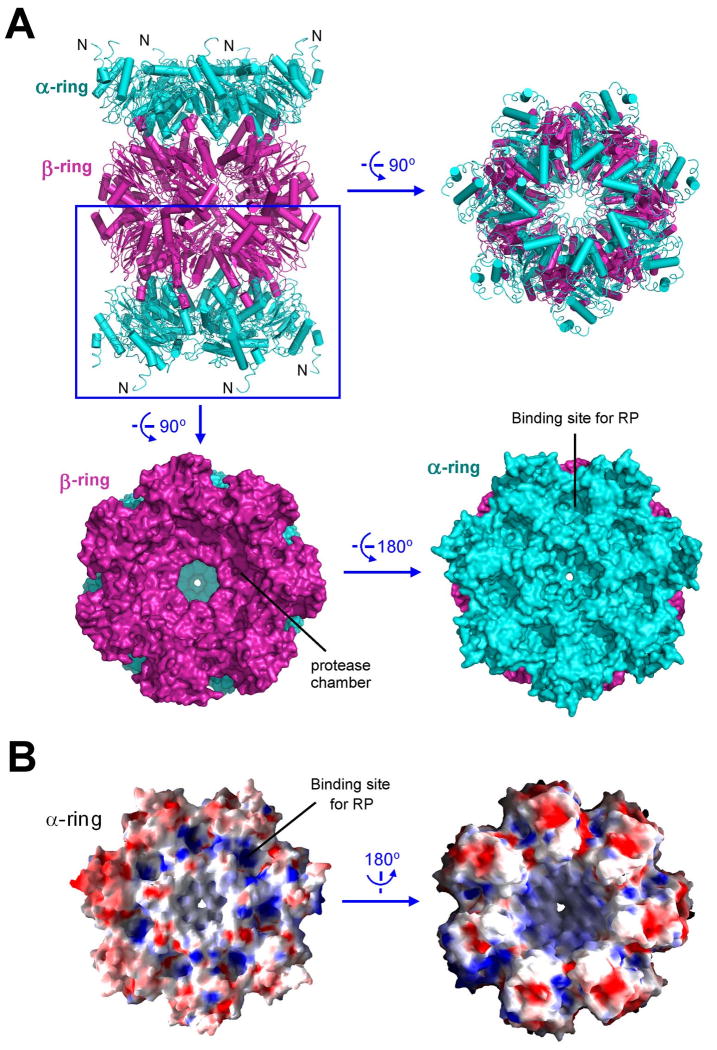
Structure of the CP
The archaeal proteasome that has been extensively characterized contains the PAN complex from M. jannaschii but the CP from T. acidophilum, in part because PAN was initially cloned from M. jannaschii (Zwickl et al., 1999), whereas structural information is available on the CP from T. acidophilum (Lowe et al., 1995) but not from M. jannaschii. To understand assembly of the complete proteasome within the same species, we crystallized the 20S core particle from M. jannaschii and determined its structure (Table 1, Supplementary Table 2). The 20S CP contains 14 α and 14 β subunits, which assemble into four stacked rings of α7-β7-β7-α7 (Figure 6A). Three chambers are formed within the CP; the central chamber formed by the two β rings is where proteolysis occurs. Delivery of an unfolded substrate peptide to the central chamber requires passage through the central channel of the α ring as well as a constriction in the β ring. The constriction in the β ring is constitutively open and large enough to allow passage of an extended polypeptide chain (Figure 6A). However, the axial channel in the α ring is thought to be closed in the absence of the RP (Forster et al., 2005; Rabl et al., 2008; Smith et al., 2007). Indeed, the constriction of the α ring of the M. jannaschii CP is too narrow to allow passage of peptide chain and exists in a closed state (Figure 6A). Opening of the gated channel of the α ring is thought to be induced by binding of the RP (Groll et al., 2000). The C-terminal residues of PAN were previously shown to bind directly to the CP (Forster et al., 2005; Rabl et al., 2008; Smith et al., 2007). Consistent with this analysis, the exposed surface of the α ring contains 7 putative binding pockets (Figure 6B). A conserved Lys residue known to play a key role in binding to RP (Smith et al., 2007) resides at the base of this pocket.
Figure 6.
Structure of the 20S core particle from M. jannaschii. (A) Structure of the 20S core particle. The α and β rings are colored cyan and magenta, respectively. Two perpendicular views of the ribbon representation are shown in the top panel. Surface representation of the α-β double ring is shown in the bottom panel. (B) Electrostatic surface potential of the α ring.
