The YidC insertase also integrates multispanning membrane proteins that had been considered to be exclusively SecYEG dependent. Only membrane proteins that require SecA can be inserted only via SecYEG. Targeting to YidC is SRP dependent, and the C-terminus of YidC cross-links to SRP, FtsY, and ribosomal subunits.
Abstract
Protein insertion into the bacterial inner membrane is facilitated by SecYEG or YidC. Although SecYEG most likely constitutes the major integration site, small membrane proteins have been shown to integrate via YidC. We show that YidC can also integrate multispanning membrane proteins such as mannitol permease or TatC, which had been considered to be exclusively integrated by SecYEG. Only SecA-dependent multispanning membrane proteins strictly require SecYEG for integration, which suggests that SecA can only interact with the SecYEG translocon, but not with the YidC insertase. Targeting of multispanning membrane proteins to YidC is mediated by signal recognition particle (SRP), and we show by site-directed cross-linking that the C-terminus of YidC is in contact with SRP, the SRP receptor, and ribosomal proteins. These findings indicate that SRP recognizes membrane proteins independent of the downstream integration site and that many membrane proteins can probably use either SecYEG or YidC for integration. Because protein synthesis is much slower than protein transport, the use of YidC as an additional integration site for multispanning membrane proteins may prevent a situation in which the majority of SecYEG complexes are occupied by translating ribosomes during cotranslational insertion, impeding the translocation of secretory proteins.
INTRODUCTION
Transport of proteins from the cytoplasm into the cytoplasmic membrane is an essential process in bacterial physiology. To facilitate protein insertion into lipid membranes, efficient transport systems have evolved, which include the membrane-embedded SecYEG translocon and the YidC insertase (Pohlschröder et al., 2005; Driessen and Nouwen, 2008). The SecYEG translocon is the major protein-conducting channel in the bacterial cytoplasmic membrane. It is involved in both membrane protein insertion and in secreting proteins across the membrane into the periplasm (Rapoport, 2007).
The transport of periplasmic proteins across the SecYEG translocon proceeds in a posttranslational manner and depends on ATP hydrolysis by the SecYEG-associated motor protein SecA. In contrast, inner membrane proteins (IMPs) need to be inserted into the membrane cotranslationally as ribosome-nascent chain complexes (RNCs). Cotranslational targeting and insertion is mediated by the signal recognition particle (SRP), which recognizes an emerging signal anchor sequence (Neumann-Haefelin et al., 2000; Bornemann et al., 2008) and targets RNCs to the membrane-bound SRP receptor FtsY. FtsY occupies the ribosome-binding site of SecY (Kuhn et al., 2011) and is probably displaced by the SRP-RNCs, which ensures efficient docking of the RNCs onto the SecYEG translocon. The hydrophobic transmembrane (TM) helices that emerge from the ribosomal tunnel exit are inserted into the SecY channel and further transferred into the lipid bilayer. The latter step is probably facilitated by a lateral opening of SecY (Van den Berg et al., 2004). SecA has also been found to cooperate with the SecYEG translocon for the translocation of large periplasmic domains of inner membrane proteins (Deitermann et al., 2005). In addition, SecYEG associates with other membrane-embedded components such as YidC (Scotti et al., 2000) to mediate the insertion of TMs. In vitro cross-linking studies using ribosome-nascent chains of IMPs such as mannitol permease (MtlA) or FtsQ have shown length-dependent contacts of TMs to SecY, YidC, and lipids (Beck et al., 2001; van der Laan et al., 2001). It was therefore suggested that YidC is located at the lateral gate of SecY, where it supports the partitioning of TMs into the lipid bilayer. In addition, YidC has been suggested to participate in the folding (Nagamori et al., 2004), assembly (Wagner et al., 2008), and quality control of membrane proteins (van Bloois et al., 2008).
YidC belongs to an evolutionarily conserved family of proteins, which include the mitochondrial homologues Oxa1 and Cox18 and the chloroplast homologues Alb3 and Alb4 (Funes et al., 2011). All members of the YidC protein family have a conserved core of five TMs that in Escherichia coli and other Gram-negative bacteria is connected via a long periplasmic loop to an additional N-terminal TM (Sääf et al., 1998; Ravaud et al., 2008). Other functional additions to the conserved core were acquired during evolution (Funes et al., 2011), such as a C-terminal extension in some YidC variants of Gram-positive bacteria, Alb3, and Oxa1 (Funes et al., 2009). This extension was shown to serve as a ribosome-binding site (Szyrach et al., 2003). YidC is essential in E. coli but can be functionally replaced by the mitochondrial Oxa1 (van Bloois et al., 2005). A complementation of Oxa1 by YidC is, however, only possible when the C-terminal ribosome-binding site of Oxa1 is attached to YidC (Preuss et al., 2005). Depletion of YidC in E. coli results in a global change in cell physiology (Price et al., 2010; Wickström et al., 2011), where chaperones such as Trigger Factor and FimC are up-regulated and stress-responsive pathways like Cpx are induced (Wang et al. 2010).
In addition to its SecYEG-associated function during membrane protein insertion, YidC can insert membrane proteins independent of SecYEG. YidC was shown to insert small proteins such as phage proteins (Samuelson et al., 2000, Serek et al., 2004; Klenner et al., 2008). YidC was also shown to insert the F0c subunit of the F0F1-ATP synthase (van der Laan et al. 2003, 2004a; Yi et al., 2003) and MscL (Facey et al., 2007), although the exact role of YidC in MscL integration is controversial (Pop et al., 2009; Berrier et al., 2011).
So far the substrates described as being inserted via YidC are small, closely spaced membrane proteins with short hydrophilic loops. How these proteins are targeted to YidC and why they evade the SecYEG translocon are unclear. A possible involvement of SRP has been proposed for the targeting of F0c (van der Laan et al., 2004a; van Bloois et al., 2004) and MscL (Facey et al., 2007; Pop et al., 2009), but a direct interaction of YidC with components of the SRP pathway has not been detected in bacteria. In contrast, the chloroplast Alb3 has been shown to interact with FtsY (Moore et al., 2003) and with cpSRP43, which replaces the 4.5S RNA in the chloroplast SRP (Falk et al., 2010; Richter et al., 2010). A recent cryo–electron microscopic (cryo-EM) study of a YidC–RNC complex indicated that the ribosome contacts YidC via the ribosomal proteins L23, L24, and L29 (Kohler et al., 2009). Because these proteins are located close to the ribosomal tunnel exit and are also involved in SecY binding (Frauenfeld et al., 2011), it was suggested that the ribosome has common binding sites for both the SecYEG translocon and the YidC insertase.
In the present study, we address the substrate specificity of YidC and demonstrate that YidC also inserts multispanning membrane proteins in vitro, which had been considered to be exclusively inserted via SecYEG. Only SecA-dependent membrane proteins cannot be inserted via YidC. Targeting to YidC is mediated by SRP, and site-directed cross-linking shows that the short C-terminus of YidC is in contact with the ribosome, SRP, and FtsY. These in vitro data suggest that the SRP pathway delivers membrane proteins cotranslationally to two distinct integration sites at the E. coli membrane.
RESULTS
Functional reconstitution of SecYEG and YidC into proteoliposomes
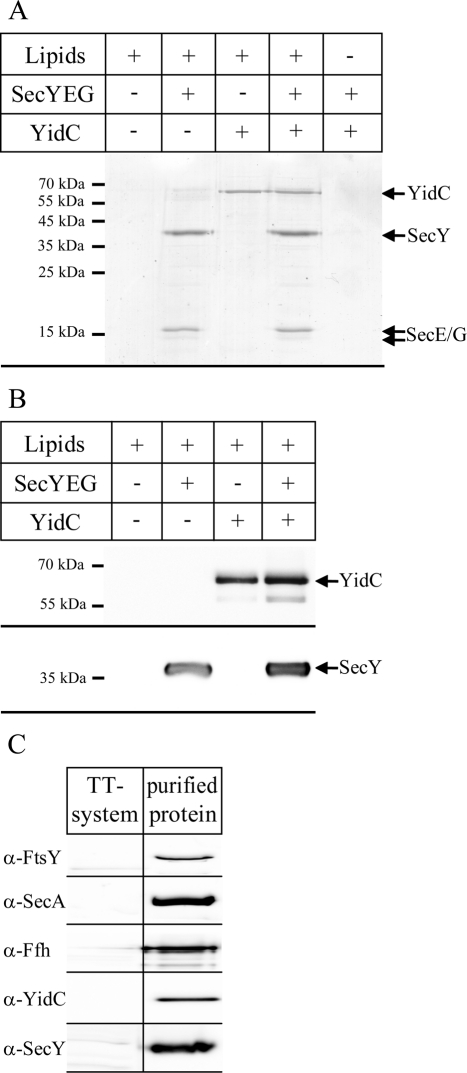
Identifying the determinants that route proteins into the YidC-only or into the SecYEG/YidC pathway by in vivo experiments has been complicated because the conditional depletion of SecY or YidC induces a multifaceted response that includes the up-regulation of proteases and chaperones (Baars et al., 2008; Price et al., 2010; Wang et al., 2010; Wickström et al., 2011). In addition, it has been impossible to generate an E. coli strain in which SecY and YidC can be inactivated individually or together (Pop et al., 2009). Finally, as the integration of YidC is SecY dependent (Koch et al., 2002) and the integration of SecY is probably influenced by YidC depletion (Price et al., 2010), experiments using the available depletion strains are extremely challenging. We therefore chose to address this question in vitro by using reconstituted proteoliposomes. Purified YidC and SecYEG were reconstituted into liposomes containing native E. coli lipids supplemented with 5% diacylglycerol (DAG), which has been shown to reduce nonphysiological spontaneous membrane protein insertion (Nishiyama et al., 2006). The proteoliposomes were then isolated by centrifugation and separated on SDS–PAGE. Coomassie blue staining revealed no protein contaminants in the pure liposome fraction (Figure 1A), whereas in SecYEG proteoliposomes, SecY and SecE were detectable. Coomassie blue only weakly stains SecG (Nishiyama et al., 2006), which explains why it was barely visible on the gel (Figure 1A). In YidC proteoliposomes, YidC was detected as a 60-kDa band, and in proteoliposomes coreconstituted with SecYEG and YidC, all proteins except SecG were detectable in comparable amounts. In the absence of lipids, neither SecYEG nor YidC was found in the pellet fraction after centrifugation, indicating that they did not aggregate. To exclude that the 60- to 70-kDa protein contaminants in purified SecYEG (Figure 1A) corresponded to YidC, we performed Western blotting, which revealed that SecYEG proteoliposomes did not contain detectable amounts of YidC. We also showed that YidC proteoliposomes were free of SecY (Figure 1B).
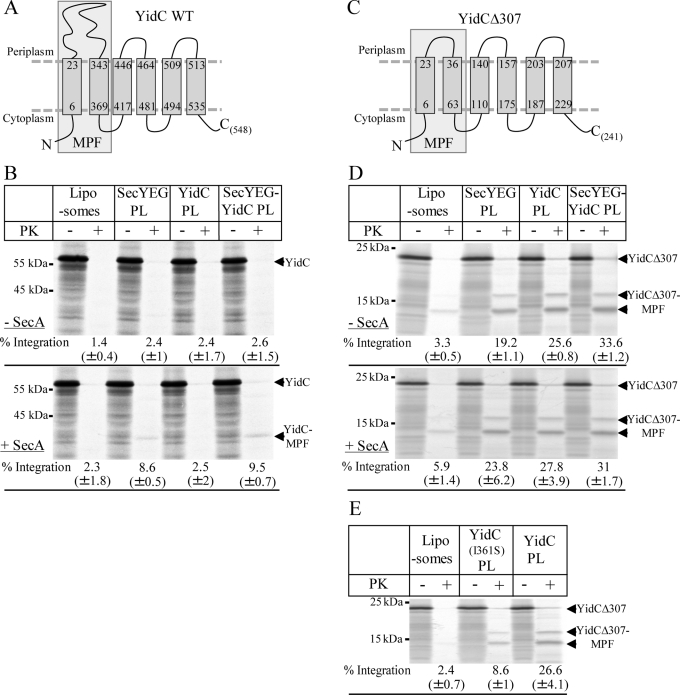
FIGURE 1:
Reconstitution of SecYEG and YidC into proteoliposomes. (A) Proteoliposomes were prepared using E. coli phospholipids supplemented with 5% DAG. Purified SecYEG or YidC were reconstituted into liposomes and pelleted by centrifugation. The pellet was resuspended and adjusted to a final concentration of 0.4 mg/ml SecY, 0.4 mg/ml YidC, or 0.4 mg/ml SecY + 0.4 mg/ml YidC. One aliquot (4 μl) was loaded onto a 15% SDS gel and Coomassie stained. Pure liposomes served as control. (B) The proteoliposomes (2 μl) shown in A were separated on SDS–PAGE and blotted onto a nitrocellulose membrane. The membrane was subsequently cut into two pieces, and the upper part was decorated with α-YidC antibodies and the lower part with α-SecY antibodies. (C) One aliquot of the purified E. coli in vitro TT system used for in vitro protein synthesis was probed with the indicated antibodies after Western blotting. Purified FtsY (0.7 μg), Ffh (0.2 μg), SecA (1.5 μg), SecY (0.8 μg), and YidC (0.8 μg) served as controls.
