Abstract
The 20th century theory of mammalian sex determination states that the embryo is sexually indifferent until the differentiation of gonads, after which sex differences in phenotype are caused by differential effects of gonadal hormones. That theory is inadequate because some sex differences precede differentiation of the gonads and/or are determined by non-gonadal effects of the sexual inequality in number and type of sex chromosomes. A general theory of sex determination is proposed, which recognizes multiple parallel primary sex-determining pathways initiated by genes or factors encoded by the sex chromosomes. The separate sex-specific pathways interact to synergize with or antagonize each other, enhancing or reducing sex differences in phenotype.
A general theory of mammalian sex determination
The theory of mammalian sex determination was recently stated as follows: “the sexual phenotype of individuals is dependent on the gonad: male and female somatic cells and tissues are initially sexually indifferent and sexual dimorphism is imposed by the type of gonad that develops” [1]. This theory was developed during the 20th century based on experiments in which the “sex of the individual” was defined by a limited group of phenotypes, predominantly the gonads and genitalia. Because sexual differentiation of the gonads was placed at the center of the sex determination universe, the foremost task in understanding sex determination was to discover the sex-specific factors that cause undifferentiated gonadal primordia to develop differently in the two sexes. The first sex-specific event in the molecular cascade leading to divergent development of the gonads in the two sexes is the expression of the Y chromosome gene Sry in the male’s undifferentiated gonadal ridge [2]. Sry promotes testicular development and suppresses ovarian differentiation, whereas autosomal or X genes in females initiate ovarian development and suppress testicular development [3,4]. Once the gonads differentiate, however, they secrete gonadal hormones that cause sex-specific patterns of development in many other tissues such as the external genitalia, internal genitalia (Wolffian and Müllerian duct structures), and brain. Although there is little controversy regarding the importance of gonadal secretions in sex determination of non-gonadal tissues, evidence in the last ~20 years indicates that the theory as stated in the first sentence is incorrect. The theory suffers on several counts. It is based on an unacceptably narrow definition of sex, it incorrectly suggests that gonadal determination precedes all other sex differences, and it misses the importance of other X and Y genes that have a logically equal position to Sry as primary sex-biased gene products. Importantly, the theory does not explain a growing list of sex differences in phenotype that are not controlled by the gonads and therefore are not downstream of differences initiated by Sry. In constructing a more accurate general theory of sex determination, therefore, gonadal differentiation should be displaced as the central event of sex determination, and replaced with whatever other condition is the unitary origin of all sex differences. In animals with heteromorphic sex chromosomes, all sex differences stem from the inherent sexual inequality of the sex chromosomes (X and Y in mammals), which are the only factors known to differ consistently in male and female zygotes. Here I summarize evidence that numerous X and Y chromosomal factors, not just Sry, are primary in determining the sex of tissues in mammals. This diverse set of primary sex-determining genes, and their downstream products such as gonadal hormones, interact with each other to produce female and male animals with sex-typical phenotypes that in the aggregate define the sex of the individual.
Limits of the 20th century theory: Why the gonads are primary and why they are not
Although the sex of the individual can be defined in many ways and assessed based on measurement of an almost unlimited number of phenotypes, from gonadal type to personality, historically gonadal differentiation was reasonably considered the premier event of mammalian sex determination (see Box 1). Firstly, the differentiation of two types of gametes is at the crux of evolution of sexual reproduction, and thus the gamete-producing tissues (gonads) and tissues involved in gamete delivery and reproduction (genitalia and brain) are fundamental to the definition of an individual’s sex. Secondly, the genetic control of the sex of the gonads was seen as mechanistically distinct from, and the developmental antecedent of, sexual differentiation of other tissues. Gonadal differentiation was viewed as genetic, caused by factors inherent to the XX vs. XY difference in the genome, whereas non-gonadal sexual differentiation was hormonal, driven by secretions of the gonads. For example, masculine differentiation occurred in XX or XY individuals as long as male hormones were present. Both historical and modern data support the idea that most sex differences in non-gonadal tissues are caused by gonadal secretions [5,6]. However, beginning before 1990 and with greater frequency since then, the sexual phenotype of specific cells or tissues has been repeatedly shown to be influenced also by sex chromosome complement (XX vs. XY) factors that are not downstream of gonadal hormones. It is not desirable to retain the old theory along with a rapidly growing list of inconvenient footnotes that document where the old theory fails, but rather to articulate a new framework that accounts for all experimental results. A new perspective, proposed here, recognizes a set of primary sex-determining factors encoded by the sex chromosomes, which are all the result of the original zygotic imbalance in the number of X and Y chromosomes. This imbalance leads to numerous inescapable sex differences in X and Y gene expression that are all primary in the causation of sex differences in tissues. The expression of Sry is among these primary sex-determining functions of the chromosomes, and is the most important of the primary factors. However, Sry is not the only or the earliest of the primary sex-determining factors, and there are likely numerous others.
Box 1. Defining the “sex of the individual” and “sex determination”.
