Septins are essential for membrane compartmentalization and remodeling. Electron tomography of yeast bud necks shows filaments perpendicular and parallel to the mother-bud axis that resemble in vitro septin arrays. Filaments are still present, although disordered, in mutants lacking a single septin, underscoring the importance of septin assembly.
Abstract
Septins are conserved GTP-binding proteins involved in membrane compartmentalization and remodeling. In budding yeast, five mitotic septins localize at the bud neck, where the plasma membrane is enriched in phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P2). We previously established the subunit organization within purified yeast septin complexes and how these hetero-octamers polymerize into filaments in solution and on PtdIns4,5P2-containing lipid monolayers. How septin ultrastructure in vitro relates to the septin-containing filaments observed at the neck in fixed cells by thin-section electron microscopy was unclear. A morphological description of these filaments in the crowded space of the cell is challenging, given their small cross section. To examine septin organization in situ, sections of dividing yeast cells were analyzed by electron tomography of freeze-substituted cells, as well as by cryo–electron tomography. We found networks of filaments both perpendicular and parallel to the mother–bud axis that resemble septin arrays on lipid monolayers, displaying a repeat pattern that mirrors the molecular dimensions of the corresponding septin preparations in vitro. Thus these in situ structures most likely represent septin filaments. In viable mutants lacking a single septin, in situ filaments are still present, although more disordered, consistent with other evidence that the in vivo function of septins requires filament formation.
INTRODUCTION
Both cytokinesis and cell morphogenesis require spatial and temporal coordination of the proteins, lipids, and enzymes that remodel the composition and shape of the plasma membrane (PM) and surrounding extracellular material. Hence the mechanisms responsible for recruitment and activation of such factors are the subject of much current research. Septins are proteins that have emerged as important contributors to PM compartmentalization and remodeling in eukaryotes (Caudron and Barral, 2009; McMurray and Thorner, 2009). Our recent studies suggest that the ability to form filaments is critical for the physiological function of septins, at least in the budding yeast Saccharomyces cerevisiae (McMurray et al., 2011). However, the actual architecture of septin assemblies in vivo has not been clearly delineated at the ultrastructural level in any cell type.
The septins were originally identified via the isolation of temperature-sensitive (ts) mutants defective in cell cycle progression (Hartwell, 1974). Mutations at four distinct loci— cdc3, cdc10, cdc11, and cdc12—shared the unique terminal phenotype of failed cell division, despite continued progress through DNA replication and nuclear division, creating multinucleated cells (Hartwell, 1971). Anisotropic bud growth occurs in these mutants, but cytokinesis fails, leading to cells with chains of highly elongated buds (Hartwell, 1971). When subsequently cloned and sequenced, the corresponding wild-type genes were found to encode a family of related proteins eventually dubbed septins (Pringle, 2008). A fifth septin-encoding gene (SEP7/SHS1), recognized on the basis of its sequence similarity to the others, is also expressed in mitotic cells (Carroll et al., 1998; Mino et al., 1998). Unlike the other four septin genes, even an shs1-null allele does not cause a ts proliferation defect, but shs1Δ cells exhibit a moderately elongated bud, and an shs1Δ mutation exacerbates the phenotype of other mutations that affect cytokinesis (Iwase et al., 2007; McMurray et al., 2011). Thus all five mitotic septins are involved in cytokinesis and cellular morphogenesis.
Biochemical studies have shown that native septins isolated from yeast cells by immunoaffinity chromatography and other fractionation methods (Frazier et al., 1998; Mortensen et al., 2002), as well as recombinant septins expressed in and purified from bacterial cells (Versele et al., 2004; Farkasovsky et al., 2005), copurify as stable complexes of defined stoichiometry, even at high salt concentration (≥200 mM; Bertin et al., 2008; Garcia et al., 2011). At low salt concentration (≤100 mM), purified yeast septin complexes self-assemble into filaments in solution (Frazier et al., 1998; Versele et al., 2004; Farkasovsky et al., 2005). Using electron microscopy (EM) and single-particle analysis, combined with physical tags, we found that complexes of the four yeast septins identified as CDC genes exist as well-ordered, linear hetero-octameric rods (4 × 4 × 32 nm) of a uniform composition: Cdc11-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Cdc11 (Bertin et al., 2008). Furthermore, filament formation at low salt concentration occurs by end-to-end association of the rods via Cdc11–Cdc11 interaction, and, as polymerization proceeds, the filaments pair in a highly cooperative manner via cross-filament association between the extended carboxy termini of Cdc3–Cdc12 dimers, yielding long, “railroad track”–like structures (Bertin et al., 2008). Fluorescence anisotropy measurements carried out in diverse organisms are compatible with the pairing of septin filaments in situ (DeMay et al., 2011). Independent studies using EM or X-ray crystallography to examine purified septin complexes from Caenorhabditis elegans (John et al., 2007) and humans (Sirajuddin et al., 2007) revealed strikingly similar linear, nonpolar rods capable of end-to-end polymerization into filaments. It is striking that when yeast septin filaments are assembled on the surface of lipid monolayers containing phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P2) to more closely mimic the PM environment believed to be present at the yeast bud neck (Prouzet-Mauléon et al., 2008; Yoshida et al., 2009), the filaments pair tightly (lack the gap seen in the “railroad tracks” formed in solution) and do not require the carboxy termini of either Cdc3 or Cdc12 to do so (Bertin et al., 2010). Moreover, the tight filament pairs are frequently cross-braced by rods oriented orthogonally to the filaments, creating a mesh-like structure (Bertin et al., 2010). Unlike hetero-octamer assembly and rod polymerization, cross-brace formation requires the carboxy-terminal extension of Cdc11 (Bertin et al., 2010).
Thin sections of glutaraldehyde- and osmium tetroxide–fixed dividing S. cerevisiae cells stained with uranyl acetate, dehydrated, and embedded in plastic resin were examined by EM and revealed an ordered array of filament-like profiles wrapped around the inside of the neck as circumferential hoops perpendicular to the mother–bud axis (or perhaps as a continuous spiral; Byers and Goetsch, 1976). In this hourglass-shaped collar, the putative filaments (spaced ∼28 nm apart) were closely apposed to the PM (apparently connected to the inner leaflet by extensions ∼12 nm long and ∼3 nm thick). In some images, each filament (∼10-nm diameter) could be resolved into two very fine spines separated by ∼5 nm (Byers and Goetsch, 1976), reminiscent of the tightly paired filaments seen on PtdIns4,5P2-containing lipid monolayers (Bertin et al., 2010). A similar pattern was observed in Candida albicans (Soll and Mitchell, 1983). When bud necks were examined by an alternative method—EM of Ta/W- or Pt-coated replicas of freeze-fractured and deep-etched fixed spheroplasts—filamentous structures resembling “gauzes” and attributed to criss-crossed, septin-containing filaments were observed (Rodal et al., 2005), reminiscent of the mesh-like networks formed by purified septin complexes on PtdIns4,5P2-containing lipid monolayers (Bertin et al., 2010).