The purified in vitro transcription/translation system (TT system) used in this study does not contain significant amounts of FtsY, Ffh, SecA, SecY, or YidC (Koch et al., 1999; Figure 1C). Thus combining an in vitro system free of membranes and targeting factors with reconstituted proteoliposomes allowed us to determine the requirements for membrane protein integration in E. coli.
The polytopic membrane protein TatC is targeted by SRP to either SecYEG or YidC
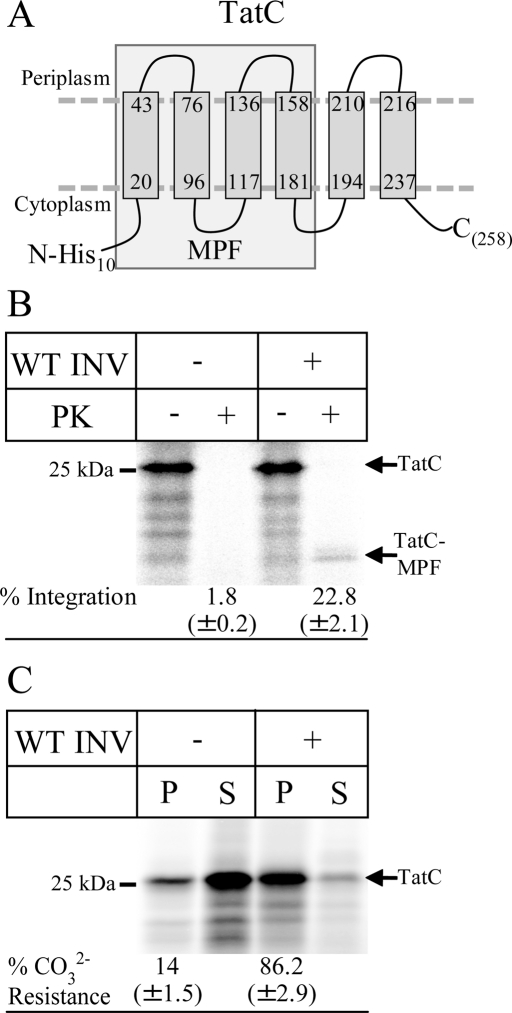
As a first substrate, we chose an N-terminally histidine (His)-tagged version of TatC, a multispanning membrane protein with a predicted molecular mass of 29 kDa. With its predicted six closely spaced TMs (Behrendt et al., 2004; Figure 2A), TatC is more complex than the so-far-identified substrates of the YidC-only pathway. When TatC was synthesized in vitro in the presence of E. coli inner membrane vesicles (INVs), we observed a membrane-protected fragment (MPF) of ∼19 kDa after proteinase K (PK) treatment (Figure 2B) that was not observed in the absence of INVs. A similar protease-protected fragment was also observed in in vivo pulse-chase experiments (Yi et al., 2003), but the exact nature of the MPF is unknown. We therefore constructed two truncated TatC derivatives, which lacked either the first two TMs (TatCΔN) or the last two TMs (TatCΔC). The migration of TatCΔC on SDS–PAGE was identical to the migration of TatC-MPF, whereas TatCΔN migrated faster than the TatC-MPF (Supplemental Figure S1). This indicates that the MPF probably corresponds to the first four TMs of TatC. However, due to the irregular migration behavior of some membrane proteins on SDS–PAGE, we cannot entirely exclude that the TatC-MPF corresponds to all six TMs. The membrane integration of TatC into E. coli INVs was further confirmed by alkaline carbonate extraction, a method that is routinely used to differentiate between membrane-inserted and soluble proteins (Fujiki et al., 1982). In the presence of INVs, almost 90% of TatC was found in the pellet fraction after carbonate extraction and centrifugation, indicating membrane insertion (Figure 2C). Carbonate treatment of in vitro–synthesized TatC in the absence of INVs resulted in >80% of TatC in the supernatant (Figure 2C). Differences in the integration rates of TatC measured by carbonate extraction or PK digestion are likely due to additional PK cleavage sites resulting in smaller PK resistant fragments, which are not included in calculating the integration rate.
FIGURE 2:
Integration of in vitro–synthesized TatC into inner membrane vesicles. (A) Predicted topology of TatC according to Behrendt et al. (2004); a His10 tag was fused to its N-terminus. The boxed portion of TatC most likely corresponds to the 19-kDa, membrane-protected fragment of TatC (TatC-MPF). (B) 35S-Labeled His10-TatC was in vitro synthesized in the absence (–INV) or presence of wild-type E. coli inner membrane vesicles (WT INV; 2 mg/ml). After synthesis, one-fourth of the reaction was precipitated with TCA, and the remainder was first treated with 0.5 mg/ml PK for 30 min at 25°C and then TCA precipitated. Full-size TatC (TatC) and the TatC-MPF are indicated. Note that wild-type E. coli INV contains sufficient amounts of SRP and FtsY (Koch et al., 1999). The percentages of PK protection was calculated using ImageQuant (GE Healthcare) by quantifying the ratio of radioactivity present in the PK-treated sample and the directly TCA-precipitated sample and are the mean values of at least three independent experiments. Note that the calculation is corrected for the loss of methionine/cysteine residues and based on the assumption that TatC-MPF corresponds to the first four TMs of TatC. (C) TatC was in vitro synthesized as in B but extracted with alkaline Na2CO3 (pH 11.3, 0.2 M final concentration). After ultracentrifugation, pellet (P) and supernatant (S) were separated by SDS–PAGE. For quantification, the amounts of radioactive material in both fractions were set as 100%, and the distribution between both fractions was calculated. The values provided are the mean values of at least three independent experiments, and the SD is indicated.
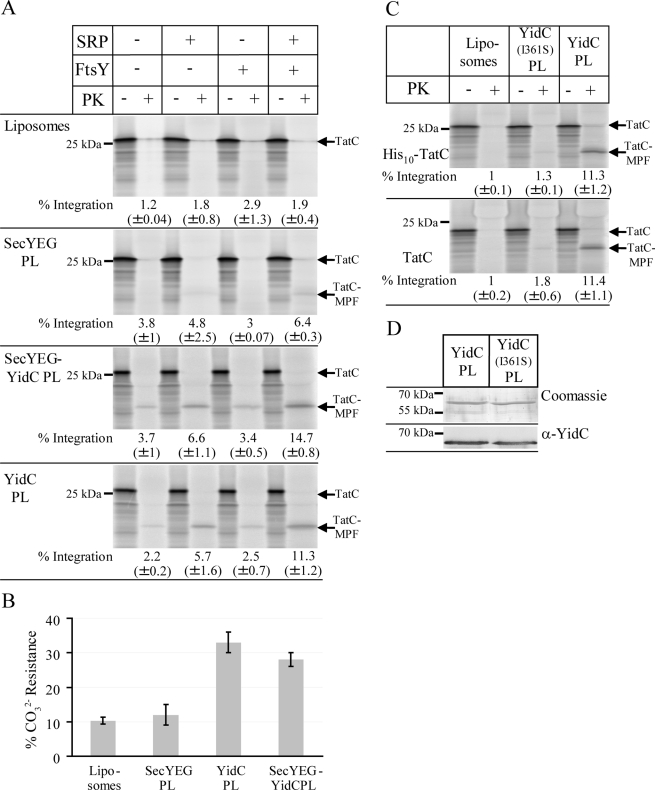
When TatC was synthesized in the presence of liposomes, the 19-kDa MPF was not detectable (Figure 3A). Instead, we noticed some protease-resistant, full-size TatC, which probably reflects TatC aggregates. In the presence of SecYEG proteoliposomes, the MPF of TatC was reproducibly detected when SRP and the bacterial SRP receptor FtsY were present (Figure 3A), but the total integration was significantly lower than in INVs (Figure 2B). Only a weak protease protection of TatC was observed in the absence of SRP/FtsY. We next analyzed whether coreconstituting YidC and SecYEG into proteoliposomes would increase the efficiency of TatC integration as would be expected if YidC and SecYEG cooperated during the integration of membrane proteins. Protease protection of TatC in the presence of SRP/FtsY was significantly stronger in SecYEG/YidC proteoliposomes than in SecYEG proteoliposomes (Figure 3A) supporting a cooperative function of YidC with SecYEG in membrane integration. We also noticed that although TatC integration into SecYEG/YidC proteoliposomes was most efficient when both SRP and FtsY were present, significant integration was also observed in their absence (Figure 3A). Owing to the intrinsic affinity of SecYEG for ribosomes (Prinz et al., 2000), a weak SRP/FtsY–independent integration is expected and was observed for other SecYEG-dependent membrane proteins in proteoliposome studies (Braig et al., 2011). Because the presence of YidC seemed to increase the SRP/FtsY–independent integration of TatC, we analyzed proteoliposomes reconstituted with YidC alone. It is striking that we observed that TatC was more efficiently integrated into YidC proteoliposomes than into SecYEG-proteoliposomes (Figure 3A). Integration of TatC via YidC was most efficient in the presence of SRP/FtsY but also occurred in the presence of SRP alone.
FIGURE 3:
TatC is targeted by SRP to either SecYEG or YidC. (A) 35S-Labeled His10-TatC was in vitro synthesized as described in Figure 1. When indicated, SRP (Ffh [150 nM] + 4.5S RNA [15 μg/ml]) and FtsY (750 nM) were present during synthesis. Synthesis was performed in the presence of 0.4 mg/ml of the liposomes or proteoliposomes (PL) shown in Figure 1. After synthesis, the samples were PK digested. (B) Carbonate resistance of TatC synthesized in the presence of liposomes or proteoliposomes. The mean values of three independent experiments are shown. (C) 35S-Labeled His10-TatC and 35S-labeled TatC were synthesized in the presence of SRP, FtsY, and proteoliposomes reconstituted with either wild-type YidC or the YidC(I361S) mutant. (D) Coomassie blue staining and immunodetection of proteoliposomes containing either wild-type YidC or the YidC(I361S) mutant. Four μl of each proteoliposome preparation were loaded for Coomassie staining and 2 μl for immunodetection.
The integration of TatC into liposomes and proteoliposomes was further analyzed by carbonate extraction, and these assays were performed in the presence of SRP/FstY. In the presence of liposomes, only ∼10% of in vitro–synthesized TatC was found in the pellet fraction (Figure 3B), which was comparable to the values observed in the absence of membranes (Figure 2C). We observed only a weak insertion of TatC into SecYEG proteoliposomes as indicated by carbonate resistance, whereas TatC was integrated very efficiently into YidC proteoliposomes (Figure 3B). Coreconstituting SecYEG together with YidC did not increase membrane insertion of TatC (Figure 3B). These data are consistent with the protease protection assays (Figure 3A), which also showed that TatC is inserted more efficiently into YidC proteoliposomes than into SecYEG proteoliposomes.