Discussion
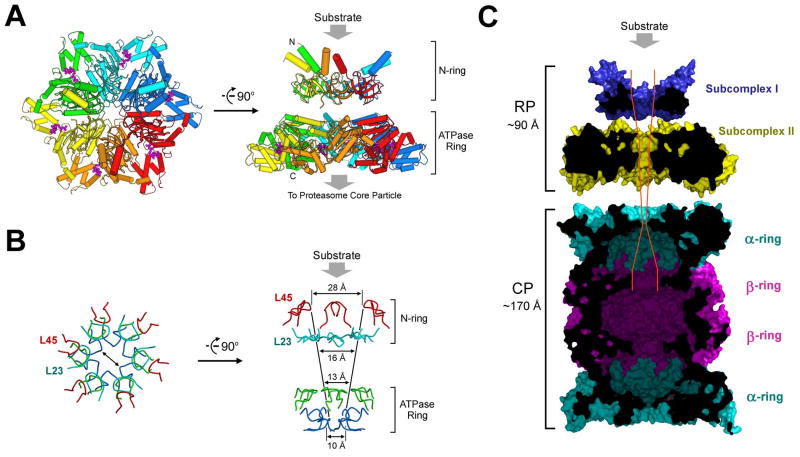
Structural information on subcomplex I and the PAN nucleotidase domain allowed modeling of the PAN regulatory particle (Figure 7A). In this model, the ATPase ring (subcomplex II) is formed by 6 nucleotidase domains, and subcomplex I consists of 6 CC-OB domains. The C-terminal residues of subcomplex I are connected to the N-termini of the ATPase ring, likely through flexible sequences (residues 151-158), because limited proteolysis of the PAN complex occurred in this region and readily generated subcomplexes I and II. Our structure-based modeling is fully consistent with previous EM study, which revealed a two-ring architecture of the PAN regulatory complex (Smith et al., 2005).
Figure 7.
Structural and functional implications on the proteasome. (A) A structure-based model of the PAN regulatory particle. Substrate protein is thought to bind to the distal face of subcomplex I as shown. (B) The constrictions are conferred by four layers of surface loops. The first two layers come from subcomplex I, involving loops L23 and L45, and the latter two layers come from the Ar-Φ and pore-2 loops in the nucleotidase domains. (C) A structural view of the complete proteasome in M. jannaschii. The modeled, complete proteasome is cut across the middle section to show the path and constrictions to the degradative chamber in the β-rings. The placement of subcomplexes I and II and their position relative to the 20S CP were based on measurement of the reported electron microscopic images of the PAN-20S complex (Smith et al., 2005).
A major finding from the present study is the existence of an unexpected domain, the OB domain, within PAN and the Rpt proteins. Six OB domains assemble to form a ring structure with an inner surface diameter of 13 Å. The constrictions, formed by loops L23 and L45, are too narrow to allow passage of folded proteins (Figure 7A, B). The positioning of this ring at the distal face of the ATPase ring indicates that it is most likely the site of substrate entry into the axial translocation channel (Figure 7A). Thus, we postulate that the requirement for substrate to pass through the OB ring is the critical barrier by which the selectivity of proteasomes for unfolded proteins is enforced. Substrates need to be translocated approximately 60 Å further to reach the proteolytic sites of the enzyme, through additional constrictions in the ATPase ring (Figure 7B, C).
In addition to functioning as a molecular sieve, we suggest that the OB domain may also serve as a fulcrum for substrate unfolding. If the substrate is unfolded by being tugged into the channel through motions of the Ar-Φ loop of the ATPase domain (Hinnerwisch et al., 2005; Inobe et al., 2008; Martin et al., 2008a; Martin et al., 2008b; Wang et al., 2001), unfolding will be mediated by an opposing force acting on the substrate, which blocks its movement. We propose that this force is exerted passively by the OB domain. The surprising stability of the OB complex may reflect this inherent resistive capacity. The presence of the OB channel distal to Ar-Φ sequesters the Ar-Φ loop from folded protein domains and allows the Ar-Φ loop to act selectively on polypeptides that have already threaded through the channel. It should be noted, however, that the structural stability of the OB ring does not necessarily imply that it cannot undergo conformational changes during substrate binding or ATP hydrolytic cycles of proteasome function. Rather, the structural observations strongly suggest that any such changes are likely to be coordinated and may impact on the overall conformation of the OB ring and the RP.