Multiple observations demonstrate that the septins are required for the formation and are integral constituents of these neck-associated filaments. The neck filaments were undetectable in cdc3, cdc10, cdc11, or cdc12 mutants shifted to the restrictive temperature but were found in all other cdc mutants examined under the same conditions (Byers and Goetsch, 1976). Whether visualized in fixed cells by antibody decoration and indirect immunofluorescence (Haarer and Pringle, 1987; Ford and Pringle, 1991; Kim et al., 1991) or as fusions to genetically encoded fluorescent tags in live cells in real time (Cid et al., 1998; Iwase et al., 2007; McMurray and Thorner, 2008b), Cdc3, Cdc10, Cdc11, Cdc12, and Shs1 all colocalize at the neck between a mother cell and its bud. As judged by photobleaching (Caviston et al., 2003; Dobbelaere et al., 2003) and fluorescence anisotropy (Vrabioiu and Mitchison, 2006, 2007; DeMay et al., 2011) of septin-GFP fusions, at the cell cycle stage when the neck filaments are observed, each of the septin subunits examined is immobilized and orientationally constrained, consistent with formation of highly ordered assemblies. Finally, concomitant with the onset of cytokinesis, the septin-containing, hourglass-shaped collar at the bud neck is split into two discrete rings that closely flank each side of the neck (Oh and Bi, 2011).
The ordered septin array at the bud neck appears to serve three functions. First, it acts as a scaffold by physically associating with other proteins that recruit factors critical for cell division, including the actin and myosin that assemble into the contractile ring (Dobbelaere and Barral, 2004), and for execution of a cell cycle checkpoint that monitors septin assembly (Shulewitz et al., 1999; Keaton and Lew, 2006). Second, it acts as a physical barricade to prevent free diffusion of plasma membrane proteins between the mother cell and its bud during cell growth (Takizawa et al., 2000; Faty et al., 2002). Third, the split rings act as gaskets or corrals to sequester the secretory vesicles, chitin-synthesizing enzymes, and other proteins required for completion of cytokinesis and septation but which the septins do not bind directly (Dobbelaere and Barral, 2004; Oh and Bi, 2011). Clearly, continuous membrane-associated, septin-containing filaments would serve as effective barriers to physically impede diffusion of integral membrane proteins, either directly or indirectly by binding to and ordering PM lipids (Bertin et al., 2010). Consistent with the conclusion that filament formation is necessary for septin function in vivo, we demonstrated that mutations that ablate filament assembly prevent septin localization to the cell cortex and block cell division, whereas deletion of certain septin subunits can be tolerated because the resulting complexes are capable of polymerizing into filaments (McMurray et al., 2011).
Despite these advances in our understanding of septin structure and function, how septin filaments are actually arranged at the bud neck and whether they correspond to the images observed before (Byers and Goetsch, 1976; Rodal et al., 2005) remained unclear. The subsequent advent of tomographic methods for EM analysis of cell components in situ allows a novel means to examine septin organization in three dimensions. Concomitantly, methods for better preservation of cellular ultrastructure, especially membranes, have also been developed. Here we used high-pressure freezing, followed by either freeze substitution or cryo-sectioning (cryo–electron microscopy of vitreous sections; Al-Amoudi et al., 2004), and coupled them with three-dimensional reconstruction by electron tomography to characterize the complex arrangement of fibrillar structures at the yeast bud neck. Of importance, we have been able to place the present in situ analysis within the context of recent in vitro studies with recombinant proteins and simple lipid systems. Relating now well-established structural features of the septin complexes and polymers formed thereof with the cellular features of the neck filaments observed in the current study strongly supports the identification of the filaments we describe here as septins.
RESULTS
Neck filaments are preserved in freeze-substituted budding yeast
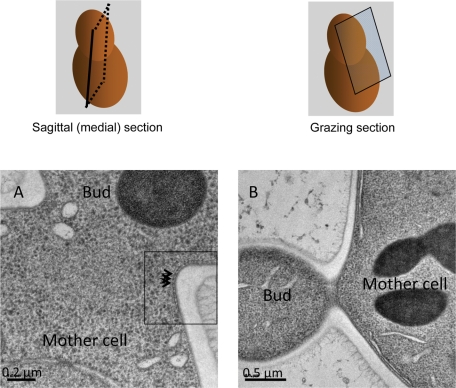
To optimize maintenance of cellular structures, S. cerevisiae cells from exponentially growing cultures were quick-frozen under high pressure and freeze substituted prior to embedding and sectioning. In sections (50 nm thick) through the middle of the isthmus that connects a mother cell to its bud (Figure 1A), a pattern of densities (resembling dark beads on a string) is present immediately adjacent to the highly convex surface of the PM at the bud neck (see inset). These dot-like profiles are very similar to those observed in medial sections of chemically fixed S. cerevisiae (Byers and Goetsch, 1976) and C. albicans (Soll and Mitchell, 1983) cells prepared by harsher techniques, which were attributed to cross sections through neck filaments. As described for the neck filaments (Byers and Goetsch, 1976), the dots in our sections are slightly removed from the PM (but appear to contact the PM via a short stem) and also seem connected by a thin, fiber-like element that runs parallel to the mother–bud axis. In sections that just graze the PM at the bud neck (Figure 1B), multiple striations (each ∼10 nm thick and separated from its neighbors by ∼30 nm) that are oriented perpendicular to the mother–bud axis are clearly observed, again very similar to the “10-nm” stripes described previously and attributed to neck filaments (Byers and Goetsch, 1976).
FIGURE 1:
Resin-embedded, 50-nm sections of wild-type S. cerevisiae bud necks. Left, sagittal (medial) section of a mother cell and its bud in schematic view (top) and an actual image (A) with the region (square) showing apparent cross sections of filament-like structures (arrowheads) closely apposed to the plasma membrane and interconnected by a continuous fibril. Scale bar, 0.2 μm. Right, grazing section of a mother cell and its bud in schematic view (top) and an actual image (B) with region (square) showing a banding pattern of apparent filament-like structures perpendicular to the mother–bud axis. Scale bar, 0.5 μm.
In addition, next to the densities that correspond to neck filaments, the inner leaflet of the PM appears significantly darker than in other areas. This additional contrast could correspond to tightly bound protein on the membrane surface or be due to preferential staining of a particular lipid enriched in this area (or both). Comparing both our medial and grazing sections and averaging over many images, we typically find at each bud neck 15–20 filaments perpendicular to the mother–bud axis, where the mean thickness for each filament is 10.2 ± 3 nm (see later discussion for a more precise estimation of filament thickness in each tomogram), and the mean distance between filaments is 30.3 ± 5 nm. Thus our observations generally agree with the qualitative conclusions reached using previous methods more prone to cause disruption (Byers and Goetsch, 1976). However, in our images the ultrastructural features of the filaments and the appearance of the membrane and cytosol are significantly improved, which provided an opportunity to undertake three-dimensional (3D) reconstruction of neck filament structure and organization in situ using electron tomography.
