Abstract
Background
Group A Streptococcus (GAS) is a human-specific pathogen responsible for a number of diseases characterized by a wide range of clinical manifestations. During host colonization GAS-cell aggregates or microcolonies are observed in tissues. GAS biofilm, which is an in vitro equivalent of tissue microcolony, has only recently been studied and little is known about the specific surface determinants that aid biofilm formation. In this study, we demonstrate that surface-associated streptococcal collagen-like protein-1 (Scl1) plays an important role in GAS biofilm formation.
Results
Biofilm formation by M1-, M3-, M28-, and M41-type GAS strains, representing an intraspecies breadth, were analyzed spectrophotometrically following crystal violet staining, and characterized using confocal and field emission scanning electron microscopy. The M41-type strain formed the most robust biofilm under static conditions, followed by M28- and M1-type strains, while the M3-type strains analyzed here did not form biofilm under the same experimental conditions. Differences in architecture and cell-surface morphology were observed in biofilms formed by the M1- and M41-wild-type strains, accompanied by varying amounts of deposited extracellular matrix and differences in cell-to-cell junctions within each biofilm. Importantly, all Scl1-negative mutants examined showed significantly decreased ability to form biofilm in vitro. Furthermore, the Scl1 protein expressed on the surface of a heterologous host, Lactococcus lactis, was sufficient to induce biofilm formation by this organism.
Conclusions
Overall, this work (i) identifies variations in biofilm formation capacity among pathogenically different GAS strains, (ii) identifies GAS surface properties that may aid in biofilm stability and, (iii) establishes that the Scl1 surface protein is an important determinant of GAS biofilm, which is sufficient to enable biofilm formation in the heterologous host Lactococcus. In summary, the GAS surface adhesin Scl1 may have an important role in biofilm-associated pathogenicity.
Background
Microbial biofilm formation is an important virulence mechanism, which allows immune evasion and survival against antibiotic treatments [1,2]. Many bacterial nosocomial infections are associated with biofilms formed on contaminated medical devices. Dispersal of biofilm has also been proposed to augment infection spread [3-8]. For group A Streptococcus (GAS), biofilm research is an emerging field and little is known about the specific surface determinants that aid in biofilm formation. GAS is characteristically associated with significant human morbidity and it is responsible for the clinically common superficial throat and skin infections, such as pharyngitis and impetigo, as well as invasive soft tissue and blood infections like necrotizing fasciitis and toxic shock syndrome [9]. Although GAS biofilm has not been associated with implanted medical devices, tissue microcolonies of GAS encased in an extracellular matrix were demonstrated in human clinical specimens [10]. Studies reported to date support the involvement of GAS surface components in biofilm formation, including the M and M-like proteins, hyaluronic acid capsule, pili and lipoteichoic acid [11-13]. As shown by Cho and Caparon [11], multiple genes are upregulated during biofilm formation and development, including the streptococcal collagen-like protein-1 (Scl1).
The scl1 gene encoding the Scl1 protein has been found in every GAS strain investigated and its transcription is positively regulated by Mga [14-18], indicating that Scl1 is co-expressed with a number of proven virulence factors. Structurally, the extracellular portion of Scl1 protein extends from the GAS surface as a homotrimeric molecule composed of distinct domains that include the most outward N-terminal variable (V) region and the adjacent collagen-like (CL) region composed of repeating GlyXaaYaa (GXY) sequence. The linker (L) region is close to the cell surface and contains a series of conserved direct repeats. The Scl1 protein can bind selected human extracellular matrix components [19] and cellular integrin receptors [20-22], as well as plasma components [23-27].
In this study, we investigated the importance of Scl1 in GAS biofilm using defined isogenic wild-type and scl1-inactivated mutant strains of GAS. We report that (i) the pathogenically diverse M41-, M28-, M3- and M1-type GAS wild-type strains have varying capacities to produce biofilm on an abiotic surface; (ii) Scl1 plays an important role during the main stages of biofilm formation with Scl1-negative mutants having an abrogated capacity for adhesion, microcolony formation and biofilm maturation; and (iii) variations in surface morphology as well as in extracellular matrix associated with bacterial cells suggest two distinct but plausible mechanisms that potentially stabilize bacterial microcolonies. We additionally show that expression of Scl1 in Lactococcus lactis is sufficient to support a biofilm phenotype. Overall, this work reveals a significant role for the Scl1 protein as a cell-surface component during GAS biofilm formation among pathogenically varying strains.
Results
Wild-type GAS strains have heterogeneous capacity for biofilm formation on abiotic surfaces
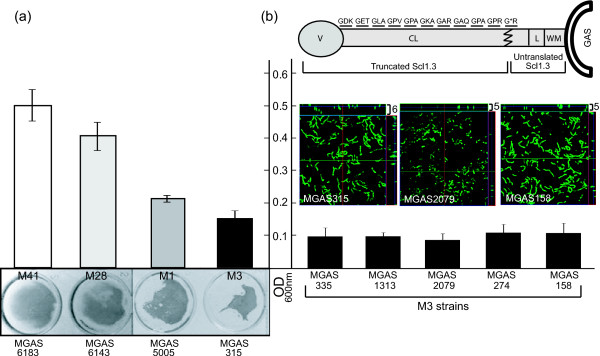
Biofilm formation was compared between M41-, M28-, M3- and M1-type GAS strains representing distinct epidemiological traits (Figure 1). To assess biofilm formation after 24 h, we used spectrophotometric measurements recorded following crystal violet staining (Figure 1a). Both the M41- and M28-type strains produced more biomass as compared with M1 strain. Furthermore, the M3-type strain produced the lowest absorbance values in a crystal violet assay, indicative of lower cell biomass, as compared with the other wild-type strains. These experiments confirm previous observations [1,28] that GAS strains have varying capacity to form biofilm in vitro.
Figure 1.
Variation in biofilm formation among GAS strains. (a) Wild type M41-, M28-, M3-, and M1-type GAS strains were grown 24 h under static conditions and analyzed spectrophotometrically following crystal violet staining (top). Visual representation of corresponding wells is shown below. (b) Schematic representation (not to scale) of Scl1.3 protein of M3-type GAS. Translated GXY repeats within the collagen-like (CL) region are shown with an asterisk representing the location of the premature stop codon resulting in a truncated protein. V, variable region; L, linker region; WM, wall-membrane associated region. Below, spectrophotometric measurements of 24-h biofilms following crystal violet staining are graphed for M3-type GAS strains. Absorbance values (OD600) are averages of at least three experiments done in triplicate wells. Corresponding confocal analyses of 24-h biofilms of MGAS315, MGAS2079, and MGAS158 are shown. Images are X-Y orthogonal Z-stack views and average vertical thickness is indicated in micrometers (top right).
The failure of M3-type strain MGAS315 to produce substantial cellular biomass in the above assay was intriguing because sequence analysis of the scl1.3 allele found in MGAS315 revealed the presence of a TAA stop codon in the 11th GXY repeat of the Scl1.3-CL region containing a total of 25 GXY triplets [29]. This premature stop codon results in a truncated Scl1.3 variant composed of 102 amino acids (~11.4 kDa), which does not contain the cell wall-membrane (WM) associated region, thus, preventing it from anchoring to the bacterial cell surface (Figure 1b). This prompted us to investigate the biofilm formation by five additional M3-type strains, all harboring the same scl1.3 allele. Five additional M3-type strains, MGAS335, MGAS1313, MGAS2079, MGAS274 and MGAS158, all harboring the same scl1.3 allele [29] also produced poor biofilm under static conditions, as measured by crystal violet staining. Confocal laser scanning microscopy (CLSM) of three representative strains (MGAS315, MGAS2079, and MGAS158) corroborated results obtained from the crystal violet assay, indicating that these M3-type strains lack the ability to form appreciable biofilm structure. Our data suggest that the lack of capacity for biofilm-formation among M3-type GAS strains examined here might be correlated, at least in part, with lack of surface-attached Scl1.3 protein.