YidC could facilitate the integration of multispanning membrane proteins like TatC by forming a protein-conducting channel like SecY, but it could also disrupt the close spacing of the phospholipid head groups in the lipid bilayer, subsequently allowing TMs to insert at the protein–lipid interface. We verified the specificity of YidC-dependent TatC integration by constructing the YidC(I361S) mutant, in which isoleucine at position 361 in TM2 of YidC was replaced by serine. This mutation has been shown to reduce the integration of the M13-coat-Lep fusion protein, a bona fide YidC substrate (Jiang et al., 2003). YidC(I361S) was purified and reconstituted into proteoliposomes in comparable amounts to YidC in YidC proteoliposomes (Figure 3D). We observed that TatC was integrated at a lower efficiency in YidC(I361S) proteoliposomes than in YidC-proteoliposomes (Figure 3D), consistent with earlier data (Jiang et al., 2003). This indicates that multispanning substrates like TatC and phage proteins follow a similar insertion path through YidC.
The TatC construct used in our experiments contained an N-terminal (His)10 tag and we wanted to exclude that the His tag influences the integration of TatC via YidC. We found that untagged TatC acquired protease protection like (His)TatC in YidC proteoliposomes but not in YidC(I361S) proteoliposomes (Figure 3E), demonstrating that the (His)10 tag did not influence YidC-dependent integration.
In summary, our data indicate that membrane insertion via the YidC-only pathway is not limited to small membrane proteins, but that YidC can also insert multispanning membrane proteins like TatC. Targeting of TatC to YidC is enhanced by SRP but appears to be less dependent on FtsY. TatC can also be inserted by SecYEG, but this pathway is less efficient. In SecYEG/YidC proteoliposomes, it is difficult to determine whether SecYEG and YidC function as distinct entities or whether they cooperate during TatC integration. The observation that FtsY-independent integration was predominantly observed in YidC proteoliposomes and SecYEG/YidC proteoliposomes could indicate that they function mainly independently.
YidC mediates integration of mannitol permease independent of the SecYEG translocon
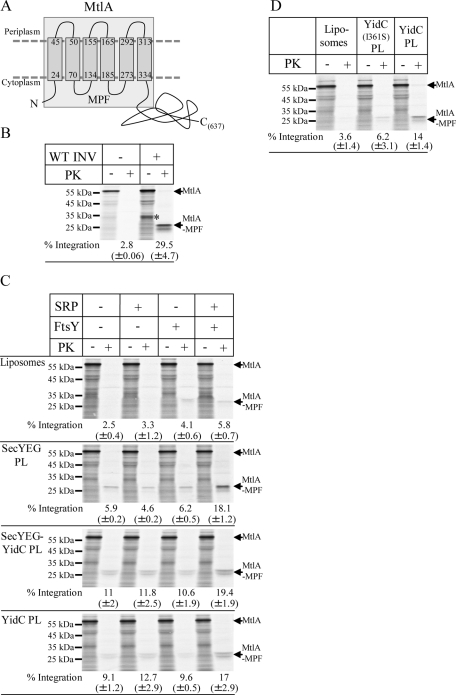
The 60-kDa membrane protein MtlA was previously used as a model substrate for studying SRP-dependent targeting to the SecYEG translocon (Koch et al., 1999; Beck et al., 2000; Braig et al., 2011). Because we observed the insertion of TatC into YidC proteoliposomes, we also analyzed MtlA insertion into YidC proteoliposomes. Like TatC, MtlA consists of a hydrophobic core of six TMs and an additional 30-kDa cytoplasmic domain (Figure 4A). Integration of MtlA into INVs is indicated by a 30-kDa MPF after PK treatment. This product corresponds to the hydrophobic six-TM core of MtlA but lacks the cytosolic domain, which is cleaved off by PK (Werner et al., 1992; Koch et al., 1999; Figure 4B). The protease-sensitive band at ∼32 kDa (Figure 4B, asterisk) is probably the result of premature termination of in vitro protein synthesis. In the presence of liposomes, we observed a weak spontaneous insertion of MtlA when SRP/FtsY was added (Figure 4C). This was observed previously and probably reflects TMs that penetrate between head groups of phospholipids (Nishiyama et al., 2006; Braig et al., 2011). In SecYEG proteoliposomes, MtlA integration was more efficient and the level of integration was strongly increased when SRP/FtsY was added (Figure 4C). The latter finding is consistent with previous studies demonstrating that efficient targeting of MtlA to the SecYEG translocon is SRP dependent (Koch et al., 1999). However, integration of MtlA into SecYEG proteoliposomes occurred also to some extent in the absence of SRP/FtsY, demonstrating that RNCs can be targeted to the SecYEG translocon independent of SRP in vitro. Different lengths of MtlA-RNCs have been shown to contact SecY and YidC sequentially, which has led to the hypothesis that YidC facilitates the release of transmembrane domains from SecYEG into the lipids (Beck et al., 2001). This SecYEG-associated function of YidC does not seem to be essential for MtlA integration, as the MPF was also observed in the absence of YidC (Figure 4C; Braig et al., 2011). Furthermore, coreconstituting SecYEG and YidC into proteoliposomes did not significantly increase MtlA integration (Figure 4C), although these proteoliposomes showed enhanced SRP-independent targeting. We also tested MtlA integration into proteoliposomes containing only YidC and found that MtlA was integrated into these proteoliposomes in the presence of SRP and FtsY (Figure 4C) but appeared to be less dependent on FtsY than on SRP, as it was also observed for TatC. MtlA was also not efficiently integrated into YidC(I361S) proteoliposomes (Figure 4D), suggesting that specific contacts between YidC and MtlA are required for integration.
FIGURE 4:
YidC is sufficient for MtlA integration. (A) Topology of MtlA according to Sugiyama et al. (1991); the cleavage site of PK is indicated. The MPF corresponds to the first six TMs. (B) 35S-Labeled MtlA was synthesized in the presence (WT INV) or absence (–INV) of INV. After synthesis, one-half of the reaction was precipitated with TCA, and the other half of the reaction was first treated with 0.5 mg/ml PK for 30 min at 25°C and then TCA precipitated. Full-length MtlA and the membrane-protected fragment (MtlA-MPF) corresponding to the hydrophobic core of MtlA are indicated. The band labeled with an asterisk probably corresponds to a product of premature termination of protein synthesis. (C) MtlA was synthesized as described in B and under the same conditions as described in the legend to Figure 3A. (D) MtlA was synthesized as described in Figure 3C.
Our data indicate that membrane proteins with multiple TMs like MtlA or TatC, which were previously believed to be exclusively inserted via the SecYEG translocon, can also be inserted by YidC. Of importance, targeting of substrates to either of the two translocases is mediated by SRP.
YidC-dependent integration is restricted to membrane proteins with small periplasmic loops
To further characterize the substrate specificity of the YidC integrase, we used YidC as a substrate. Previous studies showed that YidC is targeted by SRP to the SecYEG translocon (Koch et al., 2002; Urbanus et al., 2002) and that the translocation of its 320–amino acid long periplasmic loop requires SecA (Figure 5, A and B). This was verified in our proteoliposome approach, since protease protection of YidC in SecYEG proteoliposomes was observed only when SRP/FtsY and SecA were present (Figure 5B). The MPF of YidC corresponds to the first two TMs and the connecting periplasmic loop as determined previously (Koch et al., 2002; Serek et al., 2004; Deitermann et al., 2005). No protease protection of in vitro–synthesized YidC was observed in proteoliposomes reconstituted with YidC only (Figure 5B), even in the presence of SecA. YidC was inserted in SecYEG/YidC proteoliposomes at equivalent amounts as into SecYEG proteoliposomes (Figure 5B). This observation further supports our conclusion that the SecYEG-associated function of YidC is not essential for membrane protein integration in vitro.
FIGURE 5:
Membrane protein integration via YidC is restricted to membrane proteins with small periplasmic loops. (A) Predicted topology of YidC. The cleavage site for PK is indicated. The YidC-MPF corresponds to the first two TMs and the connecting periplasmic loop (Koch et al., 2002). (B) YidC was synthesized in presence of SRP, FtsY, and liposomes/proteoliposomes as indicated. Protease protection was analyzed in the absence or presence of 600 nM SecA. Full-length YidC and YidC-MPFs are indicated. (C) Predicted topology of YidCΔ307 (Deitermann et al., 2005). (D) Protease protection of YidCΔ307. The lower, 13-kDa MPF was used for calculating integration rates. (E) Protease protection of YidCΔ307 in the presence of proteoliposomes containing wild-type YidC or the YidC(I361S) mutant.
The observation that YidC proteoliposomes were able to integrate TatC and MtlA but not YidC suggests that the presence of large periplasmic loops impedes the integration via the YidC-only pathway. This was verified by using a YidC derivative in which the coding region for residues 31–336 was deleted, resulting in a short, 13–amino acid long periplasmic loop between TM1 and TM2 (YidCΔ307; Figure 5C; Deitermann et al., 2005). This derivative is integrated into INVs in a SecA-independent manner, yielding PK-protected MPFs of 18 and 13 kDa. The more prominent 13-kDa band represents the first two TMs plus the short periplasmic loop (Deitermann et al., 2005), whereas the exact nature of the weaker 18-kDa band is unknown. Consistent with the data obtained with INVs, the integration of YidCΔ307 into SecYEG proteoliposomes did not require the addition of SecA (Figure 5D). YidCΔ307 was also efficiently integrated into YidC proteoliposomes and SecYEG/YidC proteoliposomes independent of SecA. We also observed that integration was reduced in YidC(I361S) proteoliposomes (Figure 5E). Similar to the result for MtlA, we observed a weak spontaneous insertion of YidCΔ307 into liposomes, probably reflecting penetration of TMs between the phospholipid head groups.
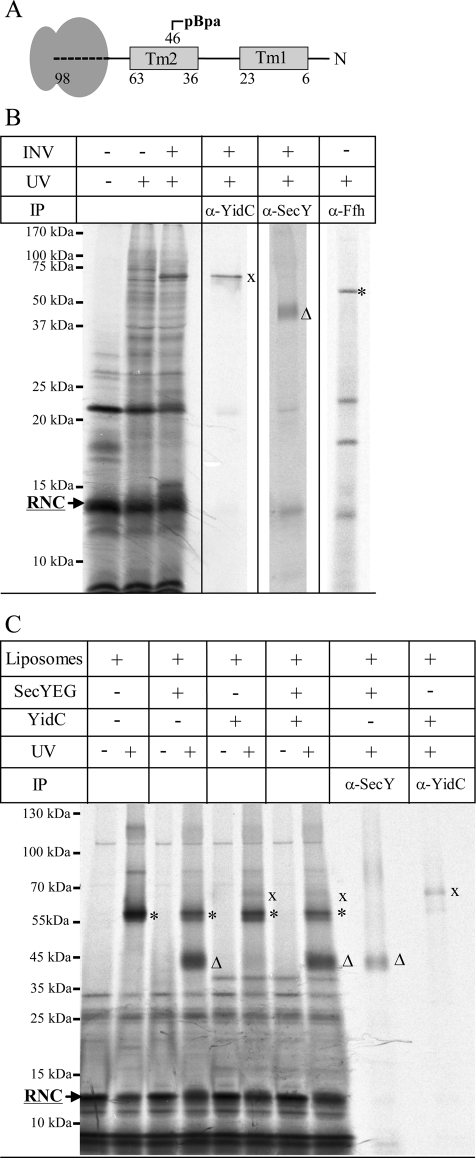
The integration of multispanning membrane proteins like YidCΔ307 by the YidC-only pathway was further analyzed by site-directed in vitro cross-linking using the UV-dependent cross-linker para-benzoyl-l-phenylalanine (pBpa). Previous INV studies showed that RNCs of membrane proteins cross-link to both SecY and YidC (Beck et al., 2001; Urbanus et al., 2001), suggesting that YidC is located in close proximity to SecY. This was also demonstrated in vitro for a 98–amino acid long RNC of YidCΔ307 (Figure 6A), which encompasses the first two TMs and harbors pBpa at position 46 within TM2. After UV exposure in the presence of INV, possible cross-links were identified by immunoprecipitations. We observed cross-links to Ffh at ∼60 kDa (Figure 6B, asterisk), supporting SRP-dependent targeting of the YidCΔ307-RNCs to the membrane. We also observed cross-links to SecY at ∼45 kDa and a strong cross-link to YidC at ∼70 kDa (Figure 6B, triangle and cross). These data support a cooperative function of YidC in SecYEG-dependent membrane insertion. However, as YidCΔ307 can also be integrated by YidC-proteoliposomes (Figure 5D), we performed cross-linking experiments with proteoliposomes containing SecYEG, YidC, or both. After the addition of purified SRP and FtsY to allow targeting, we observed a strong cross-link of YidCΔ307-RNCs to Ffh at ∼60 kDa on SDS–PAGE (Figure 6C, asterisk). In the presence of SecYEG proteoliposomes, the cross-link to Ffh became slightly weaker and an additional cross-link at ∼45 kDa that was immunoprecipitated by α-SecY antibodies appeared (Figure 6C, triangle). In the presence of YidC proteoliposomes, the cross-link to Ffh was also slightly reduced, and a cross-linking product at ∼70 kDa was detectable and recognized by α-YidC antibodies (Figure 6C, cross). However, the cross-link to YidC was significantly weaker than the cross-link to SecY. In proteoliposomes containing both YidC and SecYEG, the disappearance of the SRP cross-link was most pronounced and coincided with the appearance of both SecY and YidC cross-links.