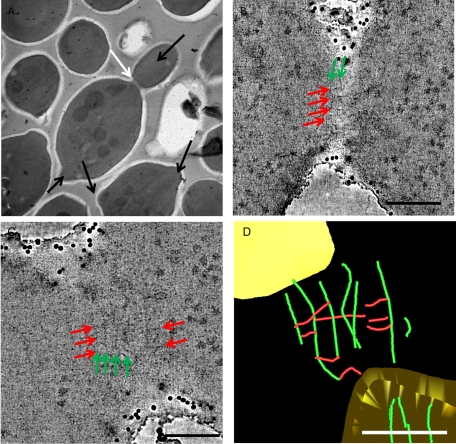
Coexistence of orthogonal filament arrays
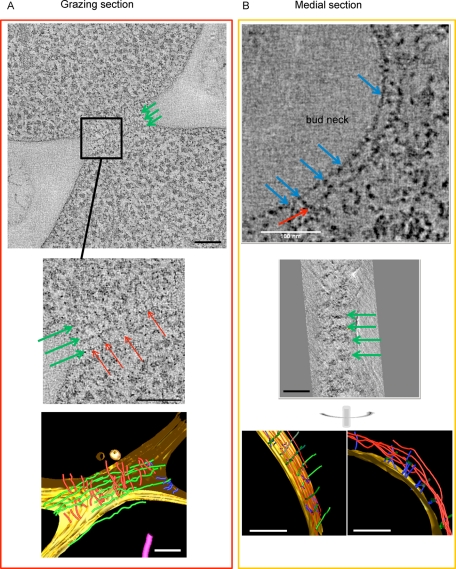
To obtain 3D reconstructions of the bud neck, we carried out dual-axis-tilted series on thicker sections (either 150 or 200 nm) of wild-type yeast cells. Examples of aligned tilted series are shown in Supplemental Movies S1 and S2). We analyzed two types of samples: medial cell sections, similar to those in Figure 1A, and grazing sections, as in Figure 1B. In total, we carried out four reconstructions from grazing sections and 16 reconstructions from medial sections (because, as expected, medial sections of dividing cells were found more frequently than grazing sections—Supplemental Movie S3 shows sequential slices through the volume). A morphological description of bud neck filaments in the crowded space of the cell is challenging, given their small cross section. As illustrated in a representative slice through the tomogram of a grazing section (Figure 2A, top), multiple filaments (in this case, at least three) clearly run around the constriction site near the PM in a direction perpendicular to the mother–bud axis (green arrows). An enlargement of the same area (Figure 2A, middle) reveals that these “circumferential” filaments seem to be interconnected by a distinct set of “axial” filaments (red arrows), which run parallel to the mother–bud axis and orthogonal to the “circumferential” filaments. When rendered using the software package IMOD (Mastronarde, 2008), a model of the full tomogram (Figure 2A, bottom) shows the following main features: 1) the circumferential profiles that run perpendicular to the mother–bud axis (green) correspond to the “10-nm” filaments first reported by Byers and Goestch (1976); 2) the circumferential filaments are intersected, and perhaps interconnected, by axial filaments (red) that run parallel to the mother–bud axis; and 3) although closely apposed to the PM (yellow surface), neither filament type appears to engage the membrane directly, but instead they are separated from the inner leaflet by ∼20 nm and connected to it via short extensions (blue).
FIGURE 2:
Tomographic visualization of resin-embedded thick sections of wild-type S. cerevisiae bud necks. (A) Grazing section. Top, slice from a tomogram showing circumferential filaments (green arrows) perpendicular to the mother–bud axis. Middle, magnified view of an area (square) from the top that shows both the circumferential filament-like densities (green arrows) and the orthogonal filament-like densities parallel to the mother–bud axis (red arrows). Bottom, segmentation model of complete tomogram with cell membrane (yellow), circumferential filaments (green), axial filaments (red), and densities connecting the circumferential filaments to the membrane (blue). (B) Medial section. Top, slice from a tomogram showing one side of the bud neck containing an axial filament (red arrow) parallel to the membrane running continuously above and periodically connected to (blue arrows) dot-like densities corresponding to cross sections through the circumferential filaments. Middle, section from the same tomogram, perpendicular to that in the top (with the common axis shown vertically), showing the double-fiber character of the circumferential filaments (green arrows). Bottom, a glancing (left) and edge-on (right) view of the cell membrane (yellow) in segmentation model of the tomogram with the circumferential filaments (green), axial filaments (red), and connecting densities from the filaments to the membrane (blue). Scale bars, 100 nm.
As illustrated in a representative slice through the tomogram of a medial section (Figure 2B, top), where the circumferential filaments are visualized in cross section (blue arrows), orthogonal filamentous structures can again be identified. Within this single slice, a continuous filament parallel to the mother–bud axis can be seen running along the PM and ∼20 nm from it (red arrow). When the reconstruction is viewed in slices perpendicular to the plane of the section, a set of parallel filaments (green arrows) that run circumferentially around the bud neck and orthogonal to the mother–bud axis is clearly present (Figure 2B, middle). These filaments correspond to the prominent circumferential 10-nm filaments seen in the grazing sections (Figure 2A), and, in some cases, they appear to be doublets (i.e., two tightly paired fibers; see also results discussed later obtained using cryo-tomography). Within the tomographic reconstruction of the medial section as a whole (Figure 2B, bottom), the circumferential filaments seem to be closer to the PM, whereas the axial filaments can be seen at different depths, are more irregularly spaced, and are separated by ∼10–20 nm (see also Supplemental Movie S4). Again, both the circumferential filaments and the axial filaments make apparent contact with the PM via short, rod-like densities (blue lines); however, these short connectors are attached primarily along the circumferential filaments (Figure 2B, bottom right) and appear to “hold” those filaments along their course, whereas these connectors to the PM are much less frequent along the axial filaments. The connectors are not systematically found where circumferential and axial filaments intersect.
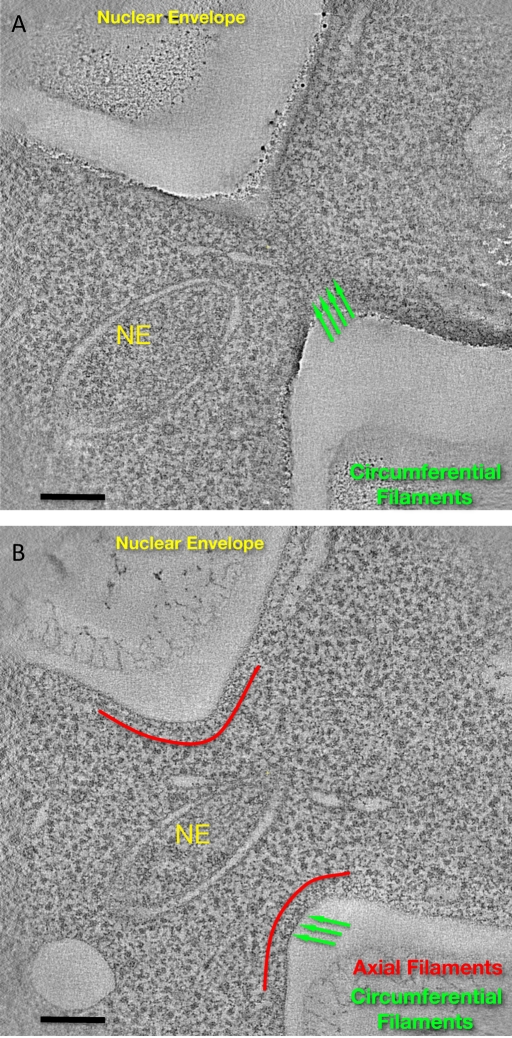
One of our tomograms, together with segmented main features, is provided as a supplementary video. The reconstruction includes a grazing slice of the plasma membrane at the bud neck (Figure 3A and Supplemental Movie S5). In this tomogram, grazing slices show clear circumferential and axial filaments, whereas the axial filaments are more clearly seen in inner slices of the cell. Within the same tomogram the nuclear envelope of the dividing nucleus is seen surrounding four mitotic microtubules and spanning the mother and daughter sides of the bud neck (Figure 3, A and B). Endoplasmic reticulum (ER) membranes are seen closely apposed to circumferential septin filaments (Supplemental Movie S5).