The sex of the individual can be defined in different ways, giving rise to different conceptual frameworks about what determines sex. Lillie [5] wrote in 1939: ‘It is clear that we must make a radical distinction between sex determination and sex differentiation. In most cases the factors of sex determination are chromosomal and subject to the laws of Mendelian inheritance…. [In] higher vertebrates, the mechanism of sex differentiation is taken over by the extracellular agents, the male and female hormones; but it is necessary to postulate that the endocrine cells producing them are first determined by the nuclear mechanism.” Lillie’s dichotomy between “sex determination” and “sexual differentiation” persists to the present because “sex determination” is still usually defined as the differentiation of the gonads (e.g., [68]). Lillie’s distinction appears to stem from the ideas that sexual differentiation of the gonads precedes and is mechanistically different from the sexual differentiation of non-gonadal cells, and that the sex-defining characteristics of the individual are determined by gonadal secretions. From this viewpoint, a person with a vagina and uterus (potential to bear children) is clearly a female, and a person with a penis is male, even if other important sex-typical traits are discordant with the sex of the individual thus defined (e.g., a male unable to produce sperm is still considered a male). Some sex-typical traits are therefore defined out of sex determination, and sex-specific expression of Xist or of genes causing sex differences in blastocysts seem to have nothing to do with sex determination or with the classic sex-defining traits. The perspective offered here is that this approach is ultimately unnecessarily constraining because the two sexes typically differ in many tissues and phenotypes, each of which can be considered to contribute to the overall sex of the individual. If “every cell has a sex” [69], meaning that XX and XY cells of many types show consistent differences, it makes sense to recognize a diverse set of primary factors (each present in the zygote) that initiate molecular cascades that bias the sexual phenotype of the cells that comprise the individual. Defining of the sex of the individual is not just a semantic exercise, but leads to a conceptual framework for categorizing and studying sex-determining factors (and their interactions) that could cause or reduce sex differences in cells and individuals.
Defining primary sex-determining genes and genomic regions
The sex of a cell, tissue, or individual is defined by consistent phenotypic differences between males vs. females. The emergent phenotype of the tissue is the result of the aggregate effects of complex interconnected molecular networks whose nodes are gene products that drive the expression of each other in the tissue. Sex differences in the network arise when specific gene products are more active in one sex than the other, driving the expression pattern in one sex away from that in the other [6]. The sex-biased genes are usually autosomal, because most of the genome is autosomal. The autosomal genes, however, cannot be primary in determining the sexual phenotype of the animal, because of the equal representation of autosomal genes in the genome of male and females zygotes. Rather, sex differences in autosomal gene expression must be downstream of primary sex differences in expression of X or Y factors resulting from the inherent sex difference in the number and type of sex chromosomes. Thus, explaining sex determination requires finding the primary sex chromosome sex-determining genes and the downstream pathways that mediate their effects. Some sex differences in expression of X genes result from the difference in number of X chromosomes, but others do not because they are downstream of sex differences in expression of other genes (for example, if they are caused by differences in the level of gonadal hormones). Thus, sex differences in expression of some X genes and all Y genes are inherent products of the imbalance in the sex chromosomes, and therefore primary in the determination of the individual’s sex. Note that “primary” here does not mean “most important”, because the Sry-initiated differentiation of the gonads is most important (because of its widespread downstream effects) among the primary sex determining genes defined in this way.
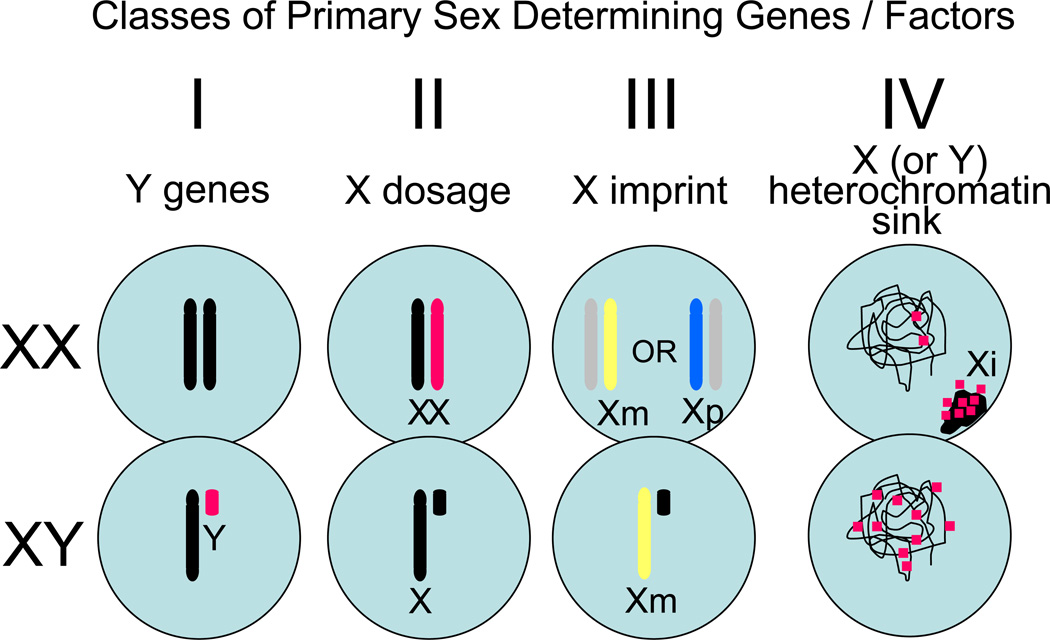
Although most primary sex-determining genes or factors have not been identified, they fall into one of several classes [7,8] (Figure 1). Class I comprises Y genes, which are expressed only in males, and thus can have only male-determining effects. Unless a Y gene is totally redundant with non-Y genes in function so that it has no male-specific effect, it qualifies as a primary sex-determining gene. Sry is the prime example of Class I, but mice possessing Sry but lacking other Y genes are not fully male, because they cannot make sperm. Several Y genes are required for normal spermatogenesis [9–11], and thus qualify as primary male-determining genes. Other Y genes likely have other male-determining functions.