FIGURE 6:
Molecular contacts of YidCΔ307 RNCs in INV and proteoliposomes. (A) Cartoon showing the 98–amino acid long YidCΔ307-RNCs used in this study. YidCΔ307-98 RNCs comprise the first 98 amino acids of YidCΔ307 (Figure 5C) and contains pBpa at position 46 in TM 2. (B) YidCΔ307-98 RNCs were synthesized in the presence of INVs. Cross-linking products were identified by immunoprecipitations after separation on 15% SDS–PAGE. (C) YidCΔ307-RNCs were synthesized in the presence of SRP, purified via a sucrose cushion, and incubated with FtsY and liposomes/proteoliposomes as indicated. Samples were loaded on a 7–18% SDS gel. Cross-links to Ffh (asterisk), YidC (cross), and SecY (triangle) are indicated.
We further analyzed whether insertion of membrane proteins via SecYEG or YidC was influenced by the membrane potential. The integration of MtlA, TatC, YidC, and YidCΔ307 into INV was not influenced by 100 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP), a protonophore that equalizes the proton concentration on both sides of the membrane (Supplemental Figure S2). We also did not observe a significant effect on the translocation of the SecA-dependent outer membrane protein OmpA (Supplemental Figure S2).These results are in line with studies showing a proton-motive force (pmf)–independent insertion of membrane proteins via SecYEG (Koch and Müller, 2000) or YidC (Yi et al., 2004; van der Laan et al., 2004b). In proteoliposomes, we found that YidC was integrated in a pmf-independent manner (Supplemental Figure S2). However, we were unable to determine a possible influence of the pmf on TatC and YidCΔ307 integration because both substrates probably aggregated in the buffer system used for energizing proteoliposomes and were not inserted.
In summary, our data demonstrate that membrane proteins like MtlA, TatC, and YidCΔ307 interact with and are integrated by YidC independent of SecYEG. However, YidC fails to insert membrane proteins with large periplasmic loops. The translocation of large periplasmic loops requires SecA. It is therefore likely that SecA-dependent membrane proteins can only be inserted by the SecYEG translocon.
YidC binds to E. coli ribosomes
The observation that YidCΔ307-RNCs can be cross-linked to YidC indicates a cotranslational interaction, which is also supported by the stimulatory effect of SRP and the recent modeling of RNCs onto a YidC dimer (Kohler et al., 2009). However, since YidC lacks the extended C-terminus that has been shown to be required for ribosome binding of Oxa1 or of the bacterial YidC2, we analyzed ribosome binding to YidC in a sedimentation assay. These assays were performed at pH 7.6 to prevent protonation of the N-terminal His tag. Sucrose gradient–purified E. coli 70S ribosomes were incubated with or without detergent-solubilized, purified, His-tagged YidC or SecY and centrifuged through a sucrose cushion. The amount of YidC or SecY in the supernatant and pellet fraction was then immunodetected using antibodies against their respective His tags. The use of antibodies against the His tag on these proteins allowed us to directly compare binding of ribosomes to YidC or SecY. Coomassie blue staining and immunodetection using antibodies against the ribosomal subunit L23 revealed that under these conditions ribosomal proteins were found in the pellet fraction (P) after centrifugation (Figure 7A). When SecY was incubated with increasing amounts of 70S ribosomes, we observed that most of SecY cosedimented with ribosomes even at a ribosome concentration of just 0.2 μM. At 0.4 μM ribosomes, SecY was almost completely bound to ribosomes and found in the pellet fraction. In contrast, SecY was exclusively recovered from the supernatant in the absence of ribosomes (Figure 7B). On the basis of these data we estimate that SecY binds to empty ribosomes with a KD of ∼0.15 μM (Figure 7C), which is in the range of the KD values determined by Prinz et al. (2000). The same experimental setup was also used for measuring binding of YidC to E. coli ribosomes. Although binding of YidC to ribosomes was detectable, the affinity of YidC to empty ribosomes appeared to be significantly weaker than that of SecY (Figure 7B). In particular, no saturation was observed under the conditions tested, suggesting nonspecific binding. A rough estimation indicates that the affinity of YidC for ribosomes is ∼5–10 times lower than the affinity of SecY for ribosomes (Figure 7C). It is also lower than the affinity of the Oxa1 C-terminus for mitochondrial ribosomes, which is in the range of 0.3–0.8 μM (Haque et al., 2010a, 2010b).
FIGURE 7:

YidC binds to E. coli ribosomes. (A) Purified and salt-washed 70S ribosomes (500 nM) were incubated in binding buffer (pH 7.6). After 30 min of incubation on ice, ribosomes were centrifuged through a sucrose cushion. Pellet (P) and supernatant fractions (S) were separated on SDS gels and analyzed by Coomassie staining (top) and immunodetection using antibodies against L23 (bottom). (B) Increasing concentrations of salt-washed ribosomes were incubated with either 100 nM detergent-purified SecYEG (top) or detergent-purified YidC (bottom) in binding buffer (pH 7.6) and subjected to the same conditions as described in A. Pellet and supernatant fractions were immunodetected using antibodies against the N-terminal His tags of SecY or YidC. The percentage of binding was calculated using ImageJ software. (C) Quantification of at least three independent ribosome-binding experiments, using varying concentrations of SecYEG or YidC, respectively. The amount of ribosome-bound SecY/YidC was plotted against the total ribosome concentration.
The YidC–ribosome interaction was further verified in vitro by cross-linking experiments using sucrose gradient–purified INVs from E. coli cells expressing YidC with the cross-linker pBpa incorporated at position L540 at its C-terminus, YidC(L540pBpa). Cross-linking experiments with these membranes were performed without additional ribosomes, and we therefore determined contacts only to proteins that were purified together with INVs. On exposing YidC(L540pBpa) INVs to UV, several additional bands compared with the –UV control were detected by α-His antibodies, indicating that cross-linking had occurred (Figure 8A). Possible cross-links to ribosomal proteins were analyzed by Western blotting. The ribosomal protein L23 is located close to the ribosomal tunnel exit and was shown to be a major contact partner for SecY in vivo (Kuhn et al., 2011). L23 was also proposed to be involved in YidC binding based on a recent cryo-EM study (Kohler et al., 2009). However, antibodies against the ribosomal protein L23 did not detect any specific cross-linking product (Figure 8B). In contrast, INVs containing SecY with pBpa incorporated within the fifth cytosolic loop, SecY(R357pBpa), cross-linked efficiently to L23 under the same experimental conditions (Figure 8B). We also tested antibodies against L24 and L29, which, like L23, are located close to the ribosomal tunnel exit, and again we did not detect any cross-linking product (data not shown).
FIGURE 8:
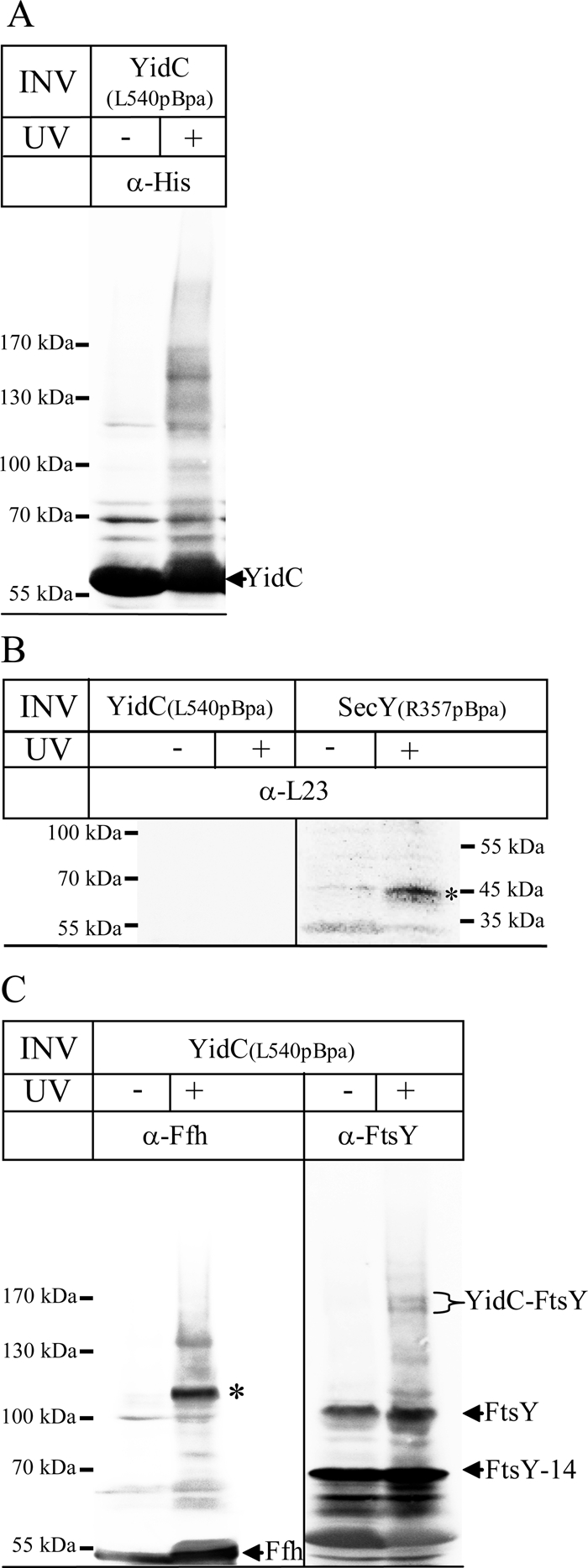
In vitro cross-links using INVs of YidC pBpa mutants show contacts to SRP and FtsY. (A) One mg of INVs from E. coli cells overexpressing YidC(L540pBpa) was resuspended in 250 μl of INV buffer and cross-linked by UV irradiation. The samples were separated on a 5–15% SDS gel, and immunodetection was performed with α-His antibodies. (B) Cross-linking with INVs purified from E. coli cells expressing YidC(L540pBpa) or the pBpa-containing SecY derivative SecY(R357pBpa), which contains pBpa within the fifth cytosolic loop of SecY. After separation on SDS–PAGE, cross-links to L23 (asterisk) were identified by immunodetection. (C) The material shown in A was also probed with α-Ffh and α-FtsY antibodies. The YidC-Ffh cross-link at 110 kDa is indicated (asterisk), as well as the Ffh that was present in the INVs. The YidC-FtsY cross-link appears in two distinct bands at ∼160 kDa, which probably reflect the two FtsY isoforms present in E. coli INV. FtsY corresponds to full-size FtsY, whereas FtsY-14 corresponds to an N-terminally–truncated FtsY derivative.