FIGURE 3:
Tomographic visualization of resin-embedded thick sections of the bud neck of wild-type S. cerevisiae undergoing mitosis. (A) Tomographic slice near the plasma membrane at the most constricted point of the bud neck, showing clear circumferential filaments (green arrows). (B) Deeper slice, clearly showing axial filaments (red lines follow their contour, at a slightly farther distance from the membrane), as well as circumferential filaments, which are seen in cross section (green arrows). The nuclear envelope is also indicated (NE). Scale bar, 100 nm. (See also corresponding Supplemental Movie.)
The thickness of an individual filament within each circumferential pair is 3.5 ± 1.4 nm, comparable to the value (∼4 nm) observed in our EM analysis of single septin filaments assembled in vitro from recombinant yeast septin complexes (Bertin et al., 2008, 2010), and the gap separating one circumferential pair from another is 30 ± 3 nm. The axial filaments have an essentially identical thickness (3.5 ±1.6 nm). As measured in reconstructions of six cells, the mean bud neck diameter was 972 ± 110 nm, quite comparable to the values for bud neck diameter (∼1000 nm) and septin ring diameter (980 ± 80 nm) in large-budded predivision cells assessed by light microscopy (Schmidt et al., 2003; Kadota et al., 2004). By contrast, we attribute the significantly smaller average value (∼500 nm) reported by Byers and Goetsch (1976) to the fixation methods they used, which are known to cause shrinkage (Murk et al., 2003).
Neck filaments in frozen, hydrated samples
The shortcomings of the dehydration, chemical embedding, and staining processes historically required for tomographic EM analysis can be avoided by preparing cryo-sections of frozen cells (Pierson et al., 2010, 2011). The samples are maintained in an unstained state, free of chemical treatments, in which ultrastructure is well preserved, but at the cost of poorer contrast in the resulting images and, hence, lower signal-to-noise ratio with regard to how well individual cellular structures stand out from the background. Nonetheless, tomography of vitreous sections (TOVIS) has been used successfully to study other major structural elements in budding yeast (Griffith et al., 2008; Pierson et al., 2009, 2011).
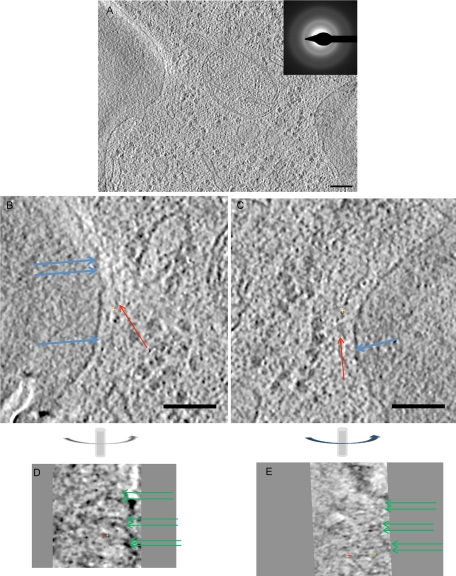
Therefore wild-type cells in the hydrated state were cryo-sectioned (50 nm thick) after the freezing procedure to carry out tomographic analysis of the yeast neck filament network. The numbers of grazing sections obtained from the limited numbers of cells examined were too few to attempt tomographic reconstructions. We were able to generate tomographic reconstructions of full (Figure 4A) and partial (Figure 4, B–E) medial sections of bud necks from individual cells (see also Supplemental Movie S6). The diffraction pattern (Figure 4A, inset) in an area of the full reconstruction indicates that such cells were fully vitrified. Because of the expected low contrast of TOVIS images as compared with those of stained, freeze-substituted samples (e.g., Figure 1), we enhanced the signal-to-noise ratio using the method of nonlinear anisotropic diffusion (Frangakis and Hegerl, 2001).
FIGURE 4:
Tomography of cryo-sections of wild-type S. cerevisiae bud necks. (A) Slice from a tomogram of a 50-nm cryo-section of the bud neck (inset, diffraction pattern confirming vitrification of the sample). (B, C) Magnified views of the left (B) and right (C) sides of the image shown in A, with axial filaments parallel to the membrane (red arrows) and connectors from the axial filaments to dot-like densities corresponding to cross sections through the circumferential filaments and to the membrane (blue arrows). (D, E) Slices perpendicular to those shown in B and C, respectively (the common axis is vertical, as indicated) showing the double-fiber nature of each circumferential filament (green arrows).
Axial filaments running along the cell membrane (red arrows), and the occasional apparent connectors linking them to the PM (blue arrows), are discernible in tomogram slices parallel to the plane of the section (Figure 4, B and C), and in slices within the section that are perpendicular to the section plane (Figure 4, D and E), regularly spaced, paired circumferential filaments (green arrows) run perpendicular to the plane of the section. The fact that the circumferential filaments are resolved into tight pairs is the most obvious difference between the tomographic reconstructions obtained from these frozen-hydrated specimens and those obtained from the resin-embedded samples. In the frozen sections, as in the fixed sections, the mean thicknesses of the individual filaments in a circumferential pair and of the axial filaments were 3.51 ± 1.8 and 4.51 ± 1.8 nm, respectively. The distance separating the two filaments in the tight pairs is 8.4 ± 1.7 nm, quite similar to the tight filament pairs seen when recombinant yeast septins are assembled on the surface of PtdIns4,5P2-containing lipid monolayers (Bertin et al., 2010) and much narrower than the spacing observed between the railroad track–like paired filaments assembled in solution (Bertin et al., 2008). It is possible that pairing of the circumferential filaments was not so readily discernible in tomograms of the freeze-substituted samples due to artifacts of preservation, resin opacity, or staining; conversely, perhaps the space between the two filaments in the circumferential pairs was exaggerated in the cryo–electron microscopy (cryo-EM) images due to the larger defocus. The gap separating each filament pair from another was 35.7 ± 3.2 nm, slightly higher than found in the resin-embedded samples (30 ± 3 nm), again because the latter have a tendency to shrink under the electron beam (and even before visualization, the other processing steps involved in preparation of the resin-embedded samples may have also contributed to this shrinkage). Similarly, the spacing between the axial filaments was ∼20 nm, slightly larger than what was seen in the resin-embedded cells (10–20 nm).