Figure 1.
Four possible classes of primary sex determining factors are recognized. Class I comprises Y genes that have a male-specific effect in one or more tissues, such as Sry and Y genes required for spermatogenesis. Class II are X genes that are expressed at a higher level in females than males by virtue of the 2:1 ratio in number of X chromosomes. Class III are X genes that receive a parental imprint. The X chromosome receiving a maternal imprint (Xm, yellow) is active in half of XX cells and all of XY cells, whereas the X chromosome receiving a paternal imprint (Xp, blue) is active in half of XX cells and no XY cells. Class IV are proposed regions of sex chromosome heterochromatin (the heterochromatic inactive X (Xi) is illustrated here) that act as sex-specific sinks for factors (red dots) that regulate the amount of euchromatin / heterochromatin at interphase and therefore affect gene expression throughout the genome. To date, specific members of only Class I have been identified (Sry and spermatogenesis genes [9,10,11]). Although evidence indicates that the number of X chromosomes leads to some sex differences in phenotype [50], the specific genes or chromosome regions that explain these X effects have not been identified. Class IV is particularly speculative at present because it is based on a limited number of studies. Future studies are likely to expand the importance of Classes II–IV.
Class II sex-determining genes are X genes that are expressed higher in females than males by virtue of the 2:1 sex ratio in number of X chromosomes. In the blastocyst, prior to onset of X-inactivation, many or most X genes appear to fall into this class. At this stage, many X genes are expressed higher in females than in males, and these genes likely cause the sex differences in expression found in a large number of autosomal genes at that stage [12–14]. Although the higher expression of most X genes at this stage could have long-lasting sex-specific effects on development and function, the differences are considerably reduced by the onset of X inactivation. The main effect of silencing of one X chromosome in each non-germline cell of females is to reduce sex differences in expression of X genes, so that the degree of sexual dimorphism in X gene expression in adults is globally similar to that found in autosomal genes on a tissue-by-tissue basis [15]. Even after X inactivation has occurred, however, some X genes continue to be expressed from both X chromosomes. Such “escapees” from inactivation are not necessarily expressed higher in females than in males, however, because other regulatory forces can make their expression more or less equivalent in the two sexes [16,17]. For example, most of the genes escaping X inactivation, as shown in studies of cell lines in vitro, do not appear to be expressed higher in females [15,16,18]. Nevertheless, some X escapees are reliably expressed at higher levels in females than in males, and these genes comprise Class II throughout most of life [19–22].
Class III genes are X genes that receive a parental imprint. X genes with a maternal imprint function under that constraint potentially in all male cells but only in about half of female cells because of inactivation of the maternal X in the other half, whereas the effect of a paternal imprint on X genes will occur in about half of female cells but never in a male cell. The sex differences in the effect of the imprint could cause sexual differentiation. The parental imprint on X genes is known to influence adult cognitive and social behaviors [23].
Other mechanisms have been proposed to explain sex differences caused by the imbalance in number of X chromosomes [8,24–26] (see Box 2). Recently evidence has emerged for Class IV sex chromosome factors, which are sex-specific regions of heterochromatin that exert effects on the epigenetic status of the rest of the genome [27,28]. In Drosophila melanogaster, the Y chromosome contains large regions of heterochromatin, which alter the epigenetic balance of heterochromatin / euchromatin at autosomal loci, and regulate the expression of hundreds to thousands of autosomal and X genes [27,29,30]. The Y effect occurs even when no Y protein-coding genes are expressed. One proposed mechanism for such sex-specific regulation is that the sex chromosome heterochromatin can alter the balance of factors (DNA methyl transferases, histone modifying enzymes, etc.) that change the epigenetic status of chromatin, serving as a sink for factors favoring heterochromatin [27–31]. The mammalian Y chromosome is also predominantly heterochromatic, but represents a much smaller proportion of the genome than in Drosophila. In mammals, however, each female cell contains an entire heterochromatic X chromosome, which could similarly alter the epigenetic status genome-wide. The production of this relatively large piece of heterochromatin is initiated by the X-linked factors that activate expression of Xist, which must be considered as a possible primary sex-determining mechanism in mammals. Although little evidence bears on this issue, recent evidence resonates with this concept. Mice differing in the number of X chromosomes show differences in expression of an autosomal gene that is sensitive to nearby heterochromatic status [28,31]. Mice with two X chromosomes show greater expression (less autosomal heterochromatin) than mice with one X chromosome, and the presence of Y chromosome appears not to affect expression. Moreover, the genome of embryonic stem cells with two X chromosomes is hypomethylated relative to those with one X chromosome, and they have reduced expression of the de novo DNA methyltransferases Dnmt3a and Dnmt3b [32]. Dnmt3a and Dnmt3b and other histone modifiers are also found to be expressed differently in male and female blastocysts, which also show differences in DNA methylation [12,33]. Thus, the heterochromatic inactive X chromosome is proposed to sequester factors regulating the epigenetic status throughout the genome in a sex-specific manner [31].
Box 2. The environment as a source of sex-specific information.
