Abstract
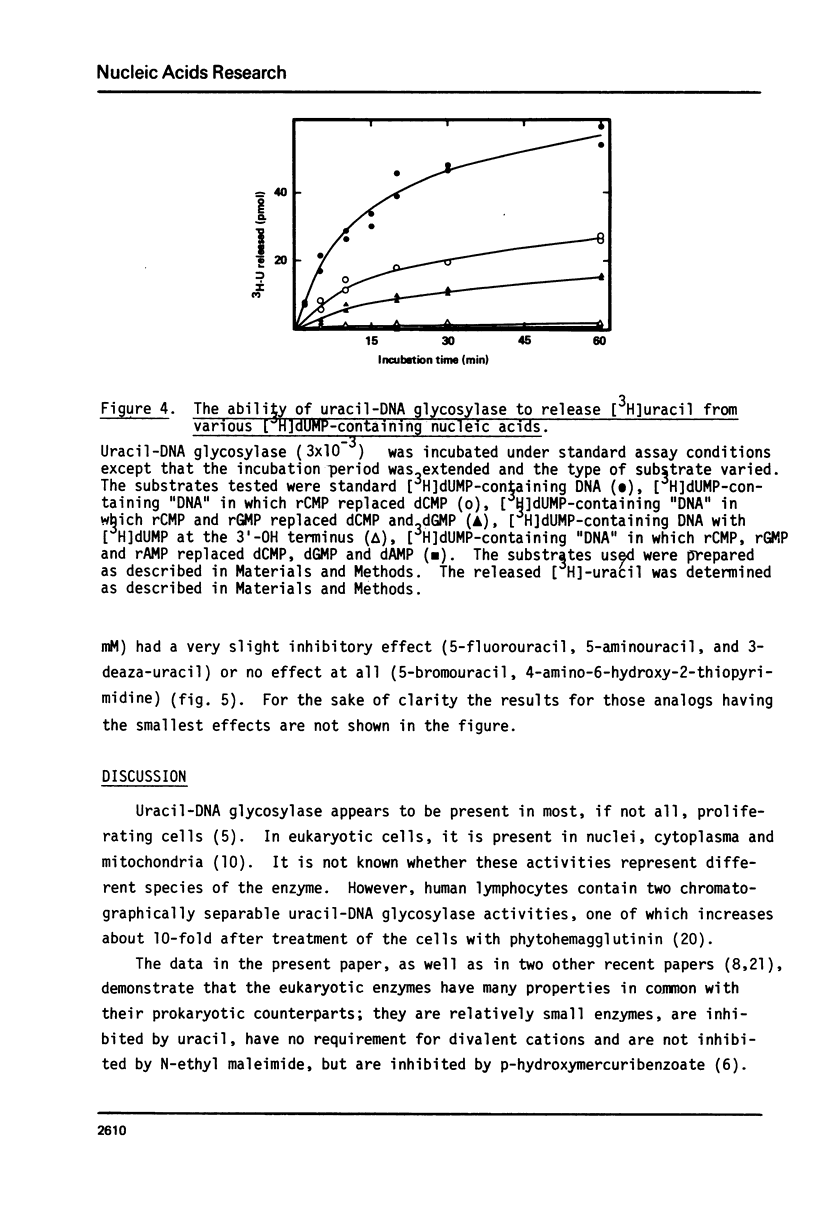
Uracil-DNA glycosylase was partially purified from HeLa cells. Various substrates containing [3H]dUMP residues were prepared by nick-translation of calf thymus DNA. The standard substrate was double-stranded DNA with [3H]dUMP located internally in the chain. Compared to the release of uracil from this substrate, a 3-fold increase in the rate was seen with single-stranded DNA, and a 20-fold reduction in the rate was observed when the [3H]dUMP-residue was located at the 3'end. The rate of [3H]uracil release decreased progressively when one, two or three of the dNMP residues were replaced by the corresponding rNMP; in the extreme case when the substrate contained [3H]dUMP in addition to rCMP, rGMP, and rAMP, the rate of [3H]uracil release was less than 3% of that of the control. The enzyme was inhibited to the same extent by uracil and the uracil analogs 6-aminouracil and 5-azauracil, but very weakly, or not at all, by 5 other analogs. Our results suggest strongly that uracil-DNA glycosylase has a high degree of selectivity for uracil in dUMP residues located internally in DNA chains and that the recognition of the correct substrate also depends on the residues flanking dUMP being deoxyribonucleotides.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. T., Friedberg E. C. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic Acids Res. 1980 Feb 25;8(4):875–888. [PMC free article] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynolf K., Eliasson R., Reichard P. Formation of Okazaki fragments in polyoma DNA synthesis caused by misincorporation of uracil. Cell. 1978 Mar;13(3):573–580. doi: 10.1016/0092-8674(78)90330-6. [DOI] [PubMed] [Google Scholar]
- Caradonna S. J., Cheng Y. C. Uracil DNA-glycosylase. Purification and properties of this enzyme isolated from blast cells of acute myelocytic leukemia patients. J Biol Chem. 1980 Mar 25;255(6):2293–2300. [PubMed] [Google Scholar]
- Cone R., Duncan J., Hamilton L., Friedberg E. C. Partial purification and characterization of a uracil DNA N-glycosidase from Bacillus subtilis. Biochemistry. 1977 Jul 12;16(14):3194–3201. doi: 10.1021/bi00633a024. [DOI] [PubMed] [Google Scholar]