The proteasomal OB domains might interact with substrate, because ligand-binding is a general feature of OB domains (Agrawal and Kishan, 2003; Arcus, 2002; Theobald et al., 2003). The face of the proteasomal OB domains that lines the substrate entry site is one that typically engages in ligand binding. Interestingly, for each eukaryotic Rpt protein, the predicted pore residues of the OB domains are highly conserved (Supplementary Figure 4). Moreover, amino acid substitutions in this region of the OB domain cause major defects in the protein unfolding and degradation by PAN (Zhang et al, to be submitted). A key question for future work will be how putative binding interactions between OB domains and specific segments of protein substrates may influence proteasome function. The OB domains might promote the initiation of substrate threading into the channel, and thus increase the likelihood of productive contacts between substrate and pore-1 loop. Alternatively, substrate binding by OB domains might promote unfolding of the substrate or help to target the substrate to the proteasome. The last scenario is more plausible for the archaeal proteasome than the eukaryotic proteasome, where ubiquitin plays a dominant role in targeting.
The OB ring may provide an interpretive framework for understanding several prior studies of proteasome function. Constitutively unfolded sequences can be used by the proteasome to drive unfolding of the remainder of the substrate (Prakash et al., 2004). The degradative “initiation sequences” may need to reach through the OB pore to the Ar-Φ loops before active unfolding can begin. The length requirement for initiation sequences may thus be related to the distance from the OB domain to Ar-Φ. On the other hand, some sequences, such as those found in the viral EBNA-1 protein, have an interfering effect on degradation (Levitskaya et al., 1997). These might physically block the translocation channel by interacting unusually strongly with the OB domain, or perhaps the downstream Ar-Φ and pore-2 loops.
The unfolding activity of PAN is linked to ATP hydrolysis (Zwickl et al., 1999). ATP-driven conformational changes within subcomplex II can be expected to underlie this unfolding activity. A question for future work is whether the CC-OB domain also undergoes ATP-dependent conformational changes. A related challenge will be to determine what role the coiled-coils of the CC-OB domain play in substrate processing. To address these questions, we have initiated systematic mutagenesis of surface-exposed residues in PAN (Zhang et al, submitted).
Whether the asymmetries and dimeric pairings of the CC-OB domain are associated with similar structural features of the nucleotidase domain remains unclear. However, it seems likely that the nucleotidase domain of the PAN hexamer is asymmetric within the assembled proteasome, if only because it is required to interface with the heptameric CP. Asymmetry within the PAN nucleotidase domain has also been suggested by EM studies (Smith et al., 2005) and by the finding that not all subunits within the PAN homohexamer can bind nucleotide simultaneously (Horwitz et al., 2007).
As the proteasome evolved from a PAN-like evolutionary precursor, contemporaneously with the introduction of ubiquitin as a degradation tag, the proteasome RP became far more complex, and specified by 19 gene products rather than one. The striking similarities between PAN and the Rpt proteins revealed in this study, however, indicate that the architectural features of this core assembly of the RP remained little altered. The recognition and manipulation of ubiquitin chains is the shared responsibility of many of the additional subunits, all of which lack apparent ATPase domains. But the addition of such new subunits is perhaps not the only means by which the proteasome acquired new functions in eukaryotes. The diversification of the PAN homohexamer into the Rpt heterohexamer may have enhanced its capacities significantly, without gross changes in architecture. In particular, the sequence differentiation from Rpt to Rpt within Ar-Φ, the OB pore, and the coiled-coil elements (Supplementary Figure 4) may all provide the Rpt hexamer with a more robust capacity for substrate recognition, unfolding, and translocation.
Experimental procedure
Protein preparation and crystallization
PAN and 20S proteasome of M. Jannaschii were cloned from genomic DNA using standard PCR procedure. The β subunit of 20S proteasome lacked six N-terminal residues (βΔ6) to gain a constitutive peptidase activity (Wilson et al., 2000). All proteins were expressed in E. coli BL21 (DE3). The C-terminally His6-tagged PAN was purified using Ni-NTA (Qiagen), followed by anion-exchange (Source-15Q, GE) and gel-filtration (Superdex-200, GE). For α subunit, the fusion protein eluted from Ni-NTA column was further treated with thrombin to release the N-terminal His6 tag prior to chromatography. For non-tagged β subunit, the crude cell lysate was heated at 85 °C for 15 min and cooled down on ice for 30 min. The supernatant was then applied to cation-exchange (Source-15S, GE) and gel-filtration for further purification. Selenomethionine-labeled CC-OB domain was generated as described (Sreenath et al., 2005).