For increasing the sensitivity of our assay system, we analyzed gel lanes of UV-treated and untreated INVs by high-resolution mass spectrometry (MS). This approach identified several ribosomal subunits as cross-linking partners of YidC (Table 1). It is important to note that all ribosomal proteins listed were reliably identified in two independent cross-linking experiments and found at the expected size of the respective YidC cross-linking product in SDS–PAGE while being absent in the control sample (–UV). Consistent with our Western blotting data, cross-links to L23, L24, and L29 were not detected by MS (Table 1). We did, however, detect cross-links to L22, which is also located close to the ribosomal tunnel exit. The ribosomal subunits detected were identified with sequence coverage in the range of 16–58% and localized to both the large and small ribosomal subunits without distinct clustering. It is important to emphasize that in our approach we only probed for YidC contacts to nontranslating ribosomes. The presence of a signal-anchor sequence within YidC could stabilize the YidC–ribosome contact and allow for a more distinct cross-linking pattern involving the tunnel exit subunits. Additional ribosomal proteins were identified in only one of the two independent experiments (ribosomal subunits L3, L4, L7, L10, L11, L15, L27, L28, L30, L31, S5, S6, S7, and S10).
TABLE 1:
Mass spectrometric analyses of YidC(L540pBpa) INV.
| Proteina | Gel molecular weight (kDa)b | Molecular weight (kDa)c | Scored | Coveragee | Peptidesf | |
|---|---|---|---|---|---|---|
| Ribosomal proteins | L5 | 76.1 | 20.3 | 120.8 | 58.1 | 11 |
| L6 | 76.1 | 18.9 | 53.8 | 36.2 | 6 | |
| L13 | 76.1 | 16 | 54.1 | 58.5 | 8 | |
| L14 | 69.2 | 13.5 | 78.9 | 28.5 | 8 | |
| L22 | 69.2 | 12.2 | 19.7 | 16.4 | 3 | |
| S11 | 72.6 | 13.8 | 23.8 | 27.9 | 3 | |
| S12 | 75 | 13.7 | 16.6 | 16.9 | 2 | |
| S13 | 72.6 | 13.1 | 28.1 | 33.1 | 3 | |
| S14 | 69.2 | 11.6 | 14.8 | 28.7 | 2 | |
| Targeting factors | Ffh | 111 | 49.8 | 144.3 | 31.6 | 13 |
| FtsYg | 161 | 54.5 | 14,84 | 3.82 | 2 |
Non–UV irradiated (not cross-linked, −UV) and UV-irradiated (cross-linked, +UV) samples of YidC(L540pBpa) INV were separated on a 5–15% SDS gel, and the proteins were visualized by Coomassie staining. The −UV and +UV lanes were cut into equal slices, followed by in-gel trypsin digestion and mass spectrometric analyses. Only ribosomal proteins that were specifically detected in two independent cross-linking experiments are listed.
aProtein identified.
bMolecular weight of gel slice determined by extrapolation.
cCalculated molecular mass.
dProtein score; in both replicates, a cut-off score of 5.5 corresponded to a protein false discovery rate of <0.01.
eSequence coverage of total sequence by detected peptides.
fNumber of peptides detected.
gFtsY/YidC-cross links were found specific in one experiment only.
Combining cross-linking with MS allowed identification of cross-links between Ffh and YidC at 111 kDa in both experiments (Table 1). A cross-linked band at 110 kDa was also strongly recognized by α-Ffh antibodies (Figure 8C), confirming the close proximity between Ffh and the C-terminus of YidC. We also detected cross-links between FtsY and YidC at ∼160 kDa (Table 1), but a corresponding band was also detected in one experiment in the –UV control. Whether this is due to the aberrant migration behavior or to some contamination is unknown. Antibodies directed against FtsY recognized two strong bands at 70 and 95 kDa in both the UV-treated and untreated samples (Figure 8C). The 95-kDa band corresponds to full-size FtsY, whereas the 70-kDa band corresponds to an N-terminally truncated FtsY derivative (FtsY-14; Weiche et al., 2008). In addition, two weak bands at ∼160 kDa were recognized by α-FtsY antibodies, which were only present in the UV-treated sample (Figure 8C). These bands most likely reflect cross-links of YidC to FtsY and FtsY-14, respectively. Additional UV-dependent bands at 110 and 130 kDa are probably nonspecifically recognized by the polyclonal α-FtsY antibody (Figure 8C). By MS, we did not detect contacts between SecA and YidC. This could explain why YidC fails to integrate SecA-dependent membrane proteins (Figure 4).
In conclusion, the ribosome-binding assays and the cross-linking approach demonstrate that ribosomes can contact the C-terminus of E. coli YidC but that this interaction appears to be much weaker than the SecY–ribosome interaction. YidC also directly interacts with SRP and to a lesser extent with FtsY. Together with the proteoliposome results, this indicates that SRP can deliver multispanning membrane proteins not only to the SecYEG translocon, but also to the YidC integrase.
DISCUSSION
Our data revealed four important functional aspects of YidC: 1) The SecYEG-associated function of YidC is not essential for the integration of the membrane proteins. 2) YidC can function independent of SecYEG as an integration site for multispanning membrane proteins. 3) Only SecA-dependent membrane proteins strictly depend on SecYEG for integration. 4) SRP delivers membrane proteins to either SecYEG or YidC.
YidC functions as an independent integrase for some membrane proteins but also assists the SecYEG translocon in membrane insertion. Proteins such as subunit K of NADH:ubiquinone oxidoreductase (Price et al., 2010), subunit A of quinol oxidase (du Plessis et al., 2006), and subunit A of ATP synthase (Yi et al., 2004) require both SecYEG and YidC for insertion. Several functions have been assigned to YidC during SecYEG-dependent membrane insertion: it is suggested to chaperone membrane proteins (Nagamori et al., 2004; Jong et al., 2010), to facilitate the release of TMs from SecYEG (Beck et al., 2001; Urbanus et al., 2001; Houben et al., 2004), to promote the formation of membrane protein complexes (Wagner et al., 2008) and to control protein quality (van Bloois et al., 2008). However, whether these SecYEG-associated functions of YidC are essential for membrane protein insertion was unknown. We now show that the integration of MtlA, TatC, and YidC via SecYEG does not require the assistance of YidC. The integration of these proteins into SecYEG proteoliposomes was almost as efficient as into proteoliposomes containing both SecYEG and YidC. The protease-protected fragments observed in the presence of SecYEG proteoliposomes were also identical to those observed in the presence of INV or SecYEG/YidC proteoliposomes. This indicates that MtlA and YidC, for which the MPFs are clearly defined (Werner et al., 1992; Koch et al., 2002; Deitermann et al., 2005), are properly inserted by SecYEG in the absence of YidC. The observation that the SecYEG-associated function of YidC is not essential for membrane protein integration is consistent with studies on the single-spanning membrane protein FtsQ (van der Laan et al., 2004b) and by studies showing that YidC depletion has only a minor effect on the integration of FtsQ or leader peptidase in vivo (Urbanus et al., 2001; Fröderberg et al., 2003).
So far, only single- or double-spanning membrane proteins with short periplasmic segments have been shown to be inserted via the YidC-only pathway, that is, independent of SecYEG (Xie and Dalbey, 2008). We now show that YidC can also insert multispanning membrane proteins like MtlA or TatC, which had been considered to be exclusively dependent on SecYEG. This is consistent with previous studies on TatC insertion in E. coli (Yi et al., 2003) and chloroplasts (Martin et al., 2009), where no clear dependence on either SecYEG or YidC for membrane insertion was demonstrated. Although YidC has been shown to improve membrane integration of TatC (Yi et al., 2003), there was no effect of YidC depletion on the transport of Tat-dependent substrates (Hatzixanthis et al., 2003). This suggests that TatC is inserted into the inner membrane either via SecYEG or YidC, as is also demonstrated by our PK protection and carbonate treatment assays. However, we cannot conclude which translocase takes precedence over the other while inserting TatC. Previous data from our lab on membrane protein insertion into SecYEG-depleted INVs show that although protein insertion is drastically reduced, there is some amount of insertion (Koch et al., 1999; Koch and Müller, 2000; Deitermann et al., 2005). This was previously attributed to membrane insertion via residual amounts of SecYEG, but considering our present data, it is likely that insertion occurred via YidC. This would also explain why SecA-dependent substrates appeared to be more sensitive toward SecYEG depletion than SecA-independent substrates (Koch et al., 2002; Beha et al., 2003), as YidC cannot transport SecA-dependent proteins. It is, however, important to mention that YidC is depleted during SecYEG depletion, since YidC has been shown to require SecYEG for its insertion (Koch et al., 2002). Thus the YidC content of SecYEG-depleted INVs is extremely difficult to control.
In conclusion, these data suggest that the YidC-only pathway could be engaged by most bacterial membrane proteins, with the exception of membrane proteins containing large periplasmic loops. Because these proteins require SecA for the translocation of the periplasmic loops across the membrane (Deitermann et al., 2005), it is likely that SecA fails to directly interact with YidC, which is also consistent with our MS data.
Targeting of MtlA or TatC to YidC is strongly enhanced by SRP/FtsY. SRP-dependent targeting to YidC is supported by our cross-linking data, which show that SRP and to a lesser extent FtsY are in close proximity to the C-terminus of YidC. SRP release from the nascent chain by FtsY is a prerequisite for protein insertion into the SecY channel (Koch et al., 1999), but insertion via YidC appears to proceed to some extent also in the absence of FtsY. It is possible that its direct interaction with YidC dissociates SRP from some nascent chains or that SRP release is induced by the RNC–YidC interaction. Finally, a possible posttargeting function of SRP has been proposed for SecY-dependent insertion (Bibi, 2012), which could also account for an SRP-dependent but FtsY-independent insertion via YidC. So far, direct interactions between SRP/FtsY and YidC homologues have only been observed for the chloroplast Alb3, which interacts with both FtsY (Moore et al., 2003) and the chloroplast-specific SRP subunit cpSRP43 (Falk et al., 2010; Lewis et al., 2010; Richter et al., 2010). Our data now show that SRP and FtsY are able to directly interact with YidC in bacteria. SRP-dependent targeting to YidC was previously also shown for MscL (Facey et al., 2007) and F0c (van Bloois et al., 2004). However, SRP targets F0c to the SecYEG translocon when YidC is absent (van Bloois et al., 2004) and leader peptidase to SecYEG, as well as to YidC proteoliposomes (Houben et al., 2002), although insertion can only occur via SecYEG. This indicates that SRP recognizes its substrates independent of downstream integration sites. Dual targeting by SRP occurs not only in bacteria but also in yeast, where SRP was shown to deliver substrates to both the Sec61 complex and the alternative Ssh1p translocon (Spiller and Stirling, 2011).
SRP-dependent targeting to SecYEG occurs cotranslationally and involves direct binding of RNCs to SecY. Whether ribosomes/RNCs can also bind to YidC has been debated, particularly because YidC lacks the extended C-terminus that was identified to be the ribosome-binding site in Oxa1 (Szyrach et al., 2003). We now show that YidC binds to nontranslating ribosomes although with a lower affinity than SecY. Ribosome binding of YidC does not show saturation, indicating nonspecific binding, which also fits with the observation that YidC does not cross-link to a single ribosomal subunit. The C-terminus of YidC can be cross-linked to ribosomal proteins, but except for L22, we did not detect proteins that are located close to the tunnel exit. This is in contrast to a recent cryo-EM reconstruction study, which implicated proteins of the tunnel exit in YidC binding (Kohler et al., 2009), very similar to the ribosome–SecY contact (Frauenfeld et al., 2011). Because our in vitro cross-linking was performed with nontranslating ribosomes, whereas the cryo-EM study was performed with translating ribosomes, it is possible that the presence of a transmembrane domain influences the ribosome–YidC contact. It is likely that SRP promotes ribosome binding to YidC in E. coli, whereas mitochondria might have adapted to the lack of the SRP pathway by extending the C-terminus and providing a SRP-independent ribosome-binding platform for cotranslational insertion of membrane proteins. This would also explain why in Streptococcus mutans, the SRP pathway is not essential, as this organism contains two YidC homologues, one containing a C-terminal ribosome binding site (as in Oxa1) that allows cotranslational integration in the absence of SRP (Hasona et al., 2005).