Microscopic evaluation reveals differences in biofilm surface morphology
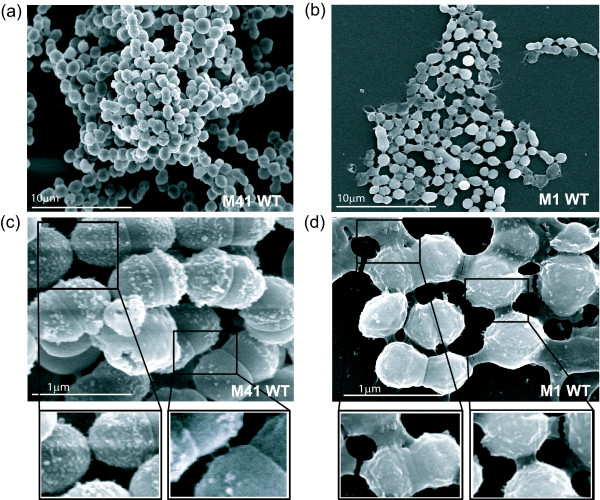
We next conducted microscopic analysis of the biofilms formed by the wild-type (WT) M41-, M28-, and M1-type GAS strains. First, we examined the overall structural characteristics of biofilms formed after 24 h using CLSM (Figure 2d-f; Additional file 1: Figure S1a-f). The average biofilm thickness (see Methods section) differed among all three strains with M1 producing considerably thinner biofilm (mean value of 9 μm) compared to M28 (12 μm) and M41 (15 μm), a result consistent with lower spectrophotometric absorbance values (Figure 1a). In addition to measured differences in biofilm thickness, closer examination of the X-Y orthogonal Z-stack views, representing biofilm cross-sections, revealed architectural differences among the M41, M28, and M1 biofilms. The M1 biofilm, although the thinnest, seems to consist of densely-packed cells that form continuous layers, while the M28 and especially M41 biofilms seem to be less dense but exhibit more elevated supracellular assembly. We therefore used field emission scanning electron microscopy (FESEM) to define more accurately these supracellular differences observed by CLSM between the biofilms produced by the WT M1 and M41 GAS (Figure 3). FESEM exposed notable architectural differences between biofilms formed by these two strains. The M41 (Figure 3, panel a) biofilm was characterized by more diverse surface architecture with the evidence of depressions or crypts, whereas the M1 biofilm (panel b) seems to lack such pronounced surface characteristics. At higher magnification, the M41 cells have a studded cell surface morphology with protrusions linking both sister cells and cells in adjacent chains (panel c). In contrast, the M1 cells had a relatively smoother appearance likely due to the rich bacterial-associated extracellular matrix (BAEM) surrounding these cells and covering their surface (panel d). BAEM material, which was clearly seen at higher resolution between the M1-type cells, was not as evident between cells of the M41-type GAS.
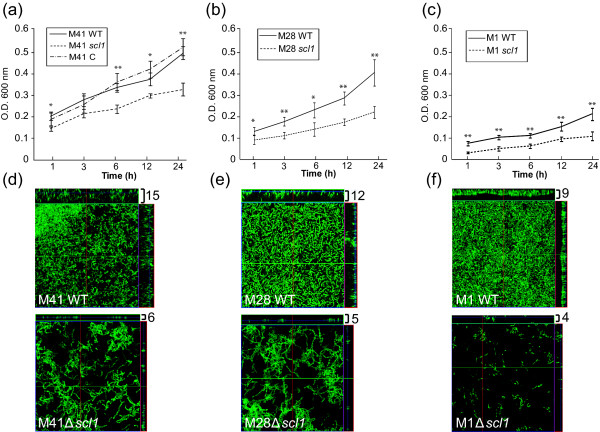
Figure 2.
Biofilm formation by wild type and scl1-inactivated isogenic mutants. Crystal violet staining and confocal laser scanning microscopy (CLSM) of the GFP-expressing GAS were used to compare biofilm formation by GAS strains. Wild type (WT) M41-, M28-, and M1-type strains, scl1-inactivated mutants (scl1), and M41 mutant complemented for Scl1.41 expression (M41 C) were used. (a-c) Isogenic GAS strains were grown under static conditions for 24 h and bacterial biomass was detected spectrophotometrically at indicated time points following crystal violet staining. Absorbance values at OD600 are representative of at least three experiments performed in quadruplicate. Statistical significance is denoted as *P ≤ 0.05 and **P ≤ 0.001. (d-f) CLSM analysis of corresponding 24 h biofilms from same experiment. Images are X-Y orthogonal Z-stack views of WT (top) and mutant (bottom) GAS strains. Views are representative of ten images within a single experiment. Average vertical biofilm thickness is indicated in micrometers (top right).
Figure 3.
Field emission scanning electron microscopy of GAS biofilms. 24-h biofilms of the M1- and M41-type GAS strains were grown on glass cover slips and analyzed by FESEM. (a-b) Architecture of GAS microcolonies shown at low magnification. (c-d) Cell surface morphology and cell-to-cell junctions observed at higher magnification. Enlargements of cell-to-cell junctions are shown below.
GAS biofilms differ in production of bacterial-associated extracellular matrix
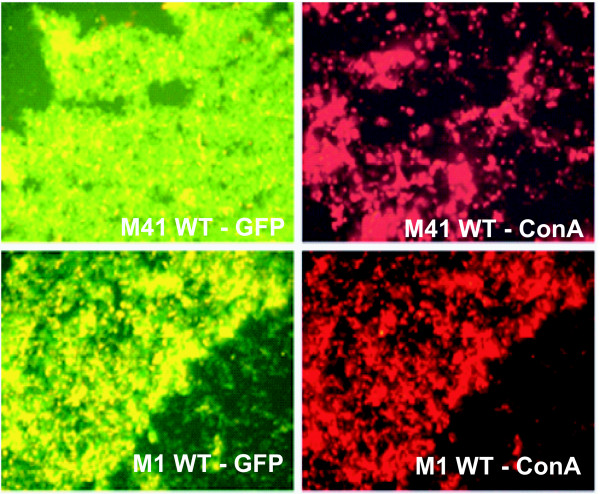
The production of BAEM has been shown to be an integral component in the structural integrity of a biofilm, imparting protection from dehydration, host immune attack, and antibiotic sensitivity [30,31]. GAS cells encased in a glycocalyx were first identified by Akiyama et al. in skin biopsies obtained from impetigo patients. We therefore compared the production of BAEM within biofilms employing GFP-expressing GAS strains of the M1 and M41 type (Figure 4). Cells were grown to form biofilms on glass cover slips for 24 h and stained with TRITC-concanavalin A (ConA), a fluorescently-labeled lectin that binds to the extracellular polysaccharides in biofilms [32]. Fluorescent microscopy was performed to compare matrix production (red staining) by GAS strains (green). Visual screening of both biofilms indicated that the M41-type strain formed a more dispersed extracellular matrix as compared to the M1 strain, which had a dense, more closely associated matrix. In addition, averages of at least 10 fields of ConA stained matrix by CLSM support our FESEM observations that more BAEM is deposited within the biofilm by the M1 GAS cells as compared to M41 GAS. This is in agreement with the report from Akiyama et al that showed a substantial FITC-ConA stained matrix associated with T1-type GAS microcolonies in vivo and in vitro [10].
Figure 4.
Production of bacterial-associated extracellular matrix. GFP-expressing wild type (WT) M41- and M1-type GAS strains were grown on glass cover slips for 24 h and stained with TRITC-conjugated concanavalin A (ConA). Confocal laser scanning microscopic (CLSM) images were separated to represent green GFP-expressing GAS cells (left images) and red ConA-TRITC staining (right images) for detection of extracellular matrix associated with each strain. Images are from one representative experiment.