Neck filaments persist but have altered organization in viable mutants lacking a septin
Although septin function is essential for yeast cell division, and mutations in cdc3, cdc10, cdc11, or cdc12 can render cells incapable of division at high temperatures, viable cells carrying cdc10Δ or cdc11Δ (complete null) mutations can be obtained under certain conditions (Frazier et al., 1998; McMurray et al., 2011), whereas cdc3Δ and cdc12Δ mutations are invariably lethal (Haarer and Pringle, 1987; Kim et al., 1991; Versele et al., 2004). In the viable Cdc10-less and Cdc11-less strains examined at the light microscope level, the remaining septins (marked by fusion to green fluorescent protein [GFP]) localize at the bud neck. However, Cdc10-less and Cdc11-less strains fail to grow at an elevated temperature (37°C) that wild-type cells tolerate well, and, for such strains, many attempts at cytokinesis are abortive at the permissive temperature, suggesting that the septin structures formed at the bud neck carry out both their scaffold and diffusion barrier functions less than optimally (McMurray et al., 2011). Indeed, using the same fixation, dehydration, resin-embedding, and staining techniques by which the neck filaments were originally observed in wild-type cells (Byers and Goetsch, 1976), others failed to detect neck filaments in cdc11Δ mutants, observed only rare neck filaments in cdc10Δ mutants, and reported that septin complexes purified from both cdc10Δ and cdc11Δ cells were incapable of self-assembly into filaments in vitro (Frazier et al., 1998). However, we and others demonstrated that recombinant septin complexes lacking Cdc10 or Cdc11 are capable of rudimentary filament formation in vitro (Versele et al., 2004; Farkasovsky et al., 2005; Bertin et al., 2010; McMurray et al., 2011), and this capacity for polymerization is greatly enhanced on the surface of PtdIns4,5P2-containing lipid monolayers (Bertin et al., 2010). Furthermore, we provided genetic, biochemical, and cytological evidence that the corresponding nonnative Cdc11-Cdc12-Cdc3-Cdc3-Cdc12-Cdc11 and Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12 hexamers form and are able to polymerize end to end (McMurray et al., 2011), albeit less efficiently than the normal octameric complex, suggesting that cdc10Δ and cdc11Δ strains are viable because septin filament assembly can occur. Accordingly, septin filament formation in vivo should be manifest as neck filaments visible in situ at the EM level in the Cdc10-less and Cdc11-less mutants. Thus we tested this prediction using EM tomography of resin-embedded, stained mutant cells.
We prepared and examined cdc10Δ cells in the same way that we did for wild-type cells. The cultures of our haploid cdc10Δ strain propagated at 30°C displayed the same abnormal growth and morphological phenotypes observed by us and others (Frazier et al., 1998; Jeong et al., 2001; Versele et al., 2004; McMurray et al., 2011), including longer-than-normal doubling time (data not shown) and nonaxial budding (Flescher et al., 1993; Figure 5A, black arrows). Considering that, normally, septins help to direct localized PM and cell wall synthesis, we were not surprised to find that the PM was generally less well preserved in the mutant cells compared with the CDC10+ parent strain after applying the same protocol (compare Figure 5, B and C, to Figure 2, A and B). Indeed, many other yeast mutants exhibit less-than-optimal preservation than wild-type cells after high-pressure freezing and freeze substitution (McDonald, 2007). One rather well-preserved bud neck (Figure 5A, white arrow) yielded distinct grazing sections (Figure 5, B and C) that clearly displayed in tomographic reconstructions the presence of both circumferential (green arrows) and axial (red arrows) neck filaments. A rendering (Figure 5D) of such a tomogram (Figure 5C) shows that these filaments are less numerous and more disorganized than those seen in wild-type cells (Figure 2). Although the reduced quantity and greater disorder observed may be due, in part, to greater fragility during sample processing of septin filaments composed of Cdc11-Cdc12-Cdc3-Cdc3-Cdc12-Cdc11 hexameric complexes, it is important to note that, either in solution (Versele et al., 2004) or on PtdIns4,5P2-containing lipid monolayers (Bertin et al., 2010), purified recombinant complexes lacking Cdc10 are similarly disordered, forming shorter and more fragile filaments than wild-type Cdc11-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Cdc11 octameric complexes. Furthermore, filament assembly in vitro by Cdc10-less complexes also requires a higher protein concentration than wild-type complexes and is promoted by the presence of guanine nucleotide (Bertin et al., 2010; McMurray et al., 2011).
FIGURE 5:
Tomographic visualization of resin-embedded thick sections of Cdc10-less mutant cells. (A) Low-magnification view of a section from a bloc of the haploid cdc10Δ strain, showing diagnostic abnormal budding patterning (black arrows) and the region from which the tomograms were collected (white arrow). (B, C) Slices through tomograms of two different grazing sections, showing circumferential filaments perpendicular to the mother–bud axis (green arrows) and axial filaments parallel to the mother–bud axis (red arrows). (D) Segmentation model from the tomogram shown in C, with circumferential filaments (green) and axial filaments (red).
It is striking that, although circumferential filaments were more difficult to see in the Cdc10-less cells, the median value for the gap between them (20.2 ± 5.8 nm) was clearly smaller than that in wild-type cells (30 ± 3 nm). The distance observed in wild-type cells is consistent with the length of a septin octamer, similar to the length of the cross-braces in the mesh-like lattices we observed on PtdIns4,5P2-containing monolayers (Bertin et al., 2010). Without the central doublet of Cdc10 subunits, the length of the resulting hexamer should be shorter by 8 nm (2 × 4 nm; Bertin et al., 2008). Thus the ∼20-nm spacing between circumferential filaments in cdc10Δ cells suggests that this spacing is dictated by a septin complex–long repeat in the axial filaments that mediates their direct contact with the circumferential filaments. In other words, the repeating pattern of subunits within septin-based axial filaments is reflected in the spacing between septin-based circumferential filaments (see Figure 6).
FIGURE 6:
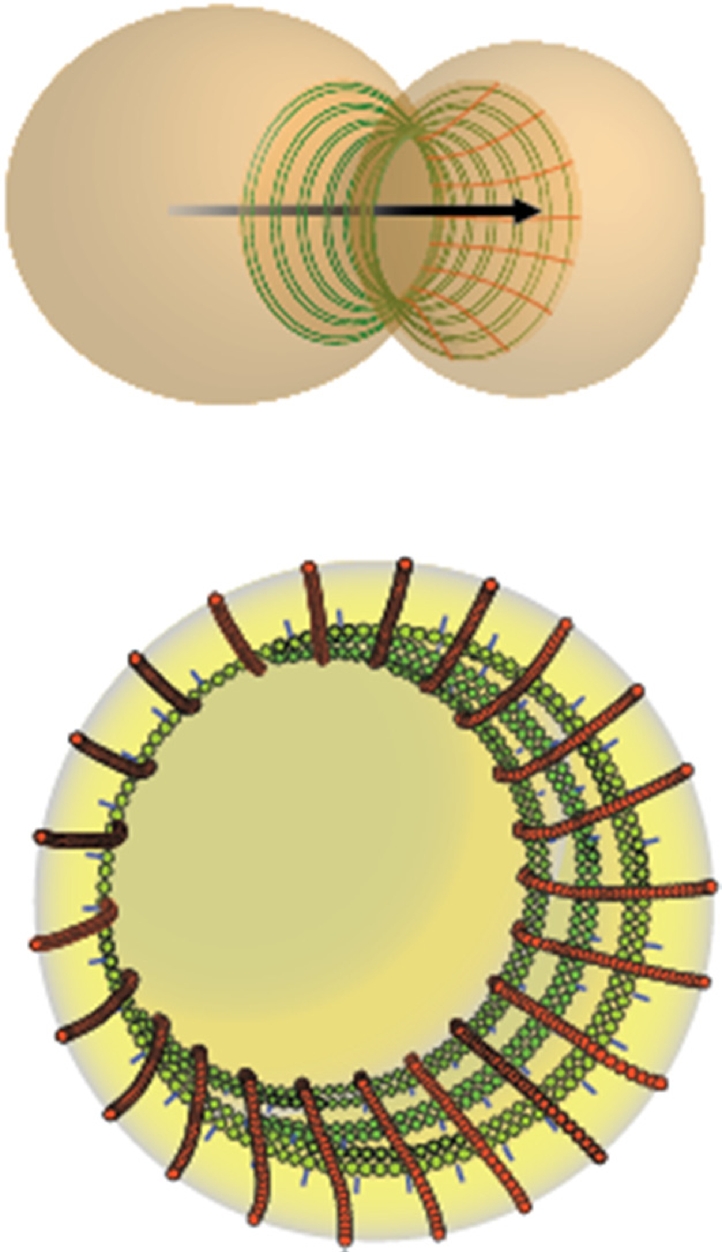
Schematic representation of the septin filament array at the yeast bud neck. Top, orientation of the lattice-like septin collar relative to the mother–bud axis (black arrow). Bottom, enlarged view depicting the 10-nm circumferential filaments composed of a pair of tightly associated septin filaments (green) with short linkers (blue) connecting them to the inner leaflet of the PM and the axial filaments (red) composed of a single septin filament that intersect the circumferential filaments with a 30-nm spacing that corresponds to the 30-nm length of each octameric repeat in the axial filaments.