YidC cannot insert IMPs with large periplasmic loops, and therefore the SRP pathway needs to selectively target these proteins to SecYEG. Because SecA and FtsY transiently associate with SecY, it is possible that the presence of two receptors at the SecYEG translocon versus only one at YidC (FtsY) provides the necessary specificity. However, this would probably require that FtsY and SecA bind nonexclusively to the SecYEG translocon, which appears unlikely (Kuhn et al., 2011). Alternatively, SecA-dependent membrane proteins that were targeted to YidC could use a mechanism similar to that proposed for CyoA insertion, which has been shown to require YidC, SecYEG, SecA, and the pmf for insertion (du Plessis et al., 2006; Celebi et al., 2006). It is possible that when YidC encounters SecA-dependent membrane proteins, it associates with the SecYEG translocon for allowing the passage of periplasmic loops. This needs to be further analyzed. It is also unknown why a small number of membrane proteins can only be inserted via YidC. This could be related to the hydrophobicity of the substrate. F0c has a mean hydrophobicity of 1.27 (calculated according to Kyte and Doolittle, 1982) and is exclusively YidC dependent, whereas TatC (0.75) and MtlA (0.33) are relatively less hydrophobic and can be inserted either via SecYEG or YidC. Dual targeting could also be dependent on charge distribution, given that it was recently shown for both NuoK (Price and Driessen, 2010) and M13 coat protein derivatives (Stiegler et al., 2011) that insertion via YidC is influenced by the presence of negatively charged residues.
It is important to emphasize that our conclusions are mainly based on proteoliposome studies, and thus it is difficult to determine to what extent dual targeting occurs in vivo. However, recent proteome analyses support the theory of dual targeting in vivo. In SecE-depleted E. coli membranes that are also depleted in SecY, it was found that almost all outer membrane proteins were reduced, whereas a large number of inner membrane proteins appeared to be unaffected (Baars et al., 2008). These inner membrane proteins lacked extended periplasmic loops, supporting our in vitro observation that multispanning membrane proteins with closely spaced TMs that are SecA independent can use YidC as an integration site. A proteome analysis of YidC-depleted membranes demonstrated that primarily the steady-state levels of membrane proteins with small soluble loops <100 amino acids are reduced (Wickström et al., 2011), which suggests that closely spaced membrane proteins are preferentially targeted to YidC. It is possible that access of these membrane proteins to YidC is stochastically favored, because E. coli contains ∼2500 YidC molecules (Drew et al., 2003) but only 300–500 SecYEG complexes that are also required for the transport of most secretory proteins. Because translation is a rather slow process (∼10 amino acids per second), most SecYEG complexes would probably be occupied by translating ribosomes if SecYEG constituted the only integration site for membrane proteins. Using YidC as an additional integration site would ensure that sufficient SecYEG complexes are accessible for the translocation of secretory proteins.
MATERIALS AND METHODS
Strains and plasmids
E. coli DH5α (Hanahan, 1983), BL21, Top10 (Invitrogen, Karlsruhe, Germany), and MC4100 (Koch et al., 1999) strains were used for amplifying plasmids and for preparation of INVs where indicated. BL21, BL21 (DE3; Novagen, Bad Soden, Germany), and TY0 (Taura et al., 1997) were used for protein purification. pTrc99a-SecYHisEG (Kuhn et al., 2011) was used to prepare SecYHisEG-overexpressing inverted membrane vesicles for in vitro cross-linking experiments and for purifying SecYHisEG used in ribosome-binding assays.
The following plasmids were used for in vitro protein synthesis: p717MtlA (MtlA; Beck et al., 2000), pDMB (OmpA; Behrmann et al., 1998), pKSM-YidC and pKSM717-YidCΔ307 (YidC, YidCΔ307; Deitermann et al., 2005), and pJM8CS7-SecY (Koch et al., 1999). tatC was amplified from the genome of E. coli MC4100 using the following primers: forward, 5′ ACGCAAGCATATGTCTGTAGAAGATACTCAACCGCTTATCACG 3′; reverse 5′ CCGCTCGAGTTATTCTTCAGTTTTTTCGCTTTCTGCTTCAG 3′. The PCR product was digested with NdeI/XhoI and integrated into the NdeI/XhoI–digested pET19b, resulting in pET19b-TatC encoding an N-terminally His-tagged TatC. The N-terminal His tag of TatC was removed using the following primers: forward, 5′ ATGTCTGTAGAAGATACTCAACCGCT 3′; reverse, 5′ GGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTC 3′. TatCΔN (Δ2-111) was constructed using 5′ phosphorylated primers: forward, 5′ GTTTCCAGCTCTCTGCTGTTTTATATCG 3′; reverse, 5′ CATGGTATATCTCCTTCTTAAAGTTAAACAAAATTATTT 3′. TatCΔC (Δ190-257) was constructed using 5′ phosphorylated primers: forward, 5′ TAACTCGAGGATCCGGCTGCTAACA 3′; reverse, 5′ TAAGTCTTCTGGCGAGGTAATCCCCAT 3′.
YidC was cloned from pROEX-HTB-YidC (Samuelson et al., 2000) into pET19b using NdeI/XhoI restriction sites. HisyidC was then cloned into pTrc99a using EcoRI/NcoI, resulting in pTrc99aHisYidC.
Mutational PCR was carried out using the Finnzymes Phusion PCR Kit (NE Biolabs, Frankfurt, Germany) and 5′ phosphorylated primers: pTrc99a-YidC(L540pBpa) (forward, 5′ GAAAAACGTGGCTAGCATAGCCG 3′; reverse, 5′ CAGACCACGGTAAATCAGCTGCTG 3′); pTrc99a-YidC(I361S) (forward, 5′ ATTATCATCAGCACCTTTATCGTTCGTG 3′; reverse, 5′ GGAGAAGCCCCAGTTACCCACA 3′). Mutations in YidCΔ307 to introduce the amber stop codon TAG were created using site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit; Agilent, Santa Clara, CA) with the following primers: pKSM717YidCΔ307(G46pBpa), forward, 5′ CCATAGCTTTGTGTAGAACTGGGGCTTC 3′; reverse 5′ GAAGCCCCAGTTCTACACAAAGCTATGG 3′.
In vitro protein synthesis, cytosolic translation factors, and salt-washed ribosomes
Proteins were synthesized in vitro using a purified transcription/translation system composed of cytosolic translation factors (CTFs) and high-salt-washed ribosomes or an S135 cell extract (Müller and Blobel, 1984; Koch et al., 1999). For in vitro cross-linking, the CTF/S135 were prepared from E. coli Top10 pSup-pBpaRS-6TRN (Ryu and Schultz, 2006). RNCs were synthesized as described (Beck et al., 2000) using the following oligonucleotide: YidCΔ307 RNC-98, 5′ CAGCGCCATCATTTCCTGGCT 3′. The [35S]methionine/[35S]cysteine labeling mix was obtained from PerkinElmer (Wiesbaden, Germany), and pBpa was obtained from Bachem (Bubendorf, Switzerland).
INVs of E. coli cells were prepared as described previously (Müller and Blobel, 1984) with the following modifications: for cells expressing pBpa-containing proteins, the INV medium was supplemented with 1 mM pBpa (Chin et al., 2002) and glucose was replaced by glycerol. Cells were cultivated at 30°C, induced at an OD600 of 1 with 1 mM isopropyl-β-d-thiogalactoside (IPTG) for 5 h, and harvested.
Protein purification
The SecYEG complex was purified from pHisEYG using a His-tagged version of SecE (Collinson et al., 2001) in TY0 (MC4100) as described in Braig et al. (2011). For ribosome-binding assays, SecYHisEG was purified following the same protocol. The proteins were visualized on an SDS gel after Coomassie staining, and the protein concentration was then adjusted to 0.4 mg/ml.
YidC and YidC(I361S) were purified from BL21 cells harboring pTrc99a-YidC or pTrc99a-YidC(I361S). Cells were grown at 180 rpm and 30°C up to an OD600 of 1 before induction with 1 mM IPTG for 5 h. Purification was carried out as described for SecYEG by Braig et al. (2011). The imidazole concentration used in the washing buffer was 40 mM. Proteins were visualized on Coomassie-stained SDS–PAGE, and the concentration was adjusted to 0.4 mg/ml.
SecA was purified of BL21 DE3 harboring pET19b SecA with an N-terminal 10xHis tag (Beha et al., 2003). Cells were grown in 3 l of LB (lysogeny broth) medium to an OD600 of 0.8 and were induced overnight with 0.5 mM IPTG. The cells were pelleted, resuspended, and broken using a French pressure cell. The lysate was centrifuged for 30 min at 30,000 × g to spin down unbroken cells. Purification was carried out as described for SecYEG by Braig et al. (2011). The protein was concentrated using a 50-kDa centrifugal filter (Amicon Ultra, Witten, Germany) and gel filtrated using an Äkta chromatography system and a Superdex 200 10/30 global column. The protein was dialyzed in CTF buffer and concentrated to 1.5 mg/ml.
pTrc99a-FtsY was purified as described previously (Braig et al., 2009). pTrc99a-Ffh was purified as described (Braig et al., 2011). Ffh was concentrated on a 10-kDa centrifugal filter (Amicon Ultra) and rebuffered in HT buffer plus 50% glycerol (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.6, 100 mM potassium acetate [KOAc], pH 7.5, 10 mM magnesium acetate [Mg(OAc)2], 1 mM dithiothreitol [DTT]) using a PD-10 column (GE Healthcare, Munich, Germany). The concentration of Ffh was 0.6 mg/ml. The protein was stored at −20°C. FtsY was rebuffered in HT buffer using a PD-10 Column (GE Healthcare). The concentration was adjusted to 1.4 mg/ml.
Preparation of proteoliposomes
E. coli phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). Preparation of DAG and of liposomes containing 5% DAG was performed as described (Nishiyama et al., 2006). Proteoliposomes were prepared by solubilizing 200 μg of 5% DAG liposomes and 14–16 μg of purified SecYEG or YidC in 150 μl of 50 mM triethanolamine acetate (TeaOAc), pH 7.5, 1 mM DTT, and 1.5% octylglycoside. The samples dialyzed with 50 mM TeaOAc, pH 7.5, and 1 mM DTT. The proteoliposomes were pelleted and resuspended in 50 mM TeaOAc, pH 7.5, and 1 mM DTT. The proteoliposomes were stored at −80°C. Before each use, proteoliposomes were briefly sonicated. The generation of energized proteoliposomes followed established protocols (Serek et al., 2004; van der Laan et al., 2004b). In brief, proteoliposomes were resuspended after dialysis in 100 mM Na2SO4 and sonicated. These proteoliposomes were added to the in vitro integration assay containing 100 mM KOAc. A transmembrane potential was established after the addition of 0.4 μM valinomycin.
Preparation of 4.5S RNA
For in vitro synthesis of 4.5S RNA, pT7/T3α19, carrying the 4.5S RNA coding sequence (Wood et al., 1992) was linearized using BamHI. In vitro transcription was performed using the AmpliScribe T7-Flash Transcription kit (Epicentre Biotechnologies, Madison, WE). The 4.5S RNA was purified using the Norgen RNA Clean-Up and Concentration Kit (Norgen Biotec, Thorold, Canada). The final concentration of the RNA was adjusted to 0.7 μg/μl and stored at −80°C.
In vitro INV pBpa cross-linking
For the cross-links with INVs, 1 mg of INVs were diluted to 4 μg/μl in INV buffer (50 mM TeaOAc, pH 7.5, 250 mM sucrose, 1 mM DTT) and incubated for 20 min on ice. INVs were cross-linked on ice by UV irradiation for 20 min. INVs were trichloroacetic acid (TCA) precipitated and visualized by Western blotting or analyzed using mass spectrometry.