Scl1 protein significantly contributes to biofilm formation by GAS
Variations in GAS pathogenicity and capacity to form biofilm are driven by specific proteins and components present on the cell surface or are secreted by the organism. It has been shown that deletion of the M and M-like surface proteins or capsule, as well as increased expression of the secreted SpeB protease decreases biofilm formation dramatically for some strains of GAS [12,33,34]. Therefore, we investigated the role of Scl1 in biofilm formation by comparing biofilms formed by GAS WT and scl1-inactivated (Δscl1) mutant strains (Figure 2; Additional file 1: Figure S1a-f). Bacterial biomass was evaluated spectrophotometrically following crystal violet staining at 1, 6, 12, and 24 h time points, representing different stages of biofilm formation, and absorbance values rendered for the WT and Δscl1 isogenic mutant strains were compared. The M41Δscl1 mutant showed a 29-35% decrease in biofilm formation (the OD600 value obtained for the WT strain at each time point was considered 100%), which was sustained throughout all time points. This reduction was statistically significant at initial adherence (1 h), as well as during biofilm development (6-12 h) and at maturation (24 h) (Figure 2a; P ≤ 0.05 at 1 and 12 h, P ≤ 0.001 at 6 and 24 h). Complementation of Scl1.41 expression in the M41Δscl1 mutant (M41 C) restored its ability to form biofilm to WT levels. Similarly, the M28Δscl1 mutant had a significantly decreased capacity for biofilm formation in the range of 29-44% as compared to WT strain (Figure 2b; P ≤ 0.05 at 1 and 6 h, P ≤ 0.001 at 3, 12 and 24 h). Likewise, there was a statistically significant decrease in M1Δscl1 biofilm biomass by ~42-75% compared to the WT strain (Figure 2c; P ≤ 0.001 at 1-24 h). CLSM analysis of corresponding 24-h biofilms of these strains confirmed our crystal violet staining results at 24 h. The Δscl1 mutants had substantially decreased average biofilm thickness by more than 50% (mean values) as compared to the parental WT organisms (Figure 2d-f). While these low average biofilm thickness values measured for the M1Δscl41 (6 μM) and M28Δscl1 (5 μM) correspond to residual biofilms made by those mutants (Additional file 1: Figure S1a-d), by comparison, the M1Δscl1 (4 μM) was shown not to produce a continuous biofilm layer under these conditions (Additional file 1: Figure S1e-f). Our data support the hypothesis that the Scl1 protein plays an important functional role during GAS biofilm formation and that Scl1 contribution varies among GAS strains with different genetic backgrounds.
Scl1 expression affects surface hydrophobicity
The surface hydrophobicity of GAS has been shown to influence the adherence to abiotic surfaces. The presence of pili [13], M and M-like proteins, and lipoteichoic acid contributes to cell surface hydrophobic properties [12,35], which in turn may influence biofilm formation by GAS. Here, we have investigated the contribution of Scl1 to surface hydrophobicity of M41-, M28-, and M1-type GAS strains using a modified hexadecane binding assay [12,36,37]. As shown in Table 1, the M28-type GAS strain MGAS6143 gave the highest actual hydrophobicity value of 94.3 ± 0.73, followed by the M41-type strain MGAS6183 (92.6 ± 0.86). In contrast, the overall surface hydrophobicity of the M1-type GAS strain MGAS5005 (80.3 ± 0.89) was significantly lower compared to both M28 and M41 strains (P ≤ 0.001 for each comparison). Inactivation of scl1.41 in M41-type GAS resulted in a modest, although statistically significant, reduction in the hydrophobicity index (100% for WT vs. 92% for mutant, P ≤ 0.001). In-trans complementation of the Scl1.41 expression in M41Δscl1-C restored the hydrophobic phenotype of the cells to WT level (hydrophobicity index ~105%). In comparison, the contribution of the Scl1.1 and Scl1.28 proteins to surface hydrophobicity is more substantial, as evidenced by a ~21% and ~22% reduction of the hydrophobicity indices of the mutants as compared to the corresponding WT strains, respectively (P ≤ 0.001 for both). Thus, the Scl1-mediated GAS-cell surface hydrophobicity reported here may contribute to the ability of this organism to form biofilm, as suggested for other cell surface components [12,35].
Table 1.
Cell surface hydrophobicity of GAS strains
| GAS Strain | M-Type | Actual Value† | Hydrophobicity Index‡ |
|---|---|---|---|
| MGAS6183 WT | M41 | 92.6 ± .86 | 100 |
| MGAS6183 Δscl1 | M41 | 85.2 ± 2.2 | **92 |
| MGAS6183 Δscl1-C | M41 | 98.0 ± .31 | 105 |
| MGAS5005 WT | M1 | 80.3 ± .89 | 100 |
| MGAS5005 Δscl1 | M1 | 63.3 ± 3.2 | **79 |
| MGAS6143 WT | M28 | 94.3 ± .73 | 100 |
| MGAS6143 Δscl1 | M28 | 72.6 ± .62 | **78 |
† Actual hydrophobicity values were calculated based on hexadecane binding as described in Methods. Values are representative of three separate experiments with ten replicates ± SD
‡ Hydrophobicity Index represents the ration of actual hydrophobicity value for each strain to that of the isogenic wild-type (WT) strain multiplied by 100
** Asterisks denote a statistically significant difference of Δscl1 mutants versus WTs at P ≤ 0.001
Scl1 is sufficient to support biofilm formation in Lactococcus lactis
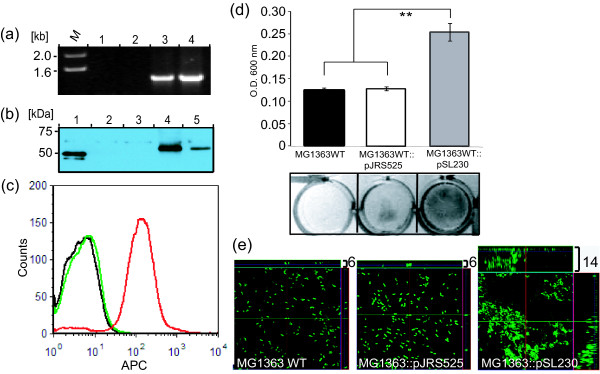
To assess whether Scl1 expression is sufficient to confer the ability for biofilm formation, we chose to express this protein in a heterologous L. lactis system [38,39]. The wild-type L. lactis strain MG1363 was transformed with plasmid pSL230 encoding the Scl1.41 protein [22] or with the shuttle vector pJRS525 alone. As shown in Figure 5a, PCR amplification of the scl1.41 gene employing specific primers yielded no product from the WT L. lactis MG1363 (lane 1) and the MG1363::pJRS525 transformant (lane 2). A product of the expected size of 1.4 kb was amplified from the pSL230 plasmid DNA control (lane 4,) as well as was amplified from the MG1363::pSL230 transformant (lane 3). Surface expression of Scl1.41 was confirmed by immunoblot analysis of cell-wall extracts prepared from L. lactis WT, and the MG1363::pJRS525 and MG1363::pSL230 transformants, as well as MGAS6163 (WT M41 GAS). As shown in Figure 5b, rabbit antiserum raised against purified recombinant Scl1.41 protein P176 lacking the WM region detected the corresponding immunogen (lane 1), and the homologous full length protein in cell-wall extracts of MGAS6183 (lane 5) as well as MG1363::pSL230 L. lactis transformant (lane 4). This band was absent in cell-wall extracts prepared from the WT L. lactis MG1363 (lane 2) and MG1363::pJRS525 transformant (lane 3). Expression of Scl1.41 at the cell surface was further established by flow cytometry. Rabbit anti-p176 antibodies stained Scl1.41 MG1363::pSL230 transformant, confirming the expression of Scl1.41 protein at the cell surface in the heterologous host L. lactis (Figure 5c, red trace). This protein was absent at the surface of WT MG1363 (black trace) and MG1363::pJRS525 transformant (green trace).
Figure 5.
Scl1 expression in L. lactis promotes biofilm formation. L. lactis was transformed with the plasmid construct pSL230 to express Scl1.41 surface protein or with pJRS525 vector. (a) PCR analysis of L. lactis transformants using scl1.41-gene-specific primers; lanes: (1) MG1363 wild-type (WT) cells; (2) MG1363::pJRS525 vector-only control; (3) MG1363::pSL230 transformant; (4) control pSL230 plasmid DNA. (b) Scl1.41 expression by western blot analysis of cell-wall extracts prepared from transformed L. lactis and control GAS strains using anti- P176 (rScl1.41) antibodies; lanes: (1) purified recombinant P176 protein (truncated Scl1.41); (2) MG1363 WT strain; (3) MG1363::pJRS525 vector; (4) MG1363::pSL230 transformant; (5) MGAS6183 (M41) control. (c) Analysis of Sc1.41 expression by flow cytometry with anti-P176 (rScl1.41) rabbit polyclonal antibodies on the surface of MGAS1363 WT strain (black trace), MGAS1363::pJRS525 vector-only control (green trace) and MG1363:pSL230 transformant (red trace). (d) Crystal violet staining of 24 h biofilms formed by L. lactis WT strain, MG1363::pJRS525 vector-only control or MG1363::pSL230 transformant (top) with visual representation of the corresponding wells (bottom). Statistical significance is denoted as **P ≤ 0.001. (e) CLSM analysis of 24 h biofilms from same experiment shown in (d). Images are X-Y orthogonal Z-stack views representative of ten images within a single experiment. Average vertical biofilm thickness is indicated in micrometers (top right).