- Englund P. T., Price S. S., Weigel P. H. The use of the T4 DNA polymerase in identification of 3' terminal nucleotide sequences of duplex DNA. Methods Enzymol. 1974;29:273–281. doi: 10.1016/0076-6879(74)29027-x. [DOI] [PubMed] [Google Scholar]
- Krokan H., Bjorklid E., Prydz H. DNA synthesis in isolated HeLa cell nuclei. Optimalization of the system and characterization of the product. Biochemistry. 1975 Sep 23;14(19):4227–4232. doi: 10.1021/bi00690a012. [DOI] [PubMed] [Google Scholar]
- Krokan H., Schaffer P., DePamphilis M. L. Involvement of eucaryotic deoxyribonucleic acid polymerases alpha and gamma in the replication of cellular and viral deoxyribonucleic acid. Biochemistry. 1979 Oct 2;18(20):4431–4443. doi: 10.1021/bi00587a025. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Little C., Aurebekk B., Otnaess A. B. Purification by affinity chromatography of phospholipase C from Bacillus cereus. FEBS Lett. 1975 Apr 1;52(2):175–179. doi: 10.1016/0014-5793(75)80800-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973 Dec;56(2):502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- Sirover M. A. Induction of the DNA repair enzyme uracil-DNA glycosylase in stimulated human lymphocytes. Cancer Res. 1979 Jun;39(6 Pt 1):2090–2095. [PubMed] [Google Scholar]
- Talpaert-Borlé M., Clerici L., Campagnari F. Isolation and characterization of a uracil-DNA glycosylase from calf thymus. J Biol Chem. 1979 Jul 25;254(14):6387–6391. [PubMed] [Google Scholar]
- Tye B. K., Nyman P. O., Lehman I. R., Hochhauser S., Weiss B. Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc Natl Acad Sci U S A. 1977 Jan;74(1):154–157. doi: 10.1073/pnas.74.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Rockstroh P. A. Incorporation and excision of 5-fluorouracil from deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1980 Feb;141(2):680–686. doi: 10.1128/jb.141.2.680-686.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wist E., Unhjem O., Krokan H. Accumulation of small fragments of DNA in isolated HeLa cell nuclei due to transient incorporation of dUMP. Biochim Biophys Acta. 1978 Sep 27;520(2):253–270. doi: 10.1016/0005-2787(78)90225-3. [DOI] [PubMed] [Google Scholar]