All diffracting crystals were grown at 22 °C using the hanging drop vapour diffusion method. For subcomplex I, the reservoir buffer contained 0.1 M NaOAc, pH 4.5, 0.2 M Li2SO4 and 34%-36% MPD (v/v). The crystals appeared overnight. Macro-seeding was used to generate single, large crystals. Crystals were slowly equilibrated in a dehydration buffer containing 0.1M NaOAc, pH 4.5, 35% MPD (v/v) and 20% PEG 400 (v/v) and flash frozen in liquid nitrogen. For the nucleotidase domain, the protein was mixed with an equal volume of the reservoir solution containing 0.1 M NaOAc, pH 4.6, and 0.65 M NH4H2PO4. For the 20S CP, the reservoir contained 0.1 M Tris, pH 6.8, 0.2 M KCl and 33-36% MPD (v/v). The α and β subunits were mixed at 1:1 molar ratio and heated at 65 °C for 30 min just before setting up tray. 2 mM ATP, 20 mM MgCl2 and 8% Sucrose-10 was added into the protein mixture as additives. The crystals grew to full size in 2-3 days. All data sets were collected at NSLS beamline X29 and processed using HKL2000 (Otwinowski and Minor, 1997).
Structure determination of subcomplex I
Subcomplex I was crystallized in space group P21212 with cell dimensions of a = 126.6 Å, b = 129.6 Å, c = 60.6 Å. The structure was determined by SeMet MAD experimental phasing using the program SHELX (Sheldrick, 2008). Initial phases were improved using SHARP (Bricogne et al., 2003), and the use of two SeMet MAD datasets resulted in an electron density map with improved interpretability, although phasing statistics are likely to be biased. Non-crystallographic symmetry was determined from inspection of the maps in the vicinity of the SeMet sites, and optimized using the program NCS6D (Jones, 1992). There are 12 molecules in the asymmetric unit (Supplementary Figure 5), with approximate 32 symmetry. Six-fold averaging significantly improved the interpretability of the electron density maps. The structure was built using the programs O (Jones et al., 1991) and COOT (Emsley and Cowtan, 2004) and refined using CNS (Brunger et al., 1998) against the high resolution SeMet peak wavelength dataset (Supplementary Table 1). Non-crystallographic symmetry restraints were used in the early stages of refinement but then released as subtle differences between the chains became evident. Three dimers form a subcomplex; two subcomplexes associate with each other through their respective coiled coils. There is a mixture of H-bonds and van der Waals contacts at the inter-subcomplex interface (Supplementary Figure 6).
Structure determination of the nucleotidase domain
The nucleotidase domain of PAN was crystallized in space group P65 with cell dimensions a = b = 116.43 Å, c = 164.17 Å, with three molecules in the asymmetric unit. The structure was determined by molecular replacement using coordinates of p97 AAA+ ATPase (Dreveny et al., 2004) and the program PHASER (Storoni et al., 2004). There were three independent molecules in the asymmetric unit. The structure was built into the model-phased electron density maps using the program O (Jones et al., 1991) and refined using the program REFMAC (Winn et al., 2003) with tightly restrained individual B-factors and six TLS groups corresponding to the N- and C- terminal domains of each of three monomers. Refinement is summarized in Supplementary Table 2.