Mass spectrometry
SDS–PAGE lanes were cut into horizontal 2-mm slices that were subjected individually to in-gel digestion by trypsin for establishing abundance profiles of identified peptides as described (Kuhn et al., 2011). Peptides were analyzed by nano high-performance liquid chromatography (HPLC)–electrospray tandem mass spectrometry using the UltiMate 3000 HPLC system (Dionex LC Packings, Idstein, Germany) online coupled to an LTQ-Orbitrap XL instrument (Thermo Fisher Scientific, Bremen, Germany) as described previously (Kaller et al., 2011). Proteins were identified by database searches against E. coli protein sequences deposited at the UniProt database (release 2011_02) using the program OMSSA (version 2.1.9; Geer et al., 2004), filtered, and quantified as described in Kuhn et al. (2011). Peptide scores were derived from P values calculated by the OMSSA program as −10 log10(P), and protein scores represent the sum of peptide scores of all different peptides identified.
In vitro transport assays
Assays in the coupled in vitro transcription/translation system have been described in detail (Koch et al., 1999). The plasmid-encoded substrate was transcribed using T7-RNA polymerase and translated in the CTF/ribosome system. INV or proteoliposomes (lipids 0.4 mg/ml, 430 nM SecYEG, 530 nM YidC) and targeting factors (SRP, 150 nM; Ffh, 4.5S RNA, 15 μg/ml; FtsY 750 nM; SecA 600 nM) were added from the beginning of the synthesis. The synthesis was carried out in a buffer with 40 mM TeaOAc, pH 7.5, 70 mM KOAc, and 10 mM Mg(OAc)2. When indicated, CCCP was added at a final concentration of 100 μM. The synthesis/transport was carried out for 30 min at 37°C. After synthesis, the transport assay was split into two parts. One part was directly TCA precipitated, and one part was digested for 25 min at 25°C using 0.5 mg/ml proteinase K, following TCA precipitation and SDS–PAGE. For carbonate extraction, in vitro–synthesized samples were incubated with 0.2 M Na2CO3, pH 11.3, for 30 min at 4°C and subsequently centrifuged for 30 min at 50,000 rpm in a Beckmann TLA 55 rotor. The supernatants were neutralized with glacial acetic acid and precipitated with one volume of 10% TCA, and the TCA pellet was resuspended in SDS loading buffer. Pellets were directly dissolved in loading buffer and, like supernatants, separated by SDS–PAGE.
In vitro proteoliposome cross-linking experiments using RNCs with incorporated pBpa
The synthesis of RNCs has been described (Behrmann et al., 1998; Beck et al., 2000). The synthesis of stalled ribosome-nascent chains with incorporated pBpa requires a purified E. coli extract from a strain with the pSup-pBpaRS-6TRN plasmid. The ribosome-nascent chain of the protein with pBpa was synthesized using DNA of the protein with an amber stop codon (TAG), an antisense oligonucleotide that hybridizes to the mRNA of the protein, thus blocking translation, RNaseH to digest the DNA/RNA hybrid, and an antisense RNA for blocking ssrA (Hanes and Plückthun, 1997) and pBpa (40 μM).
The synthesis of the RNCs was carried out for 30 min at 37°C and 300 rpm in a buffer with 40 mM TeaOAc, 70 mM KOAc, and 12 mM Mg(OAc)2. SRP was added to the synthesis for efficient binding. After synthesis, the RNCs were spun down on a sucrose cushion (580 mM sucrose, 40 mM HEPES, pH 7.5, 70 mM KOAc, 6 mM Mg(OAc)2, 1 mM DTT, 0.1 mM spermidine). RNCs were resuspended in a buffer with the ion strength of the synthesis mix, and proteoliposomes and FtsY were added. The transport was carried out for 30 min at 37°C. The transport assays were split, and one sample was cross-linked on ice by UV irradiation for 20 min (wavelength, 365 nm; VL-6.L; Vilber Lourmat, Torcy, France); the other sample was kept in the dark for the same time. The samples were TCA precipitated or used for immunoprecipitations.
Immunoprecipitations
Immunoprecipitations were carried out as described (Ahrem et al., 1989). Samples from the cross-linking reactions were denatured for 15 min at 37°C using 1% SDS for YidC and 0.5% SDS for Ffh and SecY. After denaturation, the samples were incubated for 2–14 h with antibodies bound to 10 mg of protein A–Sepharose (GE Healthcare) on a wheel at 4°C. The Sepharose was washed three times using 1 ml of detergent buffer (25 mM Tris, pH 6.8, 5 mM EDTA, 150 mM NaCl, 1% Triton-X100) and two times with 1 ml of 25 mM Tris, pH 6.8. To release the protein from the Sepharose, it was incubated for 15 min at 37°C with 35 μl of SDS loading buffer and then loaded on an SDS gel.
Ribosome-binding assays
Ribosome-binding assays were performed as described (Prinz et al., 2000). High-salt-washed ribosomes and purified SecY or YidC were incubated at 4°C for 30 min in HKM100 buffer (50 mM TeaOAc, pH 7.6, 5 mM Mg(OAc)2, 100 mM KOAc, 1 mM DTT, 0.1 mM spermidine, and 0.1% n-dodecyl-β-d-maltoside, with a total reaction volume of 30 μl). Subsequently, the samples were centrifuged for 10 min at 15,000 × g to remove aggregates, and the supernatant was loaded on a 100-μl sucrose cushion (580 mM sucrose in HKM100 buffer) and ultracentrifuged for 20 min at 70,000 rpm in a Beckmann TLA-100.3 rotor. The pellet was directly resuspended in SDS loading buffer. The supernatant was diluted twofold with HKM100 buffer and precipitated with an equal volume of 20% TCA. After 2 h of precipitation on ice, the sample was centrifuged for 15 min at 15,000 × g and the pellet was resuspended in SDS loading dye. After SDS–PAGE, gels were Coomassie blue stained or blotted and decorated with antibodies against ribosomal subunit L23, YidC, or SecY. For quantification, the protein amount in the pellet and supernatant after ultracentrifugation as seen in the immunoblots was determined using ImageJ software (National Institutes of Health, Bethesda, MD).
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (FOR929, FOR 967, and GRK1478), the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS and GSC-4 Spemann Graduate School for Biology and Medicine), the FF-Nord Foundation, and an Innovation Fellowship of the Albert-Ludwigs-University Freiburg. We thank Bettina Knapp for excellent technical assistance.
Abbreviations used:
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- IMP
inner membrane protein
- INV
inverted inner membrane vesicle
- KOAc
potassium acetate
- Mg(OAc)2
magnesium acetate
- MPF
membrane protected fragment
- MtlA
mannitol permease
- pBpa
para-benzoyl-l-phenylalanine
- PK
proteinase K
- pmf
proton-motive force
- RNC
ribosome-associated nascent chain
- SRP
signal recognition particle
- TeaOAc
triethanolamine acetate
- TM
transmembrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0590) on December 7, 2011.
REFERENCES
- Ahrem B, Hoffschulte HK, Muller M. In vitro membrane assembly of a polytopic, transmembrane protein results in an enzymatically active conformation. J Cell Biol. 1989;108:1637–1646. doi: 10.1083/jcb.108.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baars L, Wagner S, Wickstrom D, Klepsch M, Ytterberg AJ, van Wijk KJ, de Gier JW. Effects of SecE depletion on the inner and outer membrane proteomes of Escherichia coli. J Bacteriol. 2008;190:3505–3525. doi: 10.1128/JB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K, Eisner G, Trescher D, Dalbey RE, Brunner J, Muller M. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2001;2:709–714. doi: 10.1093/embo-reports/kve154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K, Wu LF, Brunner J, Muller M. Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 2000;19:134–143. doi: 10.1093/emboj/19.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beha D, Deitermann S, Muller M, Koch HG. Export of beta-lactamase is independent of the signal recognition particle. J Biol Chem. 2003;278:22161–22167. doi: 10.1074/jbc.M300929200. [DOI] [PubMed] [Google Scholar]
- Behrendt J, Standar K, Lindenstrauss U, Bruser T. Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol Lett. 2004;234:303–308. doi: 10.1016/j.femsle.2004.03.048. [DOI] [PubMed] [Google Scholar]
- Behrmann M, Koch HG, Hengelage T, Wieseler B, Hoffschulte HK, Muller M. Requirements for the translocation of elongation-arrested, ribosome-associated OmpA across the plasma membrane of Escherichia coli. J Biol Chem. 1998;273:13898–13904. doi: 10.1074/jbc.273.22.13898. [DOI] [PubMed] [Google Scholar]
- Berrier C, Guilvout I, Bayan N, Park KH, Mesneau A, Chami M, Pugsley AP, Ghazi A. Coupled cell-free synthesis and lipid vesicle insertion of a functional oligomeric channel MscL MscL does not need the insertase YidC for insertion in vitro. Biochim Biophys Acta. 2011;1808:41–46. doi: 10.1016/j.bbamem.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Bibi E. Is there a twist in the Escherichia coli signal recognition particle pathway? Trends Biochem Sci. 2012;37:1–6. doi: 10.1016/j.tibs.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Bornemann T, Jockel J, Rodnina MV, Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- Braig D, Bar C, Thumfart JO, Koch HG. Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J Mol Biol. 2009;390:401–413. doi: 10.1016/j.jmb.2009.04.061. [DOI] [PubMed] [Google Scholar]
- Braig D, Mircheva M, Sachelaru I, van der Sluis EO, Sturm L, Beckmann R, Koch HG. Signal-sequence-independent SRP-SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle. Mol Biol Cell. 2011;22:2309–2323. doi: 10.1091/mbc.E11-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celebi N, Yi L, Facey SJ, Kuhn A, Dalbey RE. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N-terminal and C-terminal domains. J Mol Biol. 2006;357:1428–1436. doi: 10.1016/j.jmb.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson I, Breyton C, Duong F, Tziatzios C, Schubert D, Or E, Rapoport T, Kuhlbrandt W. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J. 2001;20:2462–2471. doi: 10.1093/emboj/20.10.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitermann S, Sprie GS, Koch HG. A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J Biol Chem. 2005;280:39077–39085. doi: 10.1074/jbc.M509647200. [DOI] [PubMed] [Google Scholar]
- Drew D, Froderberg L, Baars L, de Gier JW. Assembly and overexpression of membrane proteins in Escherichia coli. Biochim Biophys Acta. 2003;1610:3–10. doi: 10.1016/s0005-2736(02)00707-1. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- du Plessis DJ, Nouwen N, Driessen AJ. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J Biol Chem. 2006;281:12248–12252. doi: 10.1074/jbc.M600048200. [DOI] [PubMed] [Google Scholar]
- Facey SJ, Neugebauer SA, Krauss S, Kuhn A. The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J Mol Biol. 2007;365:995–1004. doi: 10.1016/j.jmb.2006.10.083. [DOI] [PubMed] [Google Scholar]
- Falk S, Ravaud S, Koch J, Sinning I. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem. 2010;285:5954–5962. doi: 10.1074/jbc.M109.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J, et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröderberg L, Houben E, Samuelson JC, Chen M, Park SK, Phillips GJ, Dalbey R, Luirink J, De Gier JW. Versatility of inner membrane protein biogenesis in Escherichia coli. Mol Microbiol. 2003;47:1015–1027. doi: 10.1046/j.1365-2958.2003.03346.x. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S, Hasona A, Bauerschmitt H, Grubbauer C, Kauff F, Collins R, Crowley PJ, Palmer SR, Brady LJ, Herrmann JM. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci USA. 2009;106:6656–6661. doi: 10.1073/pnas.0809951106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S, Kauff F, van der Sluis EO, Ott M, Herrmann JM. Evolution of YidC/Oxa1/Alb3 insertases: three independent gene duplications followed by functional specialization in bacteria, mitochondria and chloroplasts. Biol Chem. 2011;392:13–19. doi: 10.1515/BC.2011.013. [DOI] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Elmore KB, Tripathy A, Koc H, Koc EC, Spremulli LL. Properties of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L and its interactions with mammalian mitochondrial ribosomes. J Biol Chem. 2010a;285:28353–28362. doi: 10.1074/jbc.M110.148262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Spremulli LL, Fecko CJ. Identification of protein-protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J Biol Chem. 2010b;285:34991–34998. doi: 10.1074/jbc.M110.163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasona A, Crowley PJ, Levesque CM, Mair RW, Cvitkovitch DG, Bleiweis AS, Brady LJ. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc Natl Acad Sci USA. 2005;102:17466–17471. doi: 10.1073/pnas.0508778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzixanthis K, Palmer T, Sargent F. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol. 2003;49:1377–1390. doi: 10.1046/j.1365-2958.2003.03642.x. [DOI] [PubMed] [Google Scholar]