The capacity of L. lactis expressing Scl1.41 to form biofilm was evaluated spectrophotometrically following crystal violet staining. As shown in Figure 5d, the MG1363::pSL230 transformant demonstrated a significant increase in biofilm-associated biomass at 24 h, as compared to wild type L. lactis or L. lactis-containing pJRS525 vector (P ≤ 0.001). Crystal violet stained wells were photographed for visual representation of biofilm formation prior to spectrophotometric assay. Biofilm thickness and architecture were evaluated by CLSM (Figure 5e; Additional file 1: Figure S2a-c). The MG1363::pSL230 transformant produced a substantially thicker biofilm (14 μm) as compared to both MG1363 WT (6 μm) and the vector-only transformant MG1363::pJRS525 (6 μm). The MG1363::pSL230 cells formed highly aggregated structures, thus, acquiring a phenotype consistent with biofilm formation. As shown in Table 2, the MG1363::pSL230 transformant, expressing Scl1.41 surface protein, had significantly enhanced cell surface hydrophobicity (hydrophobicity index of ~137% vs. 100% WT, P ≤ 0.001) with an actual value of 82.0 ± 2.6, when compared to the MG1363 WT (59.7 ± 7.2) and the vector-only MGAS1363::pJRS525 control (56.6 ± 5.5). These data suggest a direct relationship between Scl1 expression and cell surface hydrophobicity and establish their involvement in the microorganism's ability to form biofilm in vitro.
Table 2.
Cell surface hydrophobicity of Lactococcus strains
| Lactococcus Strain | Actual Value† | Hydrophobicity Index‡ |
|---|---|---|
| L. lactis 1363 WT | 59.7 ± 7.2 | 100 |
| L. lactis 1363::pJRS525 | 56.6 ± 5.5 | 98 |
| L. lactis 1363::pSl230 | 82.0 ± 2.6 | **137 |
† Actual hydrophobicity values were calculated based on hexadecane binding as described in Methods. Values are representative of three separate experiments with ten replicates ± SD
‡ Hydrophobicity Index represents the ration of actual hydrophobicity value for each strain to that of the isogenic wild-type (WT) strain multiplied by 100
** Asterisks denote a statistically significant difference of Δscl1 mutants versus WTs at P ≤ 0.001
Discussion
Group A Streptococcus strains vary because of the vast number of M-protein types, and this variation is associated with varying frequency of isolation and exacerbation of disease [40,41]. The M41-, M28-, M3-, and M1-type strains selected for the current study represent a significant intraspecies diversity among clinical isolates of GAS. M41 GAS was a major causative agent of superficial skin infections [42-44], and strain MGAS6183, harboring the Scl1.41 protein, has been studied extensively [19,21,22]. M28-type GAS (strain MGAS6143) has historically been associated with puerperal fever and currently is responsible for extensive human infections world-wide [45]. M1T1 GAS, represented by strain MGAS5005, is a globally disseminated clone responsible for both pharyngitis and invasive infections [46-48]. The M3-type strains of GAS cause a disproportionally large number of invasive GAS infections that are responsible for traumatic morbidity and death [49,50].
Initial studies by Lembke et al. that characterized biofilm formation among various M types of GAS typically included several strains of the same M type [1,28]. These studies reported a significant strain-to-strain variation in ability to form biofilms within each M type. Studies that followed compared biofilm formation by defined isogenic WT and mutant strains to assess the contribution of specific GAS surface components responsible for a biofilm phenotype, including M and M-like proteins, hyaluronic acid capsule, lipoteichoic acid, and pili [12,13]. In the current study, we have assessed the role and contribution of the surface protein Scl1 in the ability to support biofilm formation by GAS strains of four distinct M types.
Recent advances in molecular mega- and pathogenomics has enabled the characterization of numerous M3-type strains with a single nucleotide resolution [51,52]. Interestingly, all five M3-type strains MGAS158, 274, 315, 335, and 1313 that were originally used for scl1-gene sequencing [14], plus an additional strain MGAS2079 (not reported) harbor the same scl1.3 allele containing a null mutation that would result in secretion of a truncated Scl1.3-protein variant. Here, we demonstrate that these GAS strains do not form biofilm on an abiotic surface. Recently, bioinformatic screening of the sequences of ~250 invasive M3-type strains isolated globally, has led to the detection of this single nucleotide polymorphism that results in disruption of Scl1.3 protein (Steve Beres and Jim Musser, personal communication). Lembke et al. reported heterogeneous biofilm formation among four M3-type GAS strains examined over a 24, 48, and 72-h period [28]. Biofilm was detected for one strain at a 48 h time point, on a fibrinogen-coated surface; however, it is not known whether this clinical isolate forms biofilm on abiotic surface, whether it expresses the truncated or full-length Scl1.3 protein, and whether it produces an unknown fibrinogen-binding protein, which could augment the attachment and biofilm formation. Therefore, additional studies are necessary to define the contributions of other biofilm-formation determinants in M3-type strains.
Inasmuch as, variation in biofilm formation among GAS isolates of the same M-type has been established, the molecular basis of this phenotypic variation is not known. Several GAS surface-associated and secreted components were shown to contribute to variation in biofilm [12,13,33]. In addition, transcription regulators, such as Mga, CovR, and Srv are likely to play substantial roles in GAS biofilm formation [11,33] due to their transcriptional regulation of numerous genes. Therefore, it is logical to assume that the combination of genomic/proteomic make up, allelic polymorphisms, and transcription regulation all contribute to this phenomenon. In addition, discrepancies between in vitro data obtained with laboratory-stored strains and microcolony formation in vivo likely exist and add yet another unknown to the complexity of GAS biofilm/microcolony formation and its role in pathogenesis. Despite this complexity, the analyses involving isogenic strains of the same genetic background provide valuable information that allows assessment of the role and contribution of a given GAS component to biofilm formation.
The M1 MGAS5005 strain was shown to form biofilm in vitro and in experimental animals [8,33,53], and the present study demonstrates a significant role of Scl1.1 in this process. Likewise, the MGAS6183 strain, representing M41-type isolates often associated with pyoderma, produced a more robust biofilm biomass under the same experimental conditions and Scl1.41-deficient mutant was found to be an important determinant in this process. Similarly, Scl1.28 protein significantly contributes to a robust biofilm made by the M28-type strain MGAS6143. However, a recent study reported that another surface protein, designated AspA, found in M28-type GAS significantly contributed to biofilm formation [54]. The ΔaspA isogenic mutant showed 60% reduction in biofilm formation. The strain MGAS6180, which they used, expresses the same Scl1.28 variant present in the MGAS6143 strain we used; our Scl1.28 mutant showed ~44% reduction in 24 h biofilm. We propose that several surface proteins contribute to biofilm formation by M28-type strains including proteins AspA and Scl1.28, and potentially, proteins F1/SfbI and F2 that are also present in these strains [22]. This redundancy is likely responsible for the observed residual biofilms produced by the AspA- and Scl1.28-deficient mutants.