In addition to the primary sex determinants encoded by the sex chromosomes, the environment represents an important source of sex-specific information. From the time of birth, males and females are placed in different physical and social environments. Human males and females are taught different social attitudes so that they choose different environments throughout life. The environment’s effect is profound and contributes to sex differences in a wide variety of biological phenotypes, including aging, diet, obesity, behavior, and disease. Because the epigenetic mechanisms by which environments alter biological pathways are being investigated with greater frequency [35], the time may be ripe to inquire into the sex-specific factors in the environment that affect the sexual phenotype of individuals. The same epigenetic mechanisms (DNA methylation, histone modifications, etc.) that are influenced by sex-specific factors on the sex chromosomes and by gonadal hormones [34,35] are also sensitive to sex differences in the environment. Thus, the epigenome may be seen as an integrator of these sex-specific influences. Very little is known about this integration.
Several sex-specific processes may converge to alter phenotype via common epigenetic pathways and modification of gene expression patterns. In addition to possible role of Class IV factors, some Class II X genes encode histone demethylases (e.g., Kdm5c and Kdm6a), and are expressed differently in XX and XY mice irrespective of their gonadal sex [20.21]. These genes are candidate primary sex-determining genes that could affect gene expression throughout the genome. In addition, gonadal secretions during fetal or neonatal life cause permanent sex differences in histone modifications or DNA methylation in brain regions that correlate with the sexual differentiation of function caused by these hormones in specific brain regions [34,35]. Thus, both primary sex chromosome factors and downstream hormonal factors may have convergent effects that cause long-lasting epigenetic changes that create a sexual bias in the function of tissues (see Box 2).
Is Xist a primary sex-determining gene?
As defined here, Xist and the X factors that trigger Xist expression are primary sex-determining genes because (1) the sex-specificity of expression stems directly from the zygotic sexual imbalance in number of X chromosomes, and (2) Xist expression makes each non-germline female cell different from each male cell. Yet, Xist’s role is usually seen as the opposite, to reduce sex differences rather than produce them. The expression of Xist makes X gene expression in females globally more comparable to that of males than would otherwise be the case [15]. This example underscores the idea that the two sexes can reach the same phenotype via different routes, or that sexual equality of phenotype does not imply equality of mechanisms or of development. The existence of multiple sex-determining mechanisms means that different pathways can either be synergistic (both promoting the same male or female phenotype) or antagonistic (two sex-specific mechanisms in one sex offsetting the effects of the other) [36]. Xist initiates female-specific mechanisms that reduce the sex-specific effect of another female-specific condition, which is the presence of two X chromosomes. Although female and male cells are differentiated by the expression of sex-determining gene Xist, some end phenotypes regulated indirectly by Xist are not necessarily made sexually dimorphic.
Despite Xist’s major role of reducing sex differences in expression of X genes, there may be uninvestigated effects of X inactivation that create sex differences in adult phenotype. Among the possibilities are the proposed effects of Class IV X heterochromatin to sequester factors regulating the epigenetic status of the rest of the genome. More generally, the initiation and maintenance of X inactivation is a highly regulated process, requiring mechanisms that direct the inactivation machinery to only one chromosome in the cell even if other chromosomes are nearby [37]. It seems quite unlikely that the commitment of resources of female cells to a complex inactivation process would leave the rest of the cell untouched and equivalent to a male cell. Female cells continue throughout life to commit resources to maintaining the inactive status of one X chromosome and preventing this inactivity from having deleterious effects on other chromosomal domains. This burden can be seen as sex-determining because it is absent in males. Because the presence of two X chromosomes renders females more susceptible to some diseases [24,26], it is possible that this susceptibility is the result of the required commitment to X inactivation [26].
Sex differences in phenotypes not downstream of gonadal differentiation
What sex differences in phenotypes are influenced by sex chromosome complement irrespective of the sex of the gonads? Two types of evidence are relevant. In some cases, the sex difference appears in embryos be fore the development of gonads and hence cannot be downstream of the switch controlled by Sry. Alternatively, the difference can be shown to depend on the complement of sex chromosomes in an experimental design in which the effects of sex chromosome complement are dissociated from the effects of the gonad.
Sex differences before gonads
The tammar wallaby represents an early and iconic example of data that clearly conflict with the gonad-centric theory [39]. In this species, the scrotum and mammary tissue begin to differentiate prior to gonadal differentiation. The differentiation of the scrotum vs. pouch and mammary glands is controlled by the number of X chromosomes, not by gonadal hormones. Thus, sex chromosome complement controls important aspects of genital differentiation.
In various mammalian species, male embryos are larger than female embryos at numerous specific time points after fertilization including before implantation [12,40]. In mice, the sex difference is produced by primary sex-determining genes on both sex chromosomes, including one or more imprinted X genes [40–42]. Embryos also differ in their glucose metabolism and sensitivity to glucose [12,43]. Widespread sex differences in expression of genes have been documented prior to differentiation of the gonads [12–14,44].
When cells from rodent midbrain are harvested from day 14 embryos, before the gonads are producing strikingly different levels of gonadal hormones, the cells show sex differences in vitro, for example in their expression of tyrosine hydroxylase, the rate limited enzyme for synthesis of dopamine [45]. These differences are also found in cells harvested from XX vs. XY mice irrespective of the type of gonad in the embryos, and hence are downstream of sex-determining genes other than Sry [46].