- Houben EN, Urbanus ML, Van Der Laan M, Ten Hagen-Jongman CM, Driessen AJ, Brunner J, Oudega B, Luirink J. YidC and SecY mediate membrane insertion of a type I transmembrane domain. J Biol Chem. 2002;277:35880–35886. doi: 10.1074/jbc.M205556200. [DOI] [PubMed] [Google Scholar]
- Houben EN, ten Hagen-Jongman CM, Brunner J, Oudega B, Luirink J. The two membrane segments of leader peptidase partition one by one into the lipid bilayer via a Sec/YidC interface. EMBO Rep. 2004;5:970–975. doi: 10.1038/sj.embor.7400261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Chen M, Yi L, de Gier JW, Kuhn A, Dalbey RE. Defining the regions of Escherichia coli YidC that contribute to activity. J Biol Chem. 2003;278:48965–48972. doi: 10.1074/jbc.M307362200. [DOI] [PubMed] [Google Scholar]
- Jong WS, ten Hagen-Jongman CM, Ruijter E, Orru RV, Genevaux P, Luirink J. YidC is involved in the biogenesis of the secreted autotransporter hemoglobin protease. J Biol Chem. 2010;285:39682–39690. doi: 10.1074/jbc.M110.167650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Roh S, Hoffmann R, Warscheid B, Hermeking H. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and micro-array analysis. Mol Cell Proteomics. 2011;10:M111.010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenner C, Yuan J, Dalbey RE, Kuhn A. The Pf3 coat protein contacts TM1 and TM3 of YidC during membrane biogenesis. FEBS Lett. 2008;582:3967–3972. doi: 10.1016/j.febslet.2008.10.044. [DOI] [PubMed] [Google Scholar]
- Koch HG, Hengelage T, Neumann-Haefelin C, MacFarlane J, Hoffschulte HK, Schimz KL, Mechler B, Muller M. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol Biol Cell. 1999;10:2163–2173. doi: 10.1091/mbc.10.7.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Moser M, Schimz KL, Muller M. The integration of YidC into the cytoplasmic membrane of Escherichia coli requires the signal recognition particle, SecA and SecYEG. J Biol Chem. 2002;277:5715–5718. doi: 10.1074/jbc.C100683200. [DOI] [PubMed] [Google Scholar]
- Koch HG, Müller M. Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J Cell Biol. 2000;150:689–694. doi: 10.1083/jcb.150.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol Cell. 2009;37:344–353. doi: 10.1016/j.molcel.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Kuhn P, Weiche B, Sturm L, Sommer E, Drepper F, Warscheid B, Sourjik V, Koch HG. The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic. 2011;12:563–578. doi: 10.1111/j.1600-0854.2011.01167.x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lewis NE, Marty NJ, Kathir KM, Rajalingam D, Kight AD, Daily A, Kumar TK, Henry RL, Goforth RL. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem. 2010;285:34220–34230. doi: 10.1074/jbc.M110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JR, Harwood JH, McCaffrey M, Fernandez DE, Cline K. Localization and integration of thylakoid protein translocase subunit cpTatC. Plant J. 2009;58:831–842. doi: 10.1111/j.1365-313X.2009.03816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Goforth RL, Mori H, Henry R. Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: substrate not required. J Cell Biol. 2003;162:1245–1254. doi: 10.1083/jcb.200307067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Blobel G. In vitro translocation of bacterial proteins across the plasma membrane of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7421–7425. doi: 10.1073/pnas.81.23.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamori S, Smirnova IN, Kaback HR. Role of YidC in folding of polytopic membrane proteins. J Cell Biol. 2004;165:53–62. doi: 10.1083/jcb.200402067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin C, Schafer U, Muller M, Koch HG. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 2000;19:6419–6426. doi: 10.1093/emboj/19.23.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama K, Ikegami A, Moser M, Schiltz E, Tokuda H, Muller M. A derivative of lipid A is involved in signal recognition particle/SecYEG-dependent and -independent membrane integrations. J Biol Chem. 2006;281:35667–35676. doi: 10.1074/jbc.M608228200. [DOI] [PubMed] [Google Scholar]
- Pohlschröder M, Hartmann E, Hand NJ, Dilks K, Haddad A. Diversity and evolution of protein translocation. Annu Rev Microbiol. 2005;59:91–111. doi: 10.1146/annurev.micro.59.030804.121353. [DOI] [PubMed] [Google Scholar]
- Pop OI, Soprova Z, Koningstein G, Scheffers DJ, van Ulsen P, Wickstrom D, de Gier JW, Luirink J. YidC is required for the assembly of the MscL homopentameric pore. FEBS J. 2009;276:4891–4899. doi: 10.1111/j.1742-4658.2009.07188.x. [DOI] [PubMed] [Google Scholar]
- Preuss M, Ott M, Funes S, Luirink J, Herrmann JM. Evolution of mitochondrial oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J Biol Chem. 2005;280:13004–13011. doi: 10.1074/jbc.M414093200. [DOI] [PubMed] [Google Scholar]
- Price CE, Driessen AJ. Conserved negative charges in the transmembrane segments of subunit K of the NADH:ubiquinone oxidoreductase determine its dependence on YidC for membrane insertion. J Biol Chem. 2010;285:3575–3581. doi: 10.1074/jbc.M109.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CE, Otto A, Fusetti F, Becher D, Hecker M, Driessen AJ. Differential effect of YidC depletion on the membrane proteome of Escherichia coli under aerobic and anaerobic growth conditions. Proteomics. 2010;10:3235–3247. doi: 10.1002/pmic.201000284. [DOI] [PubMed] [Google Scholar]
- Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 2000;19:1900–1906. doi: 10.1093/emboj/19.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Ravaud S, Stjepanovic G, Wild K, Sinning I. The crystal structure of the periplasmic domain of the Escherichia coli membrane protein insertase YidC contains a substrate binding cleft. J Biol Chem. 2008;283:9350–9358. doi: 10.1074/jbc.M710493200. [DOI] [PubMed] [Google Scholar]
- Richter CV, Bals T, Schunemann D. Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur J Cell Biol. 2010;89:965–973. doi: 10.1016/j.ejcb.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- Sääf A, Monne M, de Gier JW, von Heijne G. Membrane topology of the 60-kDa Oxa1p homologue from Escherichia coli. J Biol Chem. 1998;273:30415–30418. doi: 10.1074/jbc.273.46.30415. [DOI] [PubMed] [Google Scholar]
- Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serek J, Bauer-Manz G, Struhalla G, van den Berg L, Kiefer D, Dalbey R, Kuhn A. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 2004;23:294–301. doi: 10.1038/sj.emboj.7600063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller MP, Stirling CJ. Preferential targeting of a signal recognition particle-dependent precursor to the Ssh1p translocon in yeast. J Biol Chem. 2011;286:21953–21960. doi: 10.1074/jbc.M111.219568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler N, Dalbey RE, Kuhn A. M13 procoat protein insertion into YidC and SecYEG proteoliposomes and liposomes. J Mol Biol. 2011;406:362–370. doi: 10.1016/j.jmb.2010.12.036. [DOI] [PubMed] [Google Scholar]
- Sugiyama J, Mahmoodian S, Jacobson G. Escherichia coli mannitol permease-alkaline phosphatase fusions constructed by a nested-deletion method—prediction of 6 transmembrane regions in the permease protein. FASEB J. 1991;5:A1191–A1191. [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura T, Yoshihisa T, Ito K. Protein translocation functions of Escherichia coli SecY: in vitro characterization of cold-sensitive secY mutants. Biochimie. 1997;79:517–521. doi: 10.1016/s0300-9084(97)82744-7. [DOI] [PubMed] [Google Scholar]
- Urbanus ML, Scotti PA, Froderberg L, Saaf A, de Gier JW, Brunner J, Samuelson JC, Dalbey RE, Oudega B, Luirink J. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2001;2:524–529. doi: 10.1093/embo-reports/kve108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus ML, Froderberg L, Drew D, Bjork P, de Gier JW, Brunner J, Oudega B, Luirink J. Targeting, insertion, and localization of Escherichia coli YidC. J Biol Chem. 2002;277:12718–12723. doi: 10.1074/jbc.M200311200. [DOI] [PubMed] [Google Scholar]
- van Bloois E, Dekker HL, Froderberg L, Houben EN, Urbanus ML, de Koster CG, de Gier JW, Luirink J. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 2008;582:1419–1424. doi: 10.1016/j.febslet.2008.02.082. [DOI] [PubMed] [Google Scholar]
- van Bloois E, Jan Haan G, de Gier JW, Oudega B, Luirink J. F(1)F(0) ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett. 2004;576:97–100. doi: 10.1016/j.febslet.2004.08.069. [DOI] [PubMed] [Google Scholar]
- van Bloois E, Nagamori S, Koningstein G, Ullers RS, Preuss M, Oudega B, Harms N, Kaback HR, Herrmann JM, Luirink J. The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J Biol Chem. 2005;280:12996–13003. doi: 10.1074/jbc.M414094200. [DOI] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJ. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J Cell Biol. 2004a;165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Houben EN, Nouwen N, Luirink J, Driessen AJ. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep. 2001;2:519–523. doi: 10.1093/embo-reports/kve106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Nouwen N, Driessen AJ. SecYEG proteoliposomes catalyze the Deltaphi-dependent membrane insertion of FtsQ. J Biol Chem. 2004b;279:1659–1664. doi: 10.1074/jbc.M306527200. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Urbanus ML, Ten Hagen-Jongman CM, Nouwen N, Oudega B, Harms N, Driessen AJ, Luirink J. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc Natl Acad Sci USA. 2003;100:5801–5806. doi: 10.1073/pnas.0636761100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Pop OI, Haan GJ, Baars L, Koningstein G, Klepsch MM, Genevaux P, Luirink J, de Gier JW. Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J Biol Chem. 2008;283:17881–17890. doi: 10.1074/jbc.M801481200. [DOI] [PubMed] [Google Scholar]
- Wang P, Kuhn A, Dalbey RE. Global change of gene expression and cell physiology in YidC-depleted Escherichia coli. J Bacteriol. 2010;192:2193–2209. doi: 10.1128/JB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiche B, Burk J, Angelini S, Schiltz E, Thumfart JO, Koch HG. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J Mol Biol. 2008;377:761–773. doi: 10.1016/j.jmb.2008.01.040. [DOI] [PubMed] [Google Scholar]
- Werner PK, Saier MH, Jr, Muller M. Membrane insertion of the mannitol permease of Escherichia coli occurs under conditions of impaired SecA function. J Biol Chem. 1992;267:24523–2453. [PubMed] [Google Scholar]
- Wickström D, Wagner S, Simonsson P, Pop O, Baars L, Ytterberg AJ, van Wijk KJ, Luirink J, de Gier JW. Characterization of the consequences of YidC depletion on the inner membrane proteome of E. coli using 2D blue native/SDS–PAGE. J Mol Biol. 2011;409:124–135. doi: 10.1016/j.jmb.2011.03.068. [DOI] [PubMed] [Google Scholar]
- Wood H, Luirink J, Tollervey D. Evolutionary conserved nucleotides within the E. coli 4.5S RNA are required for association with P48 in vitro and for optimal function in vivo. Nucleic Acids Res. 1992;20:5919–5925. doi: 10.1093/nar/20.22.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol. 2008;6:234–244. doi: 10.1038/nrmicro3595. [DOI] [PubMed] [Google Scholar]
- Yi L, Celebi N, Chen M, Dalbey RE. Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J Biol Chem. 2004;279:39260–39267. doi: 10.1074/jbc.M405490200. [DOI] [PubMed] [Google Scholar]
- Yi L, Jiang F, Chen M, Cain B, Bolhuis A, Dalbey RE. YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry. 2003;42:10537–10544. doi: 10.1021/bi034309h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.