The observed heterogeneity in biofilm architecture of different GAS strains was previously observed by Lembke et al. [28] and was also documented in the current study using FESEM. In addition, here we report the differences in GAS-cell surface morphology and within cell-to-cell junctions in biofilms formed by M1- and M41-type strains. The structural and genetic determination of these differences is not known since M41 genome has not been sequenced, but may be associated with the presence of additional surface proteins, such as the F2 protein [55] encoded by prtf2 gene found in this strain [22]. Even more striking was an observed difference in the amount of the extracellular material associated with each strain, referred to as BAEM (bacteria-associated extracellular matrix). It has been shown that extracellular matrix, also called glycocalyx, is produced by biofilm-forming bacteria. DNA, lipids, proteins [33], polysaccharides and dead cell debris [56] were identified in this matrix and for gram-positive bacteria, teichoic acids have also been detected [57,58]. The exopolysaccharide component of the glycocalyx is detected using carbohydrate-binding lectins, such as concanavalin A (ConA) [10]. Both FESEM analysis and ConA staining detected more BAEM associated with M1 biofilm compared to M41, which produced larger biofilm. These observations suggest that GAS biofilm is stabilized differently by different strains and that higher BAEM production does not necessarily pre-determine larger biofilm mass. Consequently, a combination of biofilm features rather than biofilm size alone may be more relevant to pathogenicity of a given GAS strain.
Diminished adherence and biofilm formation could be associated with changes in cell surface hydrophobicity [59] of the scl1 mutants. Indeed, the lack of Scl1 resulted in both decreased hydrophobicity and the ability to form biofilm, albeit in a somewhat disproportionate manner. A decrease in the hydrophobicity index by only ~8%, as compared to the wild type-strain, was measured for the M41Δscl1 mutant and this modest decrease was accompanied by a rather large reduction in biofilm formation capacity after 24 h by 30%. Greater decrease in cell-surface hydrophobicity was measured for the M1Δscl1 (~21%) and M28Δscl1 (~22%) mutants, which was accompanied by a significant loss in biofilm formation after 24 h by both isogenic strains by ~55% and ~41% (P ≤ 0.001 for each comparison), respectively. In addition, heterologous expression of Scl.41 in L. lactis increased hydrophobicity index of this organism to ~137% of the WT level, which was accompanied by significant increase in its ability to form biofilm. Similar observations have been reported for the M and M-like protein mutants that typically, but not always, exhibit concurrent loss of both biological features [12]. For example, isogenic ΔMrp49 mutant had a non-significant drop in hydrophobicity (~2%) but significantly lower biofilm formation after 48 h by ~30%, whereas ΔEmm1 mutant lost ~78% hydrophobicity and ~44% biofilm formation capacity. In summary: (i) here we report that the Scl1 adhesin is also a hydrophobin with varying contribution to the overall surface hydrophobicity among GAS strains representing different M types and (ii) Scl1-associated surface hydrophobicity is likely to contribute to Scl1-mediated biofilm formation.
To test whether Scl1 alone could support biofilm formation, we used a heterologous L. lactis strain, which provides an expression system for membrane-bound proteins of gram- positive bacteria with LPXTG cell-wall anchoring motifs [39,60-62], including the group A streptococcal M6 protein [38,63]. In a recent study by Maddocks et al. [54] it was shown that heterologous expression of AspA GAS surface protein was able to induce a biofilm phenotype in L. lactis MG1363. We were also able to achieve a gain-of-function derivative of the L. lactis WT MG1363 strain, (MG1363::pSL230), displaying an altered phenotype associated with biofilm formation, as compared to wild-type parental and vector-only controls. These data support our current model that Scl1 protein is an important determinant of GAS biofilm formation.
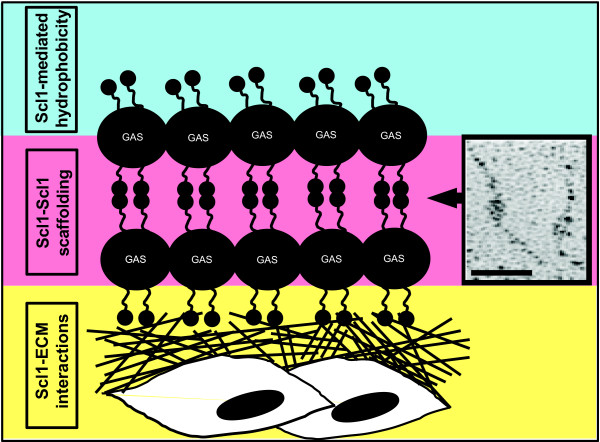
As shown by crystal violet staining and CLSM, biofilm formation by the Scl1-negative mutants was compromised during the initial stage of adherence, as well as microcolony stabilization and maturation. Consequently, their capacity for biofilm formation as compared to the respective WT controls was greatly reduced. This comparison identifies for the first time that the Scl1 protein contributes significantly to biofilm assembly and stability. Based on these observations, as well as previous work by us and others, we propose the following model of Scl1 contribution to GAS tissue microcolony formation (Figure 6). First, the Scl1 hydrophobin (current study) initiates bacterial adhesion to animate surfaces within the host [59]. Next, the Scl1 adhesin anchors the outside edge of growing microcolony in tissue by direct binding to tissue extracellular matrix components, cellular fibronectin and laminin [19]. Microcolony development is stabilized by Scl1-Scl1 scaffolding resulting from Scl1's capacity to form head-to-head dimers [64] between molecules located on adjacent chains. This model will be tested experimentally in future studies.
Figure 6.
Scl1-mediated model of GAS biofilm (not to scale). Scl1 hydrophobin (current study) initiates bacterial adhesion to animate surfaces [59] within the host (blue field). Scl1 adhesin anchors the growing microcolony by direct binding to tissue extracellular matrix (ECM) components, cellular fibronectin and laminin [19], initiating microcolony formation and anchoring the outside edge of GAS microcolony in tissue (yellow field). Microcolony scaffolding is stabilized by the formation of head-to-head dimers between Scl1 molecules on adjacent chains (pink field). Inset shows Scl1-Scl1 head-to-head dimers formed by rScl1.1 as viewed by electron microscopy after rotary shadowing [64]. Bar: 50 nm.
Conclusions
In the present work, using pathogenically differing GAS strains, we have demonstrated three concepts. First, we have confirmed previous observations that biofilm formation is an innate property of GAS strains. The M41-type strain used formed a more robust biofilm when compared to M28-type strain as well as M1-type strain. Importantly, the highly invasive M3-type strains devoid of the surface-associated Scl1 also lack the ability to form biofilm. Secondly, the absence of surface-associated Scl1 decreases GAS-cell hydrophobicity suggesting that Scl1 plays a role on the GAS surface as a hydrophobin. Thirdly, we have established that the Scl1 protein is a significant determinant for GAS biofilm formation. This concept was further tested by the heterologous expression of Scl1 in Lactococcus, an organism found in dairy fermentation environments, enabling it to form biofilm. Altogether, these data underscore the importance of Scl1 in biofilm-associated regulation of GAS pathogenicity. Recently published work has shown that the recombinant Scl1 binds to the extracellular matrix components, cellular fibronectin and laminin [19]. Our current research provides a foundation warranting additional investigation as to whether direct Scl1-ECM binding may promote GAS biofilm as a bridging mechanism within host tissues.
Methods
GAS strains and growth conditions
The wild-type GAS strains M41- MGAS6183, M1- MGAS5005, and M28-type MGAS6143, as well as their scl1-inactivated isogenic mutants and complemented M41Δscl1 mutant have been previously described [22,27,65]. In addition, a set of the wild-type M3-type GAS strains MGAS158, MGAS274, MGAS315, MGAS335, MGAS1313, and MGAS2079 was also used. GAS cultures were routinely grown on brain-heart infusion agar (BD Biosciences) and in Todd-Hewitt broth (BD Biosciences) supplemented with 0.2% yeast extract (THY medium) at 37°C in an atmosphere of 5% CO2-20% O2. Logarithmic phase cultures harvested at the optical density (600 nm) of about 0.5 (OD600 ~0.5) were used to prepare GAS inocula for biofilm analysis. Colony counts were verified by plating on tryptic soy agar with 5% sheep's blood (Remel). Lactococcus lactis subsp. cremoris strain MG1363 (provided by Dr. Anton Steen) were grown using M17 broth or agar media (Oxoid) supplemented with 0.5 M sucrose and 0.5% glucose (SGM17 media) at 30°C in an atmosphere of 5% CO2-20% O2.