Sex differences caused by sex chromosome complement, dissociated from gonadal effects
Several mouse models have been developed that allow measuring the sex-determining role of sex chromosome complement (XX vs. XY) under conditions in which gonadal hormones do not explain the sex differences. Among the informative models are mice or rats in which Sry gene expression is manipulated in non-gonadal tissues [47,48], gonadless XX and XY mice (because of knock out of Sf1) [49], and the “four core genotypes” (FCG) mouse model in which the sex of the gonads is independent of the complement of sex chromosomes. In FCG mice, Sry is “moved” from the Y chromosome and inserted onto an autosome so that the autosome is testis determining [7,50]. This model allows discovering differences in XX and XY mice of the same gonadal sex (called “sex chromosome effects”), which are examples of sex differences downstream of non-Sry X or Y genes. The model also allows discovering sex differences downstream of Sry (by comparing mice with ovaries and testes that have the same complement of sex chromosomes) [51], as well as the interaction of Sry and sex chromosome effects. The advantages and caveats for use of various models have been discussed [7,49,50,52,53].
Sex chromosome complement contributes to sex differences in a wide variety of neural and behavioral phenotypes, including distribution of vasopressin fibers in the lateral septum [7,54], brain region-specific expression of nitric oxide synthase, calbindin, and prodynorphin [55–57], and neural expression of several Class II X genes including two histone demethylases [20,21,58]. In the case of prodynorphin expression, the higher expression in XX mice relative to XY mice is caused by X genes (Classes II–IV) [57]. XX and XY mice differ in aggressive and parenting behavior [54], response to noxious stimuli [59], social and investigation behaviors [51,60,61], and formation of habits in models of drug addiction and alcohol abuse [62,63]. Sry is expressed in the midbrain substantia nigra, the origin of dopamine neurons that are targets for Parkinson’s Disease. Knockdown of Sry in the brain of adult males causes reversible male-specific effects on expression of tyrosine hydroxylase and motor behavior, providing the first identification of a Y gene that has direct male-specific effects on the brain not mediated by gonadal hormones [47].
Mouse models of disease (neural tube closure, autoimmune disease, viral infections, and hypertension) have been used to evaluate the contribution of sex chromosome complement to sex differences in physiology or response to disease. Anterior neural tube closure defects occur more often in human females than males, a sex difference mimicked in mice with a null mutation of Trp53. The sex difference is caused by the number of X chromosomes, not by gonadal sex [26]. Autoimmune diseases, such as multiple sclerosis and systemic lupus erythematosus, affect women much more than men. In mouse models of these diseases investigated with FCG mice, XX mice were more affected than XY mice, irrespective of their type of gonad [38]. XX mice also show greater myocarditis in response to infection with cocksackie virus [64]. In the studies of both autoimmune disease and viral infection, the greater immunological response in XX mice correlated with striking XX vs. XY differences in the immune system markers. Hypertension occurs more in men than women. In studies of rodents, the sex difference is explained in part by gonadal hormones and partly by sex chromosome complement. Chronic or acute treatment of FCG mice with angiotensin II causes different changes in blood pressure and heart rate in XX vs. XY mice [65,66]. In the FCG model, such XX vs. XY differences are not explained by effects of Sry. Sry itself is also proposed as a regulator of hypertension via effects on the renin-angiotensin system and sympathetic nervous system [67]. Sry is expressed in non-gonadal tissues of males, and overexpression of Sry in adrenal or kidney causes an increase in blood pressure [48]. In cell systems Sry can regulate promoter activity of tyrosine hydroxylase and genes in the renin-angiotensin network. Thus, more than one primary sex-determining factor influences sex differences in hypertension.
Concluding remarks and future perspectives
The model proposed here seeks to provide a modern conceptual framework for understanding sex determination of mammals, although the same framework is applicable to any species with heteromorphic sex chromosomes (see box 3). Because not all biological sex differences are downstream from the differentiation of gonads, the sex of the individual is no longer defined exclusively by the sex of the gonads but rather by the aggregate sexual phenotype of cells and tissues. Rather than recognizing (genetic) gonadal sexual differentiation as mechanistically distinct from (hormonal) sexual differentiation of non-gonadal tissues, the factors determining sex of all tissues are treated in the same framework. The emphasis is shifted from gonadal differentiation as the central event of sex determination, to the zygotic difference in sex chromosome complement as the primary event. Thus, any factor that has a sex-specific action, because of its unequal representation in male vs. female zygotes, is seen as a primary sex-determining factor. Based on current knowledge, the only factors that are inherently unequal in male vs. female zygotes are the sex chromosomes, so that all primary sex determinants are X or Y linked. Four major classes of primary sex-determining factors are recognized, although this list may be incomplete. Within this framework, the relative importance, in different tissues or taxa, of different classes of primary sex determining factors can be recognized. The primary sex-determining factors include Sry, but each is logically primary because of its unequal representation in the male and females genome. This framework recognizes the effect of Sry and downstream sex differences in gonadal hormones as the dominant pathway for sex determination of the large majority of non-gonadal tissues, but at the same time recognizes multiple primary sex-determining factors not downstream of the gonadal effects of Sry, which act in parallel with gonadal differentiation to influence the sex of tissues, including sex differences in the susceptibility to disease. Xist is unique as a sex-determining gene because it is expressed in virtually every non-germline cell of females. Importantly, the multiple parallel pathways can interact with each other synergistically, to sum in their sex-determining effects, or antagonistically, to reduce the sex differences that each would cause by itself.