Structure determination of the 20S CP
The 20S CP of M.jannaschii crystallized in two crystal forms in identical conditions. The larger form diffracted to 4.5 Å resolution and was in space group P212121 with a = 205.1 Å, b = 219.3 Å and c = 299.5 Å, and contained one whole and two half 20S complexes. The smaller form diffracted to 4.1 Å resolution and was in space group P21212 with cell dimensions of a = 206.7 Å, b = 219.5 Å and c = 149.0 Å, with two half complexes (α7β7) in the asymmetric unit. Structure of the larger form was first solved by molecular replacement using the structure of the Archaeoglobus fulgidus 20S proteasome (Groll et al., 2003) using the program PHASER (Storoni et al., 2004). Heptamer rings of α or β subunits were used as models, and the solution unambiguously placed the rings to form one full and two half-complexes in the asymmetric unit, comprising a total of 28 α subunits and 28 β subunits. The structure was built into 28-fold averaged model-phased 2Fo-Fc electron density maps calculated from data that had been sharpened by applying a -60 Å2 B-factor to the experimental data to give more detail in the maps. The structure was refined using the program PHENIX (Adams et al., 2002) against this sharpened data and also against the unmodified data. Final refinements were performed against unmodified structure factor amplitudes (Supplementary Table 2). Refinement was heavily restrained by 28-fold non-crystallographic symmetry with an average RMS between subunits of approximately 0.1 Å on all atoms. One B-factor per subunit was refined. Eight TLS groups, one per heptameric ring, were refined. The biasing of R-free by the high level of non-crystallographic symmetry was reduced by selecting the test set reflections in spheres of constant resolution. The refinement programs were CNS (Brunger et al., 1998) and PHENIX (Adams et al., 2002). The structure was built using COOT (Emsley and Cowtan, 2004) and O (Jones et al., 1991). Structure of the smaller form was similarly solved and refined. See Supplementary Table 2 for data collection and refinement statistics.
Supplementary Material
Acknowledgments
We thank Anand Saxena at BNL NSLS for help. The atomic coordinates of subcomplex I, the nucleotidase domain, and the core particle have been deposited in the Protein Data Bank with the accession codes XXXX, YYYY, ZZZZ, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta crystallographica. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Agrawal V, Kishan KV. OB-fold: growing bigger with functional consistency. Curr Protein Pept Sci. 2003;4:195–206. doi: 10.2174/1389203033487207. [DOI] [PubMed] [Google Scholar]
- Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends in biochemical sciences. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Lupas A. The proteasome. Current opinion in structural biology. 1997;7:273–278. doi: 10.1016/s0959-440x(97)80036-x. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annual review of biophysics and biomolecular structure. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta crystallographica. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O'Donnell M. Motors and switches: AAA+ machines within the replisome. Nature reviews. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- Dreveny I, Kondo H, Uchiyama K, Shaw A, Zhang X, Freemont PS. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. The EMBO journal. 2004;23:1030–1039. doi: 10.1038/sj.emboj.7600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nature cell biology. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annual review of biophysics and biomolecular structure. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. European journal of biochemistry / FEBS. 1992;203:9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochemical Society transactions. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Groll M, Bochtler M, Brandstetter H, Clausen T, Huber R. Molecular machines for protein degradation. Chembiochem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
- Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Investigations on the maturation and regulation of archaebacterial proteasomes. Journal of molecular biology. 2003;327:75–83. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Hanna J, Finley D. A proteasome for all occasions. FEBS letters. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Horwitz AA, Navon A, Groll M, Smith DM, Reis C, Goldberg AL. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem. 2007;282:22921–22929. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- Inobe T, Kraut DA, Matouschek A. How to pick a protein and pull at it. Nat Struct Mol Biol. 2008;15:1135–1136. doi: 10.1038/nsmb1108-1135. [DOI] [PubMed] [Google Scholar]
- Inobe T, Matouschek A. Protein targeting to ATP-dependent proteases. Current opinion in structural biology. 2008;18:43–51. doi: 10.1016/j.sbi.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA. A set of averaging programs. In: Dodson EJ, Gover S, Wolf W, editors. Molecular Replacement. SERC Daresbury Laboratory; Warrington: 1992. pp. 91–105. [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. The Journal of biological chemistry. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science (New York, NY. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008a;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008b;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science (New York, NY. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Umehara T, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem Biophys Res Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nature reviews. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin H, Jiang W, Inouye M, Heinemann U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5119–5123. doi: 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. Journal of structural biology. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, et al. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40:256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta crystallographica. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annual review of biophysics and biomolecular structure. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. Structure determination of the constitutive 20S proteasome from bovine liver at 2.75 A resolution. Journal of biochemistry. 2002a;131:171–173. doi: 10.1093/oxfordjournals.jbchem.a003084. [DOI] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002b;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- Walz J, Erdmann A, Kania M, Typke D, Koster AJ, Baumeister W. 26S proteasome structure revealed by three-dimensional electron microscopy. Journal of structural biology. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Ou MS, Aldrich HC, Maupin-Furlow J. Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J Bacteriol. 2000;182:1680–1692. doi: 10.1128/jb.182.6.1680-1692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods in enzymology. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y. Mechanism of Substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.04.022. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.