Heterologous Scl1 expression in Lactococcus lactis
Lactococcus transformation
To obtain electrocompetent cells, 500 ml of SGM17 broth supplemented with 2% glycine was inoculated with an overnight culture and grown until OD600 ~0.4 was reached. Cells were harvested and washed twice with ice-cold solution A (0.5 M sucrose, 10% glycerol); cells were then re-suspended in solution A (1/1000 of original culture volume) and stored at -80°C [66]. For transformation, cells were thawed on ice and mixed with 1 μl of DNA of the Scl1.41-expressing plasmid pSL230 or pJRS525-vector [22]; and transferred to a cold 1-mm electrode-gap cuvette. Cells were pulsed with 2.0 kV at 25 μF and 400 Ω. Immediately following, suspensions were mixed with 1 ml outgrowth medium (SGM17 broth supplemented with 20 mM MgCl2 and 2 mM CaCl2) and incubated for 2.5 h before plating on SGM17 agar supplemented with spectinomycin [62].
Molecular characterization of transformants
The pSL230 was detected in Lactococcus lactis MG1363 transformants by PCR amplification directly from bacterial colonies with scl1.41-gene specific primers 232up (5'-CTCCACAAAGAGTGATCAGTC) and 232rev (5'-TTAGTTGTTTTCTTTGCGTTT); pSL230 plasmid DNA was used as a positive control. PCR samples were analyzed on 1% agarose gel in Tris-acetate-EDTA buffer and stained with ethidium bromide. Inocula from colonies of L. lactis MG1363, as well as colonies harboring either pJRS525 vector or pSL230 construct were used in subsequent experiments.
Western blot analysis
Cell-wall extracts were prepared as previously described [22]. Briefly, cells grown to OD600 ~0.4 were harvested, washed with TES (10 mM Tris, 1 mM EDTA, 25% Sucrose), re-suspended in TES-LMR (TES containing 1 mg/ml hen egg lysozyme, 0.1 mg/ml mutanolysin, 0.1 mg/ml RNAseA and 1 mM PMSF) and incubated at 37°C for 1 h. After centrifugation at 2500 g for 10 min, the supernatants were precipitated with ice-cold TCA (16% final) at -20°C overnight. Precipitates were rinsed thoroughly with ice-cold acetone and dissolved in 1× sample buffer at 250 μl per unit OD600. Samples were subjected to 10% SDS-PAGE, transferred to nitrocellulose, and probed with anti-P176 antiserum followed by goat anti-rabbit-HRP and detected employing chemiluminescent substrate (Pierce).
Flow cytometry
Bacterial cells were grown to mid-log phase (OD600 ~0.4), washed once with filtered DPBS containing 1% FBS and re-suspended in the same buffer. Five million cells were incubated with 1:400 dilution of primary reagents, either rabbit pre-bleed (control) or rabbit anti-P176 antiserum for 30 min on ice, washed with DPBS-FBS and then incubated with 1:200 dilution of second reagent donkey anti-rabbit-APC (Jackson ImmunoResearch) for 30 min on ice. After a final wash and re-suspension in DPBS-FBS, flow cytometric data were acquired with FACSCaliber (BD Biosciences) and analyzed employing FCS Express (De Novo Software).
Analysis of biofilm formation
Crystal violet staining assay
Biofilm formation was tested using tissue culture treated polystyrene 24-well plates. 1.5 ml of logarithmic-phase GAS or Lactococcus cultures were seeded without dilution into wells and incubated at 37°C for GAS and 30°C for Lactococcus in an atmosphere of 5% CO2-20% O2 according to indicated time points upon which medium was aspirated. Wells were washed with PBS and 500 μl of 1% crystal violet was added to each well, and incubated at room temperature for 30 min. Dye was then aspirated, wells were washed with PBS, and stain was solubilized with 500 μl of 100% ethanol. Spectrophotometric readings at OD600 were recorded for each sample per time point. Samples were analyzed in triplicate in at least three experiments.
Confocal laser scanning microscopy (CLSM)
To visualize GAS and L. lactis strains by CLSM, bacterial cells were transformed with a GFP-encoding plasmid, pSB027 [67]. 15-mm glass cover slips were placed into 24-well tissue culture plate wells. Logarithmic-phase bacterial cultures were inoculated without dilution and grown for 24 h. Cover slips were rinsed with PBS and fixed with 3% paraformaldehyde at room temperature for 30 min. Biofilms present on cover slips were washed with PBS and mounted onto slides using Prolong Gold mounting media (Invitrogen). Confocal images were acquired using a 63×/1.40 Plan-Apochromat objective and a Zeiss LSM 510 laser scanning confocal on an AxioImager Z1 microscope. An orthogonal view of the Z-stacks was used to display and measure biofilm thickness using Zeiss LSM software. Ten representative images within a single experiment were used to calculate the average vertical thickness measured in micrometers.
To visualize extracellular matrix associated with GAS cells, 24-h biofilm samples were reacted with 100 μg of tetramethyl rhodamine isothiocyanate- (TRITC)-conjugated concanavalin A (TRITC-ConA) (Invitrogen) for 30 min at room temperature in the dark prior to mounting with Prolong Gold medium. An average of ten microscopic views within each sample was reviewed using the 63×/1.40 objective, as described above.
Field emission scanning electron microscopy (FESEM)
GAS biofilm samples were grown for 24 h on glass cover slips, washed with PBS, and fixed with 3% paraformaldehyde for 2 h and post-fixed in osmium tetroxide. Samples were next dehydrated in an ethanol gradient, dried using hexamethyldisalizane, mounted onto aluminum stubs and sputter-coated with gold/palladium. The samples were then imaged on a Hitachi S-4800 field emission scanning electron microscope.
Quantitation of hydrophobicity
A modified hexadecane method [12,37,68] was used to determine the cell hydrophobicity. Briefly, 5 ml of the logarithmic-phase GAS or Lactococcus cultures (OD600 ~0.5) were pelleted, washed and re-suspended in 5 ml of PBS. One ml of hexadecane was added, vortexed for 1 min and incubated for 10 min at 30°C. Mixtures were then vortexed for an additional 1 min and allowed to stand for 2 min for phase separation at room temperature. The absorbance of the lower aqueous phase was read at OD600 and compared against the PBS control. Actual hydrophobicity value was calculated using the following equation: Actual Value = [1-(A/Ao)] × 100, where A is OD600 value after hexadecane treatment and Ao is OD600 prior to hexadecane treatment.
Statistical analysis
Statistical significance was determined using a two-tailed paired Student's t-test. The results were considered statistically significant with P ≤ 0.05 (*) and P ≤ 0.001 (**).
Authors' contributions
HO-K is responsible for majority of experiments. ME characterized heterologous expression of Scl1 and BB characterized biofilm formation by M3-type strains. KHM assisted in biofilm analysis using CLSM. DS-B, BJG and HO-K performed FESEM imaging and analysis. SDR provided preliminary results and participated in helpful discussions. SL was the project leader and participated in overall design and coordination of the project. HO-K and SL drafted the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Figure S1. Biofilm formation by the isogenic wild-type and scl1-inactivated GAS strains. The figure shows gallery views and X-Y orthogonal Z-stack views of GFP-expressing GAS biofilms at 24 h rendered by confocal laser scanning microscopy (CLSM). Figure S2. Biofilm formation by the wild-type and Scl1-expressing L. lactis strains. The figure shows gallery views and X-Y orthogonal Z-stack views of GFP-expressing L. lactis biofilms at 24 h rendered by CLSM.
Contributor Information
Heaven A Oliver-Kozup, Email: holiverkozup@hsc.wvu.edu.
Meenal Elliott, Email: melliott@hsc.wvu.edu.
Beth A Bachert, Email: bbachert@mix.wvu.edu.
Karen H Martin, Email: kmartin@hsc.wvu.edu.
Sean D Reid, Email: sreid@wfubmc.edu.
Diane E Schwegler-Berry, Email: des4@cdc.gov.
Brett J Green, Email: dox6@cdc.gov.
Slawomir Lukomski, Email: slukomski@hsc.wvu.edu.