DISCUSSION
Understanding of septin structure has been advanced in recent years by the in vitro visualization of the septin complexes that serve as the building blocks of septin filaments (McMurray and Thorner, 2008a; Weirich et al., 2008; Estey et al., 2011; Oh and Bi, 2011). Less well understood is how septins organize in cells. On the basis of evidence from pioneering EM work describing the neck filaments in budding yeast (Byers and Goetsch, 1976) and C. albicans (Soll and Mitchell, 1983), it has been assumed that septins in other cell types also form arrays of PM-associated filaments. As a much-needed follow up to those valuable first observations in yeasts, we used improved EM preservation techniques and 3D tomographic reconstruction to gain new insights about the organization of the bud neck filaments. First, as shown schematically in Figure 5, our imaging indicates that each of the circumferential 10-nm neck filaments observed by Byers and Goetsch (1976) does indeed run perpendicular to the mother–bud axis. However, second, and unexpectedly, our analysis revealed a system of axial filaments that coexists with the circumferential filaments and is arranged orthogonal to them (Figure 6). The visualization of the second set of axial filaments was greatly aided by the use of three-dimensional reconstruction. Third, our observations strongly suggest that each 10-nm circumferential filament corresponds to a tightly associated pair of septin filaments (Figure 6, green), whereas each axial filament seems to correspond to a lone septin filament (Figure 6, red). In the analysis by Byers and Goetsch, the diameter of each filament was measured to be 5 nm. We believe our measurements have the added value of new and improved preparation methods and of the statistics obtained from a large number of 3D reconstructions. Furthermore, a recent study supports the conclusion that septin filaments are paired in situ (DeMay et al., 2011). Fourth, the newly identified axial filaments intersect the circumferential filaments at regular intervals on their cytosolic side and define the distance of the gap separating one circumferential filament from another.
A number of experimental and conceptual points support our assumption that both the circumferential and axial filaments are composed of septins. The 30-nm interval separating each circumferential filament from another corresponds to the length of a septin octamer, the repeating unit in a filament (Bertin et al., 2008). Thus, if the axial filaments act as a patterning scaffold to control the spacing between each circumferential filament and the next and do so via specific subunit–subunit contacts, then either Cdc10 or Cdc11 could mediate the contacts because, in a septin filament, each Cdc10–Cdc10 and Cdc11–Cdc11 pair resides ∼30 nm away from the next doublet of the same subunit. Several considerations make Cdc11 the prime candidate to be the septin subunit responsible for mediating the observed cross-filament contacts between the orthogonal filaments. First, in cdc10Δ cells, both circumferential and axial filaments are still visible. Of importance, the spacing between circumferential filaments becomes narrower in cdc10Δ cells and corresponds to the length of a hexameric rod, in agreement with the absence of Cdc10. Finally, the C-terminal extension of Cdc11 is required for formation of the orthogonal meshes we observed using recombinant septin complexes on lipid monolayers (Bertin et al., 2010).
In addition, interesting properties exhibited by purified septin complexes on PtdIns4,5P2-containing lipid monolayers (Bertin et al., 2010) are recapitulated in situ by the network of neck filaments described in the present report. Tightly paired septin filaments were similarly cross-bridged by individual octameric rods via Cdc11–Cdc11 contacts mediated by the predicted C-terminal, coiled-coil–forming sequence in this septin, creating meshworks that were uniquely observed on PtdIns4,5P2-containing lipid monolayers (Bertin et al., 2010). Moreover, these meshworks and the orthogonal array of axial and circumferential filaments we observe by tomography are reminiscent of the “gauze-like” arrangement of filaments (attributed to septins by antibody decoration) found at the bud neck when replicas of fixed yeast spheroplasts were examined (Rodal et al., 2005).
Of importance, the neck filaments defined by Byers and Goetsch persisted in cells treated with cytochalasin B, which disrupts actin filaments but were not found in cells undergoing cytokinesis (Byers and Goetsch, 1976), when the actomyosin contractile ring, but not the septin collar, remains at the neck (Oh and Bi, 2011). Hence the filaments described in the present study are unlikely to be actin or myosin.
Collectively, our findings suggest a model (Figure 6) in which circumferential filaments wrapped around the inside of the bud neck (each composed of a tight pair of septin filaments) make contact with and are spaced by axial filaments running parallel to the mother–bud axis, generating a lattice, in which Cdc11 mediates the junction at the points of intersection of these two orthogonally arranged filament arrays. The nature of the 20-nm-long connectors between the circumferential filaments and the inner leaflet of the PM is unknown. Integral membrane proteins at the bud neck known to interact with septins, such as Axl2 (Gao et al., 2007), are potential candidates.
Vrabioiu and Mitchison (2006, 2007) used fluorescence polarization spectroscopy to study the organization of septins in budding yeast through the process of cytokinesis. Of interest, their analysis suggests that the ordered ensembles in the collar and those in the split rings differ in orientation by 90°. Similarly, by a distinct polarized fluorescence microscopy method, Gladfelter and colleagues observed such an apparent rotation (DeMay et al., 2011). The tomographic studies presented here all correspond to the collar state preceding cytokinesis, which is much more common in unsynchronized cells and was basically the one sampled in our experiments. Although we cannot shed any light on changes that occur during the collar–split rings transition, our studies directly show that during the collar state two sets of filaments, perpendicular to each other, coexist. An explanation for the change in anisotropy observed by others (Vrabioiu and Mitchison, 2006, 2007; DeMay et al. 2011) could be the disappearance of one set of filaments (the axial ones) with the onset of cytokinesis, likely by disruption of the Cdc11-mediated junctions between the axial and circumferential filaments. This would allow for accordion-like collapse of the spacing between the circumferential filaments and their bundling on either side of the bud neck (the split rings).