Box 3. Primary sex-determining factors in birds and other species.
The framework proposed here is applicable with minor modifications to any group with heteromorphic sex chromosomes, such as birds (sex chromosomes ZZ male, ZW female). Indeed, research on birds has helped catalyze the increased appreciation of vertebrate sex differences that are not caused by gonadal hormones [8,70]. A critical difference between birds and mammals is that birds lack a chromosome-wide dosage compensation mechanism akin to X-inactivation, so the majority of Z genes are expressed higher in males than in females [15,71,72]. That difference places a large number of Class II primary sex-determining factors within each male and female cell, which are available for selection as primary causes of sexual dimorphism. Although a large group of studies of birds documents the profound effects of gonadal hormones to produce sex differences in birds [73], other studies suggest that sex differences are also controlled by sex chromosome effects [74]. For example, the sex of the neural circuit controlling song in zebra finches is not masculinized by testiscular tissue in genetic females, and manipulations of gonadal hormones convincingly fail to control the sex of the circuit [74]. In lateral avian gynandromorphs in which ZZ and ZW cells are present in the same individual and are in the same gonadal endocrine environment, the cells retain some of their sex differences [1,70]. Importantly, the critical sex-determining role of sex chromosome genes within somatic cells is strongly supported by experiments in which cells from chick embryos of one sex are transplanted into the other, showing that the sexual phenotype of somatic gonadal cells reflects the genetic sex of the donor rather than of the host [1]. The apparently greater influence in birds (vs. mammals) of sex chromosome factors, not regulated downstream of gonadal homones, correlates with the lack of chromosome-wide dosage compensation that leaves many more Z genes in Class II in birds, relative to the situation in mammals in which Xist reduces the number of Class II sex determinants.
The present conceptual framework can be abstracted and extended to species that lack sex chromosomes. The central task in any explanation of sex determination is to specify all of the primary factors (primary because they are independent of each other at the outset, prior to the start of sex determination) that initiate sex-specific development, and to explain the downstream molecular cascades controlled by the primary factors that lead to sex differences in phenotype.
The challenge for the future is to discover all of the sex-determining factors on the sex chromosomes, and explain the pathways downstream of each, to understand what makes males and females different. This is not just a theoretical question, but one of potential significance in medicine. The incidence and progression of many diseases differs in males and females, such that one sex is protected from the disease. In some cases, the protection offered by sex-specific factors is greater than that currently afforded by any existing treatment. One strategy for improving treatment of disease is to develop better understanding of sex-specific regulation of physiology and disease, to uncover potent protective mechanisms that could be targets for novel therapeutic interventions.
Acknowledgements
Although I am indebted to many colleagues who have taught me regarding issues discussed here, I particularly thank Paul Burgoyne, Jennifer Marshall Graves, Eric Vilain, Jake Lusis, and Karen Reue for discussions. Thanks also to Xuqi Chen, Yuichiro Itoh, Esther Melamed, Negar Ghahremani and members of my lab. Supported by NIH grants DC000217, MH059268, NS043196, and DK08356.
Glossary
- Heteromorphic sex chromosomes
Sex chromosomes that differ in gene content.
- Müllerian ducts
Embryonic structures found in both sexes that in females differentiate into oviduct, uterus, and upper vagina.
- Sf1
Steroidogenic factor 1, NR5A1. Sf1 is expressed in gonads, adrenals, brain, and pituitary, and is required for development and function in each. Mice lacking Sf1 never develop gonads or adrenals.
- Sry
sex determining region Y. Sry is a Y chromosome gene that initiates differentiation of testes in males.
- Wolffian ducts
Embryonic structures found in both sexes that in males differentiate into the sperm ducts (epididymis and vas deferens) and seminal vesicles.