Acknowledgements
We would like to thank Dr. Steen for providing the Lactococcus lactis subsp. cremoris strain MG1363. This work was supported in part by National Institutes and Health Grant AI50666 and by a research grant (RFDG) from the West Virginia University Research and Graduate Education (to S. L.). H. Oliver-Kozup was supported by a grant from the West Virginia Graduate Student Fellowship in Science, Technology, Engineering and Mathematics (STEM). Confocal microscopy experiments were performed in the West Virginia University Microscope Imaging Facility, which is supported in part by the Mary Babb Randolph Cancer Center and NIH grant P20 RR016440. We would like to acknowledge the assistance of the West Virginia University Flow Cytometry core facility which was supported in part by a grant P30 RR032138 from the National Institutes of Health. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute of Occupational Safety and Health.
References
- Conley J, Olson ME, Cook LS, Ceri H, Phan V, Davies HD. Biofilm formation by group a streptococci: is there a relationship with treatment failure? J Clin Microbiol. 2003;41(9):4043–4048. doi: 10.1128/JCM.41.9.4043-4048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Terao Y, Okuni H, Ninomiya K, Sakata H, Ikebe K, Maeda Y, Kawabata S. Biofilm formation or internalization into epithelial cells enable Streptococcus pyogenes to evade antibiotic eradication in patients with pharyngitis. Microb Pathog. 2011;51(1-2):58–68. doi: 10.1016/j.micpath.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Singh PK. Genetic variation in biofilms and the insurance effects of diversity. Microbiology. 2005;151(Pt 9):2816–2818. doi: 10.1099/mic.0.28224-0. [DOI] [PubMed] [Google Scholar]
- Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res. 2010;28(1):55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- Kaplan JB, Meyenhofer MF, Fine DH. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J Bacteriol. 2003;185(4):1399–1404. doi: 10.1128/JB.185.4.1399-1404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raad II, Fang X, Keutgen XM, Jiang Y, Sherertz R, Hachem R. The role of chelators in preventing biofilm formation and catheter-related bloodstream infections. Curr Opin Infect Dis. 2008;21(4):385–392. doi: 10.1097/QCO.0b013e32830634d8. [DOI] [PubMed] [Google Scholar]
- Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121(1):238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly KL, Roberts AL, Holder RC, Reid SD. Dispersal of group A streptococcal biofilms by the cysteine protease SpeB leads to increased disease severity in a murine model. PLoS ONE. 2011;6(4):e18984. doi: 10.1371/journal.pone.0018984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/CMR.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Morizane S, Yamasaki O, Oono T, Iwatsuki K. Assessment of Streptococcus pyogenes microcolony formation in infected skin by confocal laser scanning microscopy. J Dermatol Sci. 2003;32(3):193–199. doi: 10.1016/S0923-1811(03)00096-3. [DOI] [PubMed] [Google Scholar]
- Cho K, Caparon M. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol Microbiol. 2005;57(6):1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- Courtney HS, Ofek I, Penfound T, Nizet V, Pence MA, Kreikemeyer B, Podbielski A, Hasty DL, Dale JB. Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS ONE. 2009;4(1):e4166. doi: 10.1371/journal.pone.0004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti A, Zingaretti C, Falugi F, Capo S, Bombaci M, Bagnoli F, Gambellini G, Bensi G, Mora M, Edwards A. et al. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol. 2007;64(4):968–983. doi: 10.1111/j.1365-2958.2007.05704.x. [DOI] [PubMed] [Google Scholar]
- Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Ireland RM, Reid SD, Adams GG, Musser JM. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect Immun. 2000;68(12):6542–6553. doi: 10.1128/IAI.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Shelvin BJ, Graviss EA, Musser JM. Identification and characterization of a second extracellular collagen-like protein made by group A Streptococcus: control of production at the level of translation. Infect Immun. 2001;69(3):1729–1738. doi: 10.1128/IAI.69.3.1729-1738.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Eden A, Björck L. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect Immun. 2000;68(11):6370–6377. doi: 10.1128/IAI.68.11.6370-6377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, McIver KS. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J Bacteriol. 2004;186(23):7847–7857. doi: 10.1128/JB.186.23.7847-7857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, Walters MS, McIver KS. Mga is sufficient to activate transcription in vitro of sof-sfbX and other Mga-regulated virulence genes in the group A Streptococcus. J Bacteriol. 2006;188(6):2038–2047. doi: 10.1128/JB.188.6.2038-2047.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell CC, Oliver-Kozup H, Han R, Lukomska E, Lukomski S. Scl1, the multifunctional adhesin of group A Streptococcus, selectively binds cellular fibronectin and laminin, and mediates pathogen internalization by human cells. FEMS Microbiol Lett. 2010;303(1):61–68. doi: 10.1111/j.1574-6968.2009.01864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humtsoe JO, Kim JK, Xu Y, Keene DR, Höök M, Lukomski S, Wary KK. A streptococcal collagen-like protein interacts with the α2β1 integrin and induces intracellular signaling. J Biol Chem. 2005;280(14):13848–13857. doi: 10.1074/jbc.M410605200. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Barczyk M, Keene DR, Lukomska E, Gullberg DE, Lukomski S. Identification of the first prokaryotic collagen sequence motif that mediates binding to human collagen receptors, integrins α2β1 and α11β1. J Biol Chem. 2008;283(52):36168–36175. doi: 10.1074/jbc.M806865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell CC, Lukomska E, Seo NS, Höök M, Lukomski S. Scl1-dependent internalization of group A Streptococcus via direct interactions with the α2β1 integrin enhances pathogen survival and re-emergence. Mol Microbiol. 2007;64(5):1319–1331. doi: 10.1111/j.1365-2958.2007.05741.x. [DOI] [PubMed] [Google Scholar]
- Gao Y, Liang C, Zhao R, Lukomski S, Han R. The Scl1 of M41-type group A Streptococcus binds the high-density lipoprotein. FEMS Microbiol Lett. 2010;309(1):55–61. doi: 10.1111/j.1574-6968.2010.02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Påhlman LI, Marx PF, Morgelin M, Lukomski S, Meijers JCM, Herwald H. Thrombin-activatable fibrinolysis inhibitor binds to Streptococcus pyogenes by interacting with collagen-like proteins A and B. J Biol Chem. 2007;282(34):24873–24881. doi: 10.1074/jbc.M610015200. [DOI] [PubMed] [Google Scholar]
- Caswell C, Han R, Hovis K, Ciborowski P, Keene D, Marconi R, Lukomski S. The Scl1 protein of M6-type group A Streptococcus binds the human complement regulatory protein, factor H, and inhibits the alternative pathway of complement. Mol Microbiol. 2008;67(3):584–596. doi: 10.1111/j.1365-2958.2007.06067.x. [DOI] [PubMed] [Google Scholar]
- Reuter M, Caswell CC, Lukomski S, Zipfel PF. Binding of the human complement regulators CFHR1 and factor H by streptococcal collagen-like protein 1 (Scl1) via their conserved C termini allows control of the complement cascade at multiple levels. J Biol Chem. 2010;285(49):38473–38485. doi: 10.1074/jbc.M110.143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R, Caswell CC, Lukomska E, Keene DR, Pawlowski M, Bujnicki JM, Kim JK, Lukomski S. Binding of the low-density lipoprotein by streptococcal collagen-like protein Scl1 of Streptococcus pyogenes. Mol Microbiol. 2006;61(2):351–367. doi: 10.1111/j.1365-2958.2006.05237.x. [DOI] [PubMed] [Google Scholar]
- Lembke C, Podbielski A, Hidalgo-Grass C, Jonas L, Hanski E, Kreikemeyer B. Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl Environ Microbiol. 2006;72(4):2864–2875. doi: 10.1128/AEM.72.4.2864-2875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser JM. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania RE, Lamers GE, Vonk MJ, Huy PT, Hiemstra PS, Bloemberg GV, Grote JJ. Demonstration of bacterial cells and glycocalyx in biofilms on human tonsils. Arch Otolaryngol Head Neck Surg. 2007;133(2):115–121. doi: 10.1001/archotol.133.2.115. [DOI] [PubMed] [Google Scholar]