In our lipid monolayer in vitro experiments (Bertin et al., 2010), the filaments were associated with this surface via their direct binding to the lipids. In our tomograms, however, it appears that the septin-based filaments are situated at a distance from the inner leaflet of the PM but contact it through a ∼20-nm stem of connecting density. Of interest, in an EM image from Ashbya gossypii, septin filaments also appear to be disconnected from the plasma membrane (DeMay et al., 2011). Theoretically, it remains possible that a third set of septin filaments directly abuts the inner PM surface and comprises the intense electron density we observed on the inner leaflet of the bilayer in the bud neck region (Figure 1). However, this density may reflect known localized asymmetries in lipid content between the inner and outer leaflets of the PM at the bud neck (Chen et al., 2006; Roelants et al., 2010). A reasonable possibility is that the direct septin–lipid interaction described in the monolayer experiments may correspond to a distinct time point in the yeast cell cycle from that illustrated in our tomograms of mature buds.
Previous work by another group showed that a septin-dependent barrier restricts diffusion of ER membrane proteins at the bud neck (Luedeke et al., 2005). By lining the cortex at the yeast bud neck, and in light of our observation that the ER membrane apposes circumferential septin filaments, the filamentous septin lattice defined by our tomographic analysis is well suited to act as an efficient barrier to the diffusion of membrane-associated factors between the mother cell and its bud. By themselves, septin filaments parallel to the mother–bud axis would present a comparatively poor barrier unless they were organized into a continuous sheet with no intervening gaps, a configuration not supported by our (or previous) observations. Moreover, the new picture of septin filament ultrastructure that our study provides may also explain how septins can play an important role in controlling cortical rigidity in other cell types (Gilden and Krummel, 2010).
MATERIALS AND METHODS
High-pressure freezing
Yeast cells to be examined after freeze substitution were grown to mid-exponential phase (A600 nm = 0.4–0.6) in yeast extract–peptone–glucose (YPD) medium at 30°C. To avoid any cell damage from centrifugation, the cells were collected by vacuum filtration as described by McDonald (2007). The resulting yeast cake was transferred to 100-μm-deep membrane carriers (Leica Microsystems, Vienna, Austria) precoated with 1-hexadecene (Fluka, Sigma-Aldrich, St. Louis, MO) and frozen in a high-pressure freezing device (model EM PACT2-RTS; Leica). Yeast cells to be examined after cryo-section were grown to mid-exponential phase (A600 nm = 0.4–0.6) at 30°C in YPD medium containing 20% dextran (35–45 kDa from Leuconostoc mesenteroides; Sigma-Aldrich) and then frozen in copper tubes (Leica) using the same high-pressure freezing device.
Freeze substitution, sectioning, and poststaining
The substitution medium was acetone containing 1% osmium tetraoxide, 0.1% uranyl acetate, and 5% water, and freeze substitution was performed in a Leica AFS2 device by raising the temperature from –90°C in increments of 2°C/h up to –25°C and then in increments of 5°C/h up to 0°C. The resulting samples were rinsed in acetone three times (10 min each) and then embedded in resin by incubation in solutions containing an ascending concentration of Epon from 30 to 100% (1 h each). To remove any trace of acetone, the samples were bathed three times in pure Epon and then inserted into molds for polymerization (48 h at 60°C). Serial sections of 50-, 150-, or 200-nm thickness were carved from the blocks using a microtome (Ultracut E; Reichert, Depew, NY) equipped a 4.5-mm diamond knife (Diatome, Hatfield, PA). The resulting sections were deposited on hexagonal copper grids (mesh size of 100), covered with Formvar, and poststained with 2% uranyl acetate and lead citrate in 70% methanol. As a size standard, marker for the extracellular surface, and fiducial marks to align the tilt series, gold beads (10 nm diameter; Aurion, Wageningen, Netherlands) were deposited on both sides of the sections, and then a thin film of carbon was evaporated over the sample.
Cryo-sectioning
Ribbons (50 nm thick) of vitreous cryo-sections were obtained from the high-pressure frozen copper tubes using a cryo-ultramicrotome equipped with an anticontamination glove box or Cryosphere (Leica). The ribbons were attached to the support film of a C-Flat EM grid (Aurion) that was coated with the same fiducial 10-nm gold beads (Aurion) by electrostatic charging using the CRION ionizer (Leica; Pierson et al., 2010).
Data collection
Resin-embedded samples were placed in a dual-axis tomography holder (model 2040; Fischione Instruments, Export, PA) and preexposed to the chamber for ∼30 min prior to data collection to avoid any shrinkage or deformation of the plastic resin during the data collection process. The dual-axis-tilted series were collected using a CM200 FEG microscope (Philips Electron Optics/FEI, Eindhoven, Netherlands) and the Digital Micrograph software suite (Gatan, Pleasanton, CA; Kremer et al., 1996). In most cases, the tilted series were collected from −70 to 70°, except when the sample of interest was too close to a grid bar, which prevented a high tilt.
For vitreous cryo-sections, images and tilt series were collected on a Tecnai 12 microscope equipped with a 4k Eagle charge-coupled device camera, and the tilt series were acquired using the Xplore3D tomography software suite (all from Philips Electron Optics/FEI).
Data processing
The data were processed using IMOD (Kremer et al., 1996; Mastronarde, 2008), using the gold beads as fiducial marks for aligning the tilted series. The reconstructed volumes were obtained by weighted backprojection before the two axes were recombined. Segmentation and renderings were performed in IMOD (Mastronarde, 2008) or Amira (Stalling et al., 2005). Segmentation of tomographic reconstructions was performed by manually tracing structural features (filaments, membranes, microtubules) through sequential slices of the tomograms.
Supplementary Material
Acknowledgments
We thank Manfred Auer and Jeff Triffo (Lawrence Berkeley National Laboratory, Berkeley, CA) for their advice and technical help. This work was supported by Jane Coffin Childs Postdoctoral Research Fellowship 61-1357 (to A.B.), National Institutes of Health K99 Grant GM86603 (to M.A.M.), and National Institutes of Health R01 Grant GM21841 (to J.T.). E.N. is a Howard Hughes Medical Institute Investigator.
Abbreviations used:
- cryo-EM
cryo–electron microscopy
- EM
electron microscopy
- PM
plasma membrane
- PtdIns4,5P2
phosphatidylinositol-4,5-bisphosphate
- 3D
three dimensional
- TOVIS
tomography of vitreous sections
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-10-0850) on December 7, 2011.