- X-inactivation
Transcriptional silencing of one of the two X chromosomes in each XX female non-germline cell
- Xist
X inactivation-specific transcript. In mammalian cells with two X chromosomes, the X inactivation center encodes factors that count the number of X chromosomes and trigger expression of Xist from one X chromosome. Xist causes transcriptional silencing (inactivation) of the chromosome from which it is expressed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Zhao D, et al. Somatic sex identity is cell autonomous in the chicken. Nature. 2010;464:237–242. doi: 10.1038/nature08852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodfellow PN, Lovell-Badge R. SRY and sex determination in mammals. Annu. Rev. Genet. 1993;27:71–92. doi: 10.1146/annurev.ge.27.120193.000443. [DOI] [PubMed] [Google Scholar]
- 3.Veitia RA. FOXL2 versus SOX9: a lifelong "battle of the sexes". BioEssays. 2010;32:375–380. doi: 10.1002/bies.200900193. [DOI] [PubMed] [Google Scholar]
- 4.Koopman P. The delicate balance between male and female sex determining pathways: potential for disruption of early steps in sexual development. Int. J. Androl. 2010;33:252–258. doi: 10.1111/j.1365-2605.2009.01001.x. [DOI] [PubMed] [Google Scholar]
- 5.Lillie FR. General biological introduction. In: Allen E, et al., editors. Sex and Internal Secretions. Williams and Wilkins Co; 1939. pp. 3–14. [Google Scholar]
- 6.van Nas A, et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinol. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vries GJ, et al. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J. Neurosci. 2002;22:9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold AP. Sex chromosomes and brain gender. Nat. Rev. Neurosci. 2004;5:701–708. doi: 10.1038/nrn1494. [DOI] [PubMed] [Google Scholar]
- 9.Mazeyrat S, et al. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat. Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- 10.Vernet N, et al. The Y-encoded gene zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr. Biol. 2011;21:787–793. doi: 10.1016/j.cub.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bugoyne PS, Mitchell MJ. The role of mouse Y chromosome genes in spermatogenesis. In: Lau Y, Chan WY, editors. Y Chromosome and Male Germ Cell Biology. World Scientific Publishers; 2007. pp. 27–45. [Google Scholar]
- 12.Bermejo-Alvarez P, et al. Transcriptional sexual dimorphism during preimplantation embryo development and its consequences for developmental competence and adult health and disease. Reproduction. 2011;141:563–570. doi: 10.1530/REP-10-0482. [DOI] [PubMed] [Google Scholar]
- 13.Bermejo-Alvarez P, et al. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3394–3399. doi: 10.1073/pnas.0913843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi S, et al. Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr. Biol. 2006;16:166–172. doi: 10.1016/j.cub.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 15.Itoh Y, et al. Dosage compensation is less effective in birds than in mammals. J. BIol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 17.Berletch JB, et al. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston CM, et al. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, et al. Spatially and temporally specific expression in mouse hippocampus of Usp9x, a ubiquitin-specific protease involved in synaptic development. J. Neurosci. Res. 2005;80:47–55. doi: 10.1002/jnr.20429. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, et al. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 2008;28:4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, et al. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One. 2008;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes AM, et al. Transcriptional changes in response to X chromosome dosage in the mouse: implications for X inactivation and the molecular basis of Turner Syndrome. BMC Genomics. 2010;11:82. doi: 10.1186/1471-2164-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies W, et al. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat. Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- 24.Ryan SG, et al. Epilepsy and mental retardation limited to females: An X-linked dominant disorder with male sparing. Nat. Genet. 1997;17:92–95. doi: 10.1038/ng0997-92. [DOI] [PubMed] [Google Scholar]
- 25.Migeon BR. Females are mosaic: X inactivation and sex differences in disease. Oxford University Press; 2007. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, et al. Sex difference in neural tube defects in p53-null mice is caused by differences in the complement of X not Y genes. Dev. Neurobiol. 2008;68:265–273. doi: 10.1002/dneu.20581. [DOI] [PubMed] [Google Scholar]
- 27.Lemos B, et al. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wijchers PJ, et al. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev. Cell. 2010;19:477–484. doi: 10.1016/j.devcel.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Jiang PP, et al. Y not a dead end: epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics. 2010;186:109–118. doi: 10.1534/genetics.110.118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemos B, et al. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 31.Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–140. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Zvetkova I, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 33.Bermejo-Alvarez P, et al. Epigenetic differences between male and female bovine blastocysts produced in vitro. Physiol Genomics. 2008;32:264–272. doi: 10.1152/physiolgenomics.00234.2007. [DOI] [PubMed] [Google Scholar]
- 34.Tsai HW, et al. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy MM, et al. The epigenetics of sex differences in the brain. J. Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Vries GJ. Minireview: Sex differences in adult and developing brains: compensation, compensation, compensation. Endocrinol. 2004;145:1063–1068. doi: 10.1210/en.2003-1504. [DOI] [PubMed] [Google Scholar]
- 37.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 38.Smith-Bouvier DL, et al. A role for sex chromosome complement in the female bias in autoimmune disease. J. Exp. Med. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renfree MB, Short RV. Sex determination in marsupials: evidence for a marsupial-eutherian dichotomy. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 1988;322:41–53. doi: 10.1098/rstb.1988.0112. [DOI] [PubMed] [Google Scholar]
- 40.Burgoyne PS, et al. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos. Trans. R. Soc. Lond B Biol. Sci. 1995;350:253–260. doi: 10.1098/rstb.1995.0159. [DOI] [PubMed] [Google Scholar]
- 41.Burgoyne PS. A Y-chromosomal effect on blastocyst cell number in mice. Dev. 1993;117:341–345. doi: 10.1242/dev.117.1.341. [DOI] [PubMed] [Google Scholar]
- 42.Thornhill AR, Burgoyne PS. A paternally imprinted X chromosome retards the development of the early mouse embryo. Dev. 1993;118:171–174. doi: 10.1242/dev.118.1.171. [DOI] [PubMed] [Google Scholar]
- 43.Tiffin GJ, et al. Glucose and glutamine metabolism in pre-attachment cattle embryos in relation to sex and stage of development. J. Reprod. Fertil. 1991;93:125–132. doi: 10.1530/jrf.0.0930125. [DOI] [PubMed] [Google Scholar]
- 44.Dewing P, et al. Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res. Mol. Brain Res. 2003;118:82–90. doi: 10.1016/s0169-328x(03)00339-5. [DOI] [PubMed] [Google Scholar]
- 45.Reisert I, Pilgrim C. Sexual differentiation of monoaminergic neurons-genetic or epigenetic. Trends Neurosci. 1991;14:467–473. doi: 10.1016/0166-2236(91)90047-x. [DOI] [PubMed] [Google Scholar]
- 46.Carruth LL, et al. Sex chromosome genes directly affect brain sexual differentiation. Nat. Neurosci. 2002;5:933–934. doi: 10.1038/nn922. [DOI] [PubMed] [Google Scholar]
- 47.Dewing P, et al. Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 2006;16:415–420. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Turner ME, et al. Sry, more than testis determination? Am. J. Physiol Regul. Integr. Comp Physiol. 2011 doi: 10.1152/ajpregu.00645.2010. Epub June 15. [DOI] [PubMed] [Google Scholar]
- 49.Majdic G, Tobet S. Cooperation of sex chromosomal genes and endocrine influences for hypothalamic sexual differentiation. Front Neuroendocrinol. 2011;32:137–145. doi: 10.1016/j.yfrne.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnold AP, Chen X. What does the "four core genotypes" mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol. 2009;30:1–9. doi: 10.1016/j.yfrne.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McPhie-Lalmansingh AA, et al. Sex chromosome complement affects social interactions in mice. Horm. Behav. 2008;54:565–570. doi: 10.1016/j.yhbeh.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold AP. The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm. Behav. 2009;55:570–578. doi: 10.1016/j.yhbeh.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arnold AP. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J. Neuroendocrinol. 2009;21:377–386. doi: 10.1111/j.1365-2826.2009.01831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gatewood JD, et al. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J. Neurosci. 2006;26:2335–2342. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Budefeld T, et al. Sex differences in brain developing in the presence or absence of gonads. Dev. Neurobiol. 2008;68:981–995. doi: 10.1002/dneu.20638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abel JM, et al. Sex Differences in the Cerebellum and Frontal Cortex: Roles of Estrogen Receptor Alpha and Sex Chromosome Genes. Neuroendocrinol. 2011;93:230–240. doi: 10.1159/000324402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X, et al. X chromosome number causes sex differences in gene expression in adult mouse striatum. Eur. J. Neurosci. 2009;29:768–776. doi: 10.1111/j.1460-9568.2009.06610.x. [DOI] [PubMed] [Google Scholar]
- 58.Xu J, et al. Sexually dimorphic expression of Usp9x is related to sex chromosome complement in adult mouse brain. Eur. J. Neurosci. 2005;21:3017–3022. doi: 10.1111/j.1460-9568.2005.04134.x. [DOI] [PubMed] [Google Scholar]
- 59.Gioiosa L, et al. Sex chromosome complement affects nociception in tests of acute and chronic exposure to morphine in mice. Horm. Behav. 2008;53:124–130. doi: 10.1016/j.yhbeh.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cox KH, Rissman EF. Sex differences in juvenile mouse social behavior are influenced by sex chromosomes and social context. Genes Brain Behav. 2011;10:465–472. doi: 10.1111/j.1601-183X.2011.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grgurevic N, et al. Aggressive behaviors in adult SF-1 knockout mice that are not exposed to gonadal steroids during development. Behav. Neurosci. 2008;122:876–884. doi: 10.1037/0735-7044.122.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinn JJ, et al. Sex chromosome complement regulates habit formation. Nat. Neurosci. 2007;10:1398–1400. doi: 10.1038/nn1994. [DOI] [PubMed] [Google Scholar]
- 63.Barker JM, et al. Dissociation of genetic and hormonal influences on sex differences in alcoholism-related behaviors. J. Neurosci. 2010;30:9140–9144. doi: 10.1523/JNEUROSCI.0548-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson DP, et al. Sex chromosome complement contributes to sex differences in Coxsackievirus B3 but not Influenza A virus pathogenesis. Biol. Sex Differ. 2011;2:8. doi: 10.1186/2042-6410-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ji H, et al. Sex Chromosome Effects Unmasked in Angiotensin II-Induced Hypertension. Hypertension. 2010;55:1275–1282. doi: 10.1161/HYPERTENSIONAHA.109.144949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caeiro XE, et al. Sex Chromosome Complement Contributes to Sex Differences in Bradycardic Baroreflex Response. Hypertension. 2011;58:505–511. doi: 10.1161/HYPERTENSIONAHA.111.175661. [DOI] [PubMed] [Google Scholar]
- 67.Ely D, et al. Review of the Y chromosome, Sry and hypertension. Steroids. 2010;75:747–753. doi: 10.1016/j.steroids.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Dev. 2010;137:3921–3930. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- 69.Wizeman TM, Pardue M-L, editors. US National Institute of Medicine Committee on Understanding the Biology of Sex and Gender Disorders. Exploring the Biological Contributions to Human Health: Does Sex Matter? US National Academy Press; 2001. [PubMed] [Google Scholar]
- 70.Agate RJ, et al. Neural not gonadal origin of brain sex differences in a gynandromorphic finch. Proc. Natl. Acad. Sci. U. S A. 2003;100:4873–4878. doi: 10.1073/pnas.0636925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ellegren H, et al. Faced with inequality: Chicken does not have a general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arnold AP, et al. A bird's-eye view of sex chromosome dosage compensation. Annu. Rev. Genomics Hum. Genet. 2008;9:109–127. doi: 10.1146/annurev.genom.9.081307.164220. [DOI] [PubMed] [Google Scholar]
- 73.Arnold AP, Itoh Y. Factors causing sex differences in birds. Avian Biology Research. 2011;4:44–51. doi: 10.3184/175815511X13070045977959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wade J, Arnold AP. Sexual differentiation of the zebra finch song system. Ann. N. Y. Acad. Sci. 2004;1016:540–559. doi: 10.1196/annals.1298.015. [DOI] [PubMed] [Google Scholar]