- Maeyama R, Mizunoe Y, Anderson JM, Tanaka M, Matsuda T. Confocal imaging of biofilm formation process using fluoroprobed Escherichia coli and fluoro-stained exopolysaccharide. J Biomed Mater Res A. 2004;70(2):274–282. doi: 10.1002/jbm.a.30077. [DOI] [PubMed] [Google Scholar]
- Doern CD, Roberts AL, Hong W, Nelson J, Lukomski S, Swords WE, Reid SD. Biofilm formation by group A Streptococcus: a role for the streptococcal regulator of virulence (Srv) and streptococcal cysteine protease (SpeB) Microbiology. 2009;155(Pt 1):46–52. doi: 10.1099/mic.0.021048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buznach N, Dagan R, Greenberg D. Clinical and bacterial characteristics of acute bacterial conjunctivitis in children in the antibiotic resistance era. Pediatr Infect Dis J. 2005;24(9):823–828. doi: 10.1097/01.inf.0000178066.24569.98. [DOI] [PubMed] [Google Scholar]
- Wadstrom T, Schmidt KH, Kuhnemund O, Havlicek J, Köhler W. Comparative studies on surface hydrophobicity of streptococcal strains of group-a, group-B, group-C, group-D and group-G. J Gen Microbiol. 1984;130(Mar):657–664. doi: 10.1099/00221287-130-3-657. [DOI] [PubMed] [Google Scholar]
- Grivetti LE, Ogle BM. Value of traditional foods in meeting macro- and micronutrient needs: the wild plant connection. Nutr Res Rev. 2000;13(1):31–46. doi: 10.1079/095442200108728990. [DOI] [PubMed] [Google Scholar]
- Pan WH, Li PL, Liu Z. The correlation between surface hydrophobicity and adherence of Bifidobacterium strains from centenarians' faeces. Anaerobe. 2006;12(3):148–152. doi: 10.1016/j.anaerobe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Piard JC, Hautefort I, Fischetti VA, Ehrlich SD, Fons M, Gruss A. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J Bacteriol. 1997;179(9):3068–3072. doi: 10.1128/jb.179.9.3068-3072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares DM, Kok J, Poolman B. Genome sequences of Lactococcus lactis MG1363 (Revised) and NZ9000 and comparative physiological studies. J Bacteriol. 2010;192(21):5806–5812. doi: 10.1128/JB.00533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore AM, Kapur V, Sullivan DJ, Musser JM, Kehoe MA. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol. 1994;14(4):619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- Bessen DE, Sotir CM, Readdy TL, Hollingshead SK. Genetic correlates of throat and skin isolates of group A streptococci. J Infect Dis. 1996;173:896–900. doi: 10.1093/infdis/173.4.896. [DOI] [PubMed] [Google Scholar]
- Anthony BF. In: Streptococcal Infections. Stevens DL, Kaplan EL, editor. New York: Oxford University Press; 2000. Streptococcal pyoderma; pp. 144–151. [Google Scholar]
- Anthony BF, Perlman LV, Wannamaker LW. Skin infections and acute nephritis in American Indian children. Pediatrics. 1967;39(2):263–279. [PubMed] [Google Scholar]
- Top FH Jr, Wannamaker LW, Maxted WR, Anthony BF. M antigens among group A streptococci isolated from skin lesions. J Exp Med. 1967;126(4):667–685. doi: 10.1084/jem.126.4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, Beres SB, Graviss EA, Allison JE, McGeer AJ, Vuopio-Varkila J, LeFebvre RB, Musser JM. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J Clin Microbiol. 2005;43(8):4083–4091. doi: 10.1128/JCM.43.8.4083-4091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R, Kotb M. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg Infect Dis. 2008;14(10):1511–1517. doi: 10.3201/eid1410.071660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J Bacteriol. 2005;187(10):3311–3318. doi: 10.1128/JB.187.10.3311-3318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, Martin DR. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63(3):994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul R, McGeer A, Low DE, Green K, Schwartz B, Study OGAS, Simor AE. Population-based surveillance for group A streptococcal necrotizing fasciitis: clinical features, prognostic indicators, and microbiologic analysis of 77 cases. Am J Med. 1997;103:18–24. doi: 10.1016/S0002-9343(97)00160-5. [DOI] [PubMed] [Google Scholar]
- Sharkawy A, Low DE, Saginur R, Gregson D, Schwartz B, Jessamine P, Green K, McGeer A. Severe group a streptococcal soft-tissue infections in Ontario: 1992-1996. Clin Infect Dis. 2002;34(4):454–460. doi: 10.1086/338466. [DOI] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu M-Y, Smoot JC, Porcella SF, Parkins LD. et al. Genome sequence of a serotype M3 strain of group A Streptococcus: Phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002;99(15):10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Sturdevant DE, Granville CN, Liu M, Ricklefs SM, Whitney AR, Parkins LD, Hoe NP, Adams GJ. et al. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc Natl Acad Sci USA. 2004;101(32):11833–11838. doi: 10.1073/pnas.0404163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AL, Connolly KL, Doern CD, Holder RC, Reid SD. Loss of the group A Streptococcus regulator Srv decreases biofilm formation in vivo in an otitis media model of infection. Infect Immun. 2010;78(11):4800–4808. doi: 10.1128/IAI.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks SE, Wright CJ, Nobbs AH, Brittan JL, Franklin L, Stromberg N, Kadioglu A, Jepson MA, Jenkinson HF. Streptococcus pyogenes antigen I/II-family polypeptide AspA shows differential ligand-binding properties and mediates biofilm formation. Mol Microbiol. 2011;81(4):1034–1049. doi: 10.1111/j.1365-2958.2011.07749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe J, Natanson-Yaron S, Caparon MG, Hanski E. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two domains. Mol Microbiol. 1996;21:373–384. doi: 10.1046/j.1365-2958.1996.6331356.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186(12):3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43(6):1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33(1):206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- Courtney HS, Dale JB, Hasty DL. In: Handbook of Bacterial Adhesion: Principles, Methods, and Applications. An YH, Friedman RJ, editor. Totowa: Humana Press, Inc.; 2000. Strategies for preventing group A streptococcal adhesion and infection; pp. 553–579. [Google Scholar]
- Luo HL, Wan K, Wang HH. High-frequency conjugation system facilitates biofilm formation and pAM β1 transmission by Lactococcus lactis. Appl Environ Microb. 2005;71(6):2970–2978. doi: 10.1128/AEM.71.6.2970-2978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber SD, Solioz M. Efficient transformation of Lactococcus lactis IL1403 and generation of knock-out mutants by homologous recombination. J Basic Microb. 2007;47(3):281–286. doi: 10.1002/jobm.200610297. [DOI] [PubMed] [Google Scholar]
- Que YA, Haefliger JA, Francioli P, Moreillon P. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect Immun. 2000;68(6):3516–3522. doi: 10.1128/IAI.68.6.3516-3522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piard JC, JimenezDiaz R, Fischetti VA, Ehrlich SD, Gruss A. The M6 protein of Streptococcus pyogenes and its potential as a tool to anchor biologically active molecules at the surface of lactic acid, bacteria. Streptococci and the Host. 1997;418:545–550. doi: 10.1007/978-1-4899-1825-3_126. [DOI] [PubMed] [Google Scholar]
- Xu Y, Keene DR, Bujnicki JM, Höök M, Lukomski S. Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J Biol Chem. 2002;277(30):27312–27318. doi: 10.1074/jbc.M201163200. [DOI] [PubMed] [Google Scholar]
- Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68(2):535–542. doi: 10.1128/IAI.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V. et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. 2003;112(5):645–657. doi: 10.1016/S0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet M, Morrier JJ, Benay G, Barsotti O. Effect of hydrophobicity on in vitro streptococcal adhesion to dental alloys. J Mater Sci Mater Med. 2000;11(10):637–642. doi: 10.1023/A:1008913915399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Biofilm formation by the isogenic wild-type and scl1-inactivated GAS strains. The figure shows gallery views and X-Y orthogonal Z-stack views of GFP-expressing GAS biofilms at 24 h rendered by confocal laser scanning microscopy (CLSM). Figure S2. Biofilm formation by the wild-type and Scl1-expressing L. lactis strains. The figure shows gallery views and X-Y orthogonal Z-stack views of GFP-expressing L. lactis biofilms at 24 h rendered by CLSM.