REFERENCES
- Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlen LP, Richter K, Blanc NS, Studer D, Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park SS, Garcia G 3rd, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Thai L, Garcia G 3rd, Votin V, Grob P, Allyn T, Thorner J, Nogales E. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404:711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Altman R, Schieltz D, Yates JR, Kellogg D. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Longtine M, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang J, Muthusamy BP, Liu K, Zare S, Andersen RJ, Graham TR. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- Cid VJ, Adamíková L, Cenamor R, Molina M, Sánchez M, Nombela C. Cell integrity and morphogenesis in a budding yeast septin mutant. Microbiology. 1998;144:3463–3474. doi: 10.1099/00221287-144-12-3463. [DOI] [PubMed] [Google Scholar]
- DeMay BS, Bai X, Howard L, Occhipinti P, Meseroll RA, Spiliotis ET, Oldenbourg R, Gladfelter AS. Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J Cell Biol. 2011;193:1065–1081. doi: 10.1083/jcb.201012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- Estey MP, Kim MS, Trimble WS. Septins. Curr Biol. 2011;21:R384–R387. doi: 10.1016/j.cub.2011.03.067. [DOI] [PubMed] [Google Scholar]
- Farkasovsky M, Herter P, Voss B, Wittinghofer A. Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol Chem. 2005;386:643–656. doi: 10.1515/BC.2005.075. [DOI] [PubMed] [Google Scholar]
- Faty M, Fink M, Barral Y. Septins: a ring to part mother and daughter. Curr Genet. 2002;41:123–131. doi: 10.1007/s00294-002-0304-0. [DOI] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford SK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Wong ML, Longtine MS, Pringle JR, Mann M, Mitchison TJ, Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Sperber LM, Kane SA, Tong Z, Tong AH, Boone C, Bi E. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol Biol Cell. 2007;18:2542–2560. doi: 10.1091/mbc.E06-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G, III, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol. 2011 doi: 10.1083/jcb.201107123. 195, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilden J, Krummel MF. Control of cortical rigidity by the cytoskeleton: emerging roles for septins. Cytoskeleton (Hoboken) 2010;67:477–486. doi: 10.1002/cm.20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J, Mari M, De Mazièr A, Reggiori F. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 2008;9:1060–1072. doi: 10.1111/j.1600-0854.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Pringle JR. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol Cell Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M, Luo J, Bi E, Toh-e A. Shs1 plays separable roles in septin organization and cytokinesis in Saccharomyces cerevisiae. Genetics. 2007;177:215–229. doi: 10.1534/genetics.107.073007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JW, Kim DH, Choi SY, Kim HB. Characterization of the CDC10 product and the timing of events of the budding site of Saccharomyces cerevisiae. Mol Cells. 2001;12:77–83. [PubMed] [Google Scholar]
- John CM, et al. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 2007;26:3296–3307. doi: 10.1038/sj.emboj.7601775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton MA, Lew DJ. Eavesdropping on the cytoskeleton: progress and controversy in the yeast morphogenesis checkpoint. Curr Opin Microbiol. 2006;9:540–546. doi: 10.1016/j.mib.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Luedeke C, Frei SB, Sbalzarini I, Schwarz H, Spang A, Barral Y. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J Cell Biol. 2005;169:897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc. 2008;230:212–217. doi: 10.1111/j.1365-2818.2008.01977.x. [DOI] [PubMed] [Google Scholar]
- McDonald K. Cryopreparation methods for electron microscopy of selected model systems. Methods Cell Biol. 2007;79:23–56. doi: 10.1016/S0091-679X(06)79002-1. [DOI] [PubMed] [Google Scholar]
- McMurray MA, Bertin A, Garcia G 3rd, Lam L, Nogales E, Thorner J. Septin filament formation is essential in budding yeast. Dev Cell. 2011;20:540–549. doi: 10.1016/j.devcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Thorner J. Biochemical properties and supramolecular architecture of septin hetero-oligomers and septin filaments. In: Hall PA, Russell SEG, Pringle JR, editors. The Septins. Chichester, United Kingdom: John Wiley & Sons; 2008a. pp. 49–100. [Google Scholar]
- McMurray MA, Thorner J. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr Biol. 2008b;18:1203–1208. doi: 10.1016/j.cub.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Thorner J. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 2009;4:18.11–18.40. doi: 10.1186/1747-1028-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino A, Tanaka K, Kamei T, Umikawa M, Fujiwara T, Takai Y. Shs1p: a novel member of septin that interacts with spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;251:732–736. doi: 10.1006/bbrc.1998.9541. [DOI] [PubMed] [Google Scholar]
- Mortensen EM, McDonald H, Yates JR, Kellogg DR. Cell cycle-dependent assembly of a Gin4-septin complex. Mol Biol Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murk JL, Posthuma G, Koster AJ, Geuze HJ, Verkleij AJ, Kleijmeer MJ, Humbel BM. Influence of aldehyde fixation on the morphology of endosomes and lysosomes: quantitative analysis and electron tomography. J Microsc. 2003;212:81–90. doi: 10.1046/j.1365-2818.2003.01238.x. [DOI] [PubMed] [Google Scholar]
- Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2011;21:141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson J, Fernández JJ, Bos E, Amini S, Gnaegi H, Vos M, Bel B, Adolfsen F, Carrascosa JL, Peters PJ. Improving the technique of vitreous cryo-sectioning for cryo-electron tomography: electrostatic charging for section attachment and implementation of an anti-contamination glove box. J Struct Biol. 2010;169:219–225. doi: 10.1016/j.jsb.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Pierson J, Sani M, Tomova C, Godsave S, Peters PJ. Toward visualization of nanomachines in their native cellular environment. Histochem Cell Biol. 2009;132:253–262. doi: 10.1007/s00418-009-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson J, Ziese U, Sani M, Peters PJ. Exploring vitreous cryo-section-induced compression at the macromolecular level using electron cryo-tomography; 80S yeast ribosomes appear unaffected. J Struct Biol. 2011;173:345–349. doi: 10.1016/j.jsb.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Pringle JR. Origins and development of the septin field. In: Hall PA, Russell SEG, Pringle JR, editors. The Septins. Chichester, United Kingdom: John Wiley & Sons; 2008. pp. 7–34. [Google Scholar]
- Prouzet-Mauléon V, Lefebvre F, Thoraval D, Crouzet M, Doignon F. Phosphoinositides affect both the cellular distribution and activity of the F-BAR-containing RhoGAP Rgd1p in yeast. J Biol Chem. 2008;283:33249–33257. doi: 10.1074/jbc.M805161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Kozubowski L, Goode BL, Drubin DG, Hartwig JH. Actin and septin ultrastructures at the budding yeast cell cortex. Mol Biol Cell. 2005;16:372–384. doi: 10.1091/mbc.E04-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci USA. 2010;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Varma A, Drgon T, Bowers B, Cabib E. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol Biol Cell. 2003;14:2128–2141. doi: 10.1091/mbc.E02-08-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kühlmann D, Macara IG, Weyand M, Stark H, Wittinghofer A. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- Soll DR, Mitchell LH. Filament ring formation in the dimorphic yeast Candida albicans. J Cell Biol. 1983;96:486–493. doi: 10.1083/jcb.96.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalling D, Westerhoff M, Hege H-C. Amira: a highly interactive system for visual data analysis. In: Hansen CD, Johnson CR, editors. The Visualization Handbook. New York: Elsevier; 20052005. pp. 749–767. [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Versele M, Gullbrand B, Shulewitz MJ, Cid VJ, Bahmanyar S, Chen RE, Barth P, Alber T, Thorner J. Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:4568–4583. doi: 10.1091/mbc.E04-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443:466–469. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- Vrabioiu AM, Mitchison TJ. Symmetry of septin hourglass and ring structures. J Mol Biol. 2007;372:37–49. doi: 10.1016/j.jmb.2007.05.100. [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: architecture and dynamics. Nature Rev Mol Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Bartolini S, Pellman D. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23:810–